Abstract
We have identified a ligand (CO) accommodation cavity in the signal transducer sensor protein HemAT (heme-based aerotactic transducer) that allows us to gain single-molecule insights into the mechanism of gas sensor proteins. Specific mutations that are distal and proximal to the heme were designed to perturb the electrostatic field near the ligand that is bound to the heme and near the accommodated ligand in the cavity. We report the detection of a second site in heme proteins in which the exogenous ligand is accommodated in an internal cavity. The conformational gate that directs the ligand-migration pathway from the distal to the proximal site of the heme, where the ligand is trapped, has been identified. The data provide evidence that the heme pocket is the specific ligand trap and suggest that the regulatory mechanism may be tackled starting from more than one position in the protein. Based on the results, we propose a dynamic coupling between the two distinct binding sites as the underlying allosteric mechanism for gas recognition/discrimination that triggers a conformational switch for signaling by the oxygen sensor protein HemAT.
Keywords: docking site, FTIR spectroscopy, gas sensor proteins
The recognition and discrimination of diatomic gases that affect biological processes through the perturbation of protein function is a formidable challenge toward our understanding of the molecular mechanisms involved in signal transduction by heme-based sensor proteins (1, 2). Based on extensive studies of myoglobin (Mb) and Hb, there is a general consensus now that internal cavities in proteins are involved in controlling the dynamics and reactivity of the protein reactions with small ligands, such as O2, CO, and NO, usually through ligand accommodation (3–5). These cavities serve as a local storehouse for ligands near the active site, thereby increasing the effective concentration of the ligand by many times. In addition to having a functional role in ligand binding, they are important for determining relative affinities. In these studies, it has been assumed that the proteins have preexisting cavities that are modestly perturbed upon photodissociation of the ligand. However, the accommodation of the ligand in the cavities before its photodissociation has never been observed. The internal cavities and their potential fields determine the discrimination in favor of or against ligand binding; therefore, the measurement of unligated CO can be used to predict the parameters of physiological ligands. In the heme-based oxygen sensors such as HemAT (heme-based aerotactic transducer), the recognition and discrimination of the specific gas leads to either activation or inhibition of a regulated domain (6–11). Although the heart of signal transduction is the switching of proteins between active and inactive states, little is known about the molecular pathways involved in intramolecular signal transduction by gas sensory proteins.
In HemAT, the heme domain has a recognizable sequence homology with the oxygen-storage (SWMb, sperm-whale Mb) and oxygen-transport (Hb) proteins, in which extensive studies on the internal cavities have revealed profound information regarding the reequilibration among conformational substrates that are known to play an essential role in controlling protein function (7). In an effort to explore the recognition and discrimination of diatomic gases by HemAT, we have used FTIR spectroscopy to characterize the CO-bound complex of the sensor domain protein, as well as the distal-to-the-heme mutants R91A, T95A, Y70F, L92A, L92V, and L92F and the proximal-to-the-heme mutant Y133F (Fig. 1). The results show the accommodation of an exogenous ligand in a protein cavity of a heme protein. In HemAT, the cavity is located in the proximal environment of the heme, near the Y133 residue. The gateway of the ligand from the heme Fe to the cavity is through the distal L92 residue, providing strong evidence of the presence of a large channel (in contrast to Mb and Hb) that facilitates ligand passage. The characterization of such a conformational gate that directs the ligand-migration pathway from the heme Fe into the cavity provides key information for understanding the correlated motions in individual molecules. The concerted or sequential conformational changes induced to the protein upon ligand binding suggest a dynamic cascade of events that are essential for function. Given the highly dynamic behavior of the chemoreceptors, the present data identify functionally important protein motions in HemAT for understanding the molecular mechanisms involved in gas recognition/discrimination and regulation by heme-based sensors.
