Abstract
A collagen-degrading thermophile, Geobacillus collagenovorans MO-1, extracellularly produces a collagenolytic protease with a large molecular mass. Complete nucleotide sequencing of this gene after gene cloning revealed that the collagenolytic protease is a member of the subtilisin family of serine proteases and consists of a signal sequence for secretion, a prosequence for maturation, a catalytic region, 14 direct repeats of 20 amino acids at the C terminus, and a region with unknown function intervening between the catalytic region and the numerous repeats. Since the unusual repeats are most likely to be cleaved in the secreted form of the enzyme, the intervening region was investigated to determine whether it participates in collagen binding to facilitate collagen degradation. It was found that the mature collagenolytic protease containing the intervening region at the C terminus bound collagen but not the other insoluble proteins, elastin and keratin. Furthermore, the intervening region fused with glutathione S-transferase showed a collagen-binding ability comparable to that of the mature collagenolytic protease. The collagen-binding ability was finally attributed to two-thirds of the intervening region which is rich in β-strands and is approximately 35 kDa in molecular mass. In the collagenolytic protease from strain MO-1, hydrogen bonds most likely predominate over the hydrophobic interaction for collagen binding, since a higher concentration of NaCl released collagen from the enzyme surface but a nonionic detergent could not. To the best of our knowledge, this is the first report of a thermophilic collagenolytic protease containing the collagen-binding segment.
Collagens are ubiquitously and abundantly present in the extracellular matrix of mammalian and insect cells (9, 20). Due to the unique structure of collagen, three helically wound polypeptides including numerous repeats of tripeptide Gly-Pro-X, the characteristic proteins are difficult to degrade. Nevertheless, collagens are prospective biomaterials for applications because of their significant involvement in many cellular events (9, 17, 20). Furthermore, pathological and biochemical studies on collagen degradation by microorganisms have demonstrated enormous potential for applications (15, 