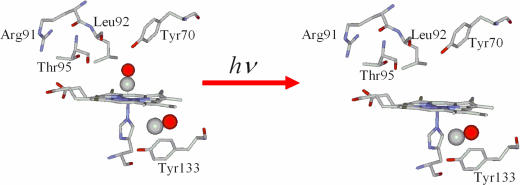
Fig. 1.
Schematic representation of the docking site before and after ligand photodetachment.
Results
The process of ligand docking in a cavity upon photolysis of the ligand from the heme Fe and, thus, the direct observation of intermediate states have been reported in Mb, Hb, and ba3-oxidase (3–5, 12–16). The mid-IR dynamic experiments on Mb have revealed the initial locations of photodissociated CO within a distal docking site through the identification of the B1 (νCO = 2,130 cm−1) and B2 (νCO = 2,120 cm−1) states (3–5). In the photorelaxed state, the docked CO is located in the proximal site (14–16).
FTIR Spectroscopy.
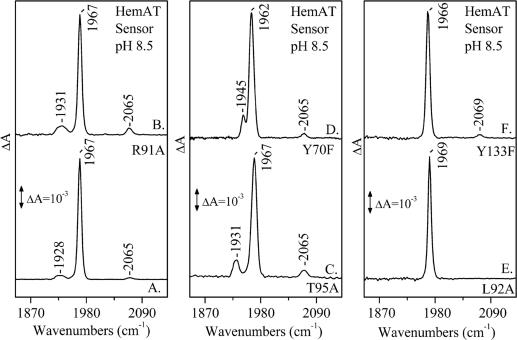
In addition to the open (no interaction with the distal protein environment) and closed (interaction with the distal protein environment, including H bonding) protein forms of the CO-bound sensor domain HemAT detected at 1,967 and 1,928 cm−1, respectively, another mode is observed, at 2,065 cm−1 (Fig. 2, trace A). The same mode has also been observed in all distal mutant proteins (Fig. 2, traces B–D) that we have examined, with the exception of L92A (Fig. 2, trace E), L92V, and L92F (Fig. 6a, which is published as supporting information on the PNAS web site). The 13CO sensitivity (ν13CO = 2,018 cm−1) of this band and its insensitivity to H/D exchanges is shown in Figs. 7 and 8, which are published as supporting information on the PNAS web site. In Y133F (Fig. 2, trace F), this mode is up-shifted, and it is observed at 2,069 cm−1. The frequency of the CO that we have detected under static conditions is close to that detected under photolytic conditions (see below); therefore, we assign it to a thermally dissociated state of HemAT in which the ligand is docked in a cavity of the protein. It is has been shown recently that the frequency of the “docked” CO depends markedly on the structure and charge of the amino acid side chains surrounding the ligand (5). In the photolysis experiments of Mb, the νCO at 2,130 cm−1 in the B1 state has been attributed to a direct H-bonding interaction of His-64 Nε···C–O, whereas the B2 state at 2,120 cm−1 has been attributed to a N–H···O–C interaction (5).
Fig. 2.
FTIR spectra of CO-bound sensor domain HemAT proteins at pH 8.5 and T = 298 K. Labeling is as follows: trace A, WT; trace B, R91A; trace C, T95A; trace D, Y70F; trace E, L92A; trace F, Y133F.
Density Functional Theory.
We have applied density functional theory to investigate the effect of charged residues on νCO of a docked CO molecule in a protein cavity. Because of the nature of these cavities, theoretical calculations have to be performed on simplified models with a restricted number of atoms. In these models, the charged residues in the protein cavity are represented by NH4+ (e.g., the arginine side chain) and HCOO− (e.g., aspartic acid).
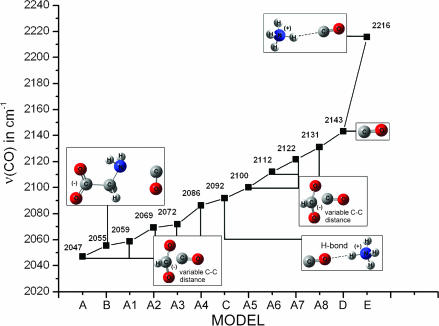
Fig. 3 shows the models in which the free CO interacts with (i) HCOO− (model A, HCOO–CO), (ii) a simple molecule that provides both carboxyl and amino groups, such as Gly (model B), and (iii) NH4+ (model E, NH4+-CO; model C, CO-NH4+). In the case of two NH4+ molecules (model not shown), two unconstrained NH4+ molecules strongly repel each other, whereas a constrained model (with fixed N···CO···N distances) exhibits a large positive binding energy. Models A1–A8 show the effect of the C–C distance (negative charge near CO) on νCO. In model A, the C–C distance is optimized at 2.98 Å without using constraints, whereas models A1–A8 have constrained C–C distances of 3.08, 3.18, 3.58, 3.78, 3.98, 4.18, 4.38, and 4.58 Å, respectively. All models except for A1–A8 are optimized without constraints. In model B, in which both the amino and carboxyl groups interact with the CO, the carboxyl group is deprotonated, whereas the amino group is neutral. Model C contains hydrogen-bonding interaction in addition to the positive charge effect on CO.
Fig. 3.
Empirical diagram showing the effect of positive and negative charges to νCO.
Fig. 3 shows the calculated νCO trend versus the model. Positive charges around CO increase νCO, whereas negative charges decrease it. Straub and Karplus (17) have calculated the νCO shifts in different CO–imidazole dimmer complexes at the 4–21G basis set level to vary between −17 cm−1 (H bonding C–O···H–N) to +36 cm−1 (N–H···C–O). Moreover, Nutt and Meuwly (18) have implemented molecular dynamics simulations on the photodissociated state of carboxymyoglobin based on a three-site charge model for CO to calculate the IR spectra of the free CO molecule in the heme pocket (18). The IR spectrum obtained exhibits peaks between 2,170 and 2,200 cm−1, corresponding to the signal from a single CO molecule docked in Mb. These signals are sensitive to the precise position and orientation of the CO molecule, as well as to the effect of the environment of the protein matrix (Mb) on the CO molecule. In general, the stretching frequencies νCO of heme-bound and photodissociated CO serve as powerful tools to probe the electrostatic fields and accessible space in the vicinity of the CO molecule inside the protein matrix. The docked, photodissociated CO ligands in the protein matrix display IR peaks, providing strong evidence for the existence of structurally well defined docking site(s). In analogy to the bound CO forms, the νCO of the docked CO is affected by the local environment through Stark effects of the local electric field acting on the CO dipole (5). In the photoproduct state, only a percentage of the ligand can be trapped in the docking site(s) in the time it takes to collect an FTIR spectrum (≈100 s), and the photodissociated ligands produce much less intense peaks than those observed for the heme Fe-CO bound forms. Because we do not have evidence for the detailed protein environment of the docked CO in HemAT (in contrast to Mb), we have constructed an empirical diagram of νCO to investigate the effect of positive or negative charges and that of H-bonding interactions to different orientations of CO. The empirical diagram shows a significant variability in νCO ranging from 2,047 cm−1 (interaction with only a negatively charged COO− group) to 2,216 cm−1 (interaction with only a positive charge). The calculations also show that νCO appears in the range of 2,047–2131 cm−1 at ≈10-cm−1 steps when the CO distance to a negatively charged carboxyl group is varied. The combined interaction of carboxyl and amino groups to CO has a strong effect on νCO, as shown by the 88-cm−1 down-shift from 2,143 cm−1 of gas CO, even though CO is not interacting directly with the deprotonated carboxyl. Our calculated vibrational frequencies are consistent with the Badger’s rule: re = cij(1/νe2/3) + dij, where re is the equilibrium distance C–O and νe refers to νCO (Fig. 9, which is published as supporting information on the PNAS web site). The empirical parameters cij and dij were calculated to be 61.535 and 0.767, respectively.
Although we do not have evidence for the orientation of the docked CO in HemAT, the νCO at 2,065 cm−1 indicates that it exhibits interactions that substantially weaken the C–O bond. We exclude the possibility of H-bonding interactions of the docked CO with H2O because the 2,065-cm−1 mode is insensitive to H/D exchanges (Figs. 7 and 8). The observation of νCO = 2,069 cm−1 in the Y133F mutant indicates that the frequency of the ligand is slightly disturbed from its value in the WT and in the distal protein mutants. The small but detectable shift in the frequency of CO in the Y133F mutant indicates the lack of direct interaction of the ligand with the F133 residue. It appears that the Y133F mutation induces a conformational change near the environment of the accommodated ligand, resulting in the small frequency shift we have observed. This finding provides unprecedented evidence of a preexisting cavity that accommodates a ligand in a protein. The absence of the 2,065-cm−1 mode in the L92A, L92V, and L92F mutants shows that if this residue is mutated, it blocks the thermally dissociated ligand from the heme to the cavity. Evidently, conformational fluctuations induced by the L92A, L92V, and L92F mutations create a high thermal barrier to dissociation and consequently slow the ligand off rate. Taken together, the static FTIR data indicate the presence of a conformational gate in the distal-to-the-heme L92 environment that directs the thermally dissociated CO to the protein cavity.
Photolysis FTIR Spectroscopy of the CO-Bound HemAT Proteins.
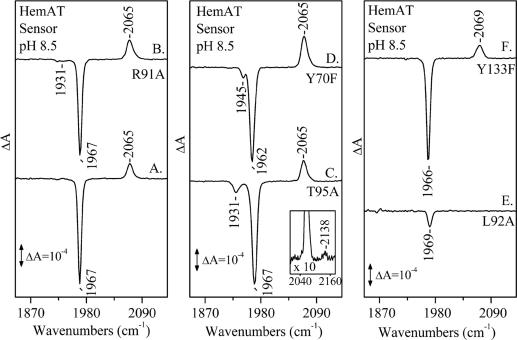
Fig. 4 shows the light (416 nm)-minus-dark FTIR difference spectra of the WT sensor domain HemAT and R91A, T95A, Y70F, L92A, and Y133F mutant proteins. The difference spectrum of the WT HemAT (Fig. 4, trace A) shows that, upon CO photolysis (negative band at 1,967 cm−1), the 2,065-cm−1 mode has gained intensity as compared with that obtained under static conditions (Fig. 2, trace A). This observation indicates that the photolyzed CO from the heme Fe is trapped within the same cavity that is located near the Y133 residue. The observation of the 2,065-cm−1 mode in the R91A (Fig. 4, trace B) and Y70F (Fig. 4, trace D) mutants indicates that these residues have no direct control in the pathway of the photolyzed CO from the heme Fe to the cavity. In Y133F (Fig. 4, trace F), the observation of the 2,069-cm−1 mode upon photolysis further supports our assignment that Y133 induces a conformational change to the environment that accommodates CO. In T95A (Fig. 4, trace C), in addition to the 2,065-cm−1 mode, a weaker mode is observed at 2,138 cm−1. The frequency of the 2,138-cm−1 mode is close to the free-gas value of CO (2,143.3 cm−1) and very similar to that found in Mb, characterizing the B1 state in which the CO is trapped into a docking site located above the pyrrole ring C of the heme (3–5). Accordingly, we assign the 2,138-cm−1 mode to the B1 state in which the CO is funneled into a docking site in the vicinity of the A95 residue. In contrast to the photolytic activity and properties of the WT and all other mutant proteins that we have examined, in the L92A (Fig. 4, trace E), L92V, and L92F (Fig. 6b) mutants, the 2,065-cm−1 mode is absent. This finding further supports our conclusions that L92 mutations keep the gateway closed for thermal dissociation, as judged from the absence of the 2,065-cm−1 mode (Fig. 2, trace E; see also Fig. 6a). It also provides unprecedented evidence that, in heme proteins, the gateway for ligand photodissociation is the same as that for the thermal dissociated ligand. In the most extensive photolytic studies of Mb and Hb, it has been always assumed that the proteins have preexisting cavities that are modestly perturbed by the photodissociated ligand (3–5). A schematic representation of the docking site in HemAT before and after ligand photodetachment is shown in Fig. 1.
Fig. 4.
Light (416 nm)-minus-dark FTIR difference spectra of CO-bound sensor domain HemAT proteins at ambient temperatures. Labeling is as follows: trace A, WT; trace B, R91A; trace C, T95A; trace D, Y70F; trace E, L92A; trace F, Y133F. Inset shows the 2,040- to 2,160-cm−1 region ×10.
We suggest that the long-lived docked CO causes equilibrium (under static conditions) and transient (under photolytic conditions) large-scale protein fluctuations near the Y133 residue. Protein fluctuations create exit channels through which ligands migrate into other internal cavities, from which they finally escape from the protein. Such protein fluctuations in the distal and proximal environment upon ligand binding have been proposed to initiate a cascade of events toward signal transduction in HemAT (7, 8, 11).
Discussion
Conformational Gating of Ligand Migration and Accommodation.
The ligand accommodation to an internal cavity of HemAT at physiological temperature provides direct confirmation for the functional relevance of such a cavity. The results on static CO-bound HemAT complex reveal an obvious protein channel that is large enough to facilitate ligand passage to the cavity. The migration of the photolyzed CO to the same cavity demonstrates the limited role of the diatomic ligand in forcing its way by pushing side chains to occupy different sites/cavities in the protein. The conformational gate that directs the ligand-migration pathway of both thermal-dissociated and photodissociated CO is L92. Our observations do not distinguish between synchronous or asynchronous internal motions of the distal and proximal conformational gates. It is certain, however, that in the case of the migrated photodissociated ligand to the cavity, the structure and conformational flexibility of the amino acid residues in the ligand pathway are tuned to optimize the reversible binding of the ligand to the heme Fe. Similarly, the distal-to-the-heme amino acids also retain conformational flexibility to allow the rebinding of the ligand to the heme Fe. These observations unveil functional protein motions and illustrate at an atomic level the relationship among protein structure, dynamics, and function.
The static structures of thousands of proteins are known, but function requires motion. It is important to know that there is not a single protein whose function is fully understood at an atomistic level. T95A HemAT demonstrates how structural changes near the active site can have large functional effects. Replacing T95 with A95 decreases the volume of the distal cavity in which the ligand becomes transiently trapped after dissociation. The picture that emerges from L92A, L92V, and L92F shows how a relatively small structural change near the active site can have large functional effects. Indeed, in these mutants, we have not observed CO in the cavity. In the work presented here, the interplay between two conformational gates of ligand migration and ligand accommodation has been characterized. The first one describes the gating from the heme Fe through the distal amino acid residues to the cavity, whereas the second describes the gating of the ligand to the environment of the cavity. It remains to be established whether concerted or sequential motions of the protein residues are necessary for such gating and whether there is a coupling between them that leads to an extended internal conformational change. Given the similar size and polarity of O2 and CO, O2 migration to the docking site is expected. The same docking site that shelters CO and the photodissociated CO from the heme Fe is expected to trap O2. This way, protein fluctuations in both the distal and proximal-to-the-heme environments are expected to occur in the presence of the physiological O2 ligand. Structural information on the communication pathway between the heme upon ligand binding and the protein is necessary for a better determination of the molecular pathways involved in the intramolecular signal transduction.
Physiological Relevance.
Why does HemAT contain a well defined docking site? Dissolved O2 must compete with water to bind to HemAT, and oxy-HemAT has a water molecule in the distal ligand-binding site. Because the concentration of water is much higher than that of O2 in oxygen-saturated solutions, the cavity (second binding site) may serve as a local storehouse of O2 near the heme Fe site, thereby increasing the effective oxygen concentration of the first binding site (heme Fe) by many times. Thus, the cavity in HemAT that is located near Y133 appears to have a functional role in ligand binding and may be important not only for the kinetics of ligand binding but also for determining relative affinities. Because the fluctuations connecting the cavity and the heme Fe-binding site determine accessibility, fast fluctuations of side chains may have an evolutionarily conserved survival value. The role of hydrophobic cavities as a means of controlling protein function may be a general feature in gas sensory proteins.
It is anticipated that both binding sites are responsible for the kinetic control of ligand motion/binding and escape. The simultaneous observation of the binding sites can also be used to evaluate the ligand input and escape pathways and suggests a pathway connecting the two binding sites. The data presented here, and those previously reported, suggest that although the heme cavity recognizes ligands such as O2, CO, and NO, the conformational changes induced upon ligand binding are not similar, and, thus, only the binding of O2 induces the necessary conformational changes that are subsequently transmitted to the effector domain (7–8, 10–11, 19, 20). We have recently shown (19) that R91 is not involved in any kind of structural changes that influence the ligand recognition and discrimination. The behavior of T95 and Y70 upon binding of O2, CO, and NO to the heme Fe produces distinct conformations, indicating that these residues are the major contributors toward ligand recognition and discrimination. On the other hand, L92 induces the necessary structural changes to T95 and Y70 for the formation of the NO-bound conformations, exhibiting interaction with the distal residues and with the conformations exhibiting no interaction. For the CO-bound ligand, L92 generates the necessary structural changes to the distal environment to maintain the H-bonded conformer. These data demonstrate the unique specificity and tuning of the distal amino acid residues of the E helix in constructing the necessary conformational changes toward ligand recognition.
We suggest a dynamic population shift between two preexisting conformations (allosteric activation) in the protein that involves the E helix (Y70 and T95) shown in Fig. 5 and Y133 as the underlying mechanism of activation, and we suggest that only oxygen, which regulates signal transduction pathways, triggers the conformational switch. This way, upon ligand binding to the heme Fe, the induced conformational changes are transmitted to the E helix and, in conjunction with the conformational changes produced by the presence of the ligand in the docking site, are communicated to the effector domain. Our observation of the docking site near Y133 in both the sensor domain and full-length HemAT (Fig. 10, which is published as supporting information on the PNAS web site) further supports our proposal for the role of the second binding site in intramolecular signal transduction in HemAT. The stabilization of such conformations may be a fundamental paradigm for specific recognition/discrimination of diatomic gases by heme sensor proteins. Further studies on the dynamics in defined structural states, combined with the knowledge of the active and inactive conformations, are necessary for the direct observation of the functional conformational-equilibrium process. Time-dependent conformational changes to unveil the structural basis of the resultant conformational relaxation governing ligand migration through the matrix will help to elucidate this process.
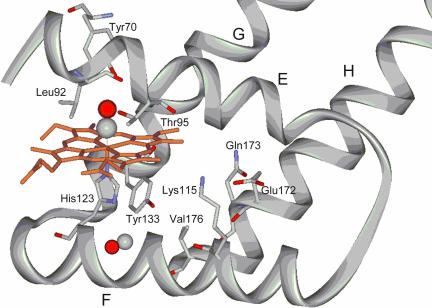
Fig. 5.
X-ray protein structure according to ref. 7. A ribbon model of HemAT with the proximal-to-the-heme and distal-to-the-heme residues is shown.
Methods
Expression and Purification of HemAT.
HemAT was expressed in Escherichia coli BL21 (DE3) under the control of the T7 promoter in the pET24(+) vector (Novagen, Madison, WI). HemAT was obtained as a C-terminal 6×His-tagged protein. All mutants were prepared by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The E. coli BL21 (DE3) harboring the expression vector was grown aerobically at 37°C for 4 h in Terrific Broth medium containing 30 μg/ml kanamycin. The expression was induced by adding 1 mM isopropyl-1-thio-d-galactopyranoside, and then the cultivation continued at 22°C for 18 h. The cells were harvested by centrifugation at 4,000 × g and stored at −78°C until use. Purification of HemAT was carried out as follows. The cells were thawed and resuspended in 50 mM Tris·HCl buffer (pH 8.0) containing 15 mM glycine and 1 M NaCl and then were broken by sonication. The resulting suspension was centrifuged at 100,000 × g for 20 min, and the supernatant was loaded on a Ni-charged HiTrap Chelating column (GE Healthcare, Piscataway, NJ). After washing the column with 50 mM Tris·HCl buffer (pH 8.0) containing 15 mM glycine and then with 1 M NaCl and 50 mM Tris·HCl buffer (pH 8.0), the adsorbed proteins were eluted by 50 mM Tris·HCl buffer (pH 8.0) containing 100 mM imidazole. The fractions containing HemAT were combined and loaded on a HiTrap Q HP column (Amersham). The column was washed with 50 mM Tris·HCl buffer (pH 8.0) containing 100 mM NaCl; then, HemAT was eluted by increasing the concentration of NaCl in 50 mM Tris·HCl buffer (pH 8.0).
FTIR Spectroscopy.
Dithionite-reduced samples were exposed to 1 atmosphere (atm) of CO (1 atm = 101.3 kPa) (1 mM) in an anaerobic cell to prepare the carbonmonoxy adduct and loaded anaerobically into a cell with CaF2 windows and a 0.015-mm spacer. CO gas was obtained from Messer (Krefeld, Germany), and isotopic CO (13CO) was purchased from Isotec (Miamisburg, OH). The pD (pH + 0.4) solutions prepared in D2O buffers were measured by using a pH meter and assuming pD = pH (observed) + 0.4. FTIR spectra were obtained from 1 mM samples with an Equinox 55 FTIR spectrometer (Bruker, Ettlingen, Germany) equipped with a liquid-nitrogen-cooled mercury cadmium telluride detector. The FTIR spectra were obtained as differences, using the buffer as a background, and each spectrum is the average of 1,000 scans. The photolysis light (416 nm, 10 mW) was provided by a diode laser (TOptica, Munich, Germany). The light (416 nm)-minus-dark FTIR difference spectra are the average of 5,000 scans. Optical absorbance spectra were recorded before and after FTIR measurements to assess sample stability with a Lamda 20 UV-visible spectrometer (Perkin-Elmer, Bodenseewerk, Germany).
Density Functional Theory.
For each structure considered, a full geometry optimization was performed by using the density functional B3LYP method. We used the 6–31g (d,p) double-ζ valence basis set augmented with p-PGTOs on H atoms, as implemented in the Gaussian 03 software package (21). When vibrational frequencies are calculated by electronic structure theory, they can often be improved by scaling, and it is useful to have general scaling factors. Such factors depend on the level of electronic structure theory and the one-electron basis set. It has been established that calculated frequencies may be scaled in various ways (22). For example, one scaling factor is applied to reproduce the true harmonic frequencies, the true fundamental frequencies, or the zero point energy. To accurately reproduce the experimental νCO of interaction-free gas CO, a scaling factor of ×0.9703 was used for the theoretically calculated νCO. A scan for νCO using different basis sets starting from 6–31g (d,p) through 6–311g++(2df, 2pd) showed identical trends (±10 cm−1) for νCO concerning the environmental effect. Solvation effects count for only a 5-cm−1 theoretical shift and, thus, were not taken into account.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Education of Greece (Pythagoras II) and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan (B16370065 to S.A.).
Abbreviation
- Mb
myoglobin.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rodgers KR. Curr Opin Chem Biol. 1999;3:158–167. doi: 10.1016/S1367-5931(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 2.Chan MK. Curr Opin Chem Biol. 2001;5:216–222. doi: 10.1016/s1367-5931(00)00193-9. [DOI] [PubMed] [Google Scholar]
- 3.Lim M, Jackson TA, Anfirud PA. Science. 1995;269:962–966. doi: 10.1126/science.7638619. [DOI] [PubMed] [Google Scholar]
- 4.Frauendfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 5.Nienhaus K, Olson JS, Franzen S, Nienhaus GU. J Am Chem Soc. 2005;127:40–41. doi: 10.1021/ja0466917. [DOI] [PubMed] [Google Scholar]
- 6.Aono S, Kato T, Matsuki M, Nakajima H, Ohta T, Uchida T, Kitagawa T. J Biol Chem. 2002;277:13528–13538. doi: 10.1074/jbc.M112256200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Philips G. Structure (London) 2003;11:1097–1110. doi: 10.1016/s0969-2126(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W, Olson JS, Philips G. Biophys J. 2005;88:2801–2814. doi: 10.1529/biophysj.104.047936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aono S, Nakajima H, Ohta T, Kitagawa T. Methods Enzymol. 2004;381:618–628. doi: 10.1016/S0076-6879(04)81040-1. [DOI] [PubMed] [Google Scholar]
- 10.Ohta T, Yoshimura H, Yoshioka S, Aono S, Kitagawa T. J Am Chem Soc. 2004;126:15000–15001. doi: 10.1021/ja046896f. [DOI] [PubMed] [Google Scholar]
- 11.Ohta T, Kitagawa T. Inorg Chem. 2005;44:758–769. doi: 10.1021/ic0486318. [DOI] [PubMed] [Google Scholar]
- 12.Koutsoupakis C, Soulimane T, Varotsis C. J Biol Chem. 2003;278:36806–36809. doi: 10.1074/jbc.M307117200. [DOI] [PubMed] [Google Scholar]
- 13.Koutsoupakis C, Soulimane T, Varotsis C. J Am Chem Soc. 2003;125:14728–1473. doi: 10.1021/ja036107e. [DOI] [PubMed] [Google Scholar]
- 14.Srajer V, Teng T, Ursby T, Pradervand C, Ren Z, Adachi S, Schildkamp W, Bourgeois D, Wulff M, Moffat K. Science. 1996;274:1726–1729. doi: 10.1126/science.274.5293.1726. [DOI] [PubMed] [Google Scholar]
- 15.Chu K, Vojtchovsky J, MacMahon BH, Sweet RM, Berendzen J, Schlichting I. Nature. 2000;403:921–923. doi: 10.1038/35002641. [DOI] [PubMed] [Google Scholar]
- 16.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 17.Straub E, Karplus M. Chem Phys. 1991;158:221–248. [Google Scholar]
- 18.Nutt DR, Meuwly M. Biophys J. 2003;85:3612–3623. doi: 10.1016/S0006-3495(03)74779-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinakoulaki E, Yoshimura H, Yoshioka S, Aono S, Varotsis C. Biochemistry. 2006;45:7763–7766. doi: 10.1021/bi0604072. [DOI] [PubMed] [Google Scholar]
- 20.Yoshimura H, Yoshioka S, Kobayashi K, Ohta T, Uchida T, Kubo M, Kitagawa T, Aono S. Biochemistry. 2006;45:8301–8307. doi: 10.1021/bi060315c. [DOI] [PubMed] [Google Scholar]
- 21.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, et al. Gaussian 03. Wallingford, CT: Gaussian; 2004. Revision B.03. [Google Scholar]
- 22.Pople JA, Scott AP, Wong MW, Radom L. Isr J Chem. 1993;33:345–350. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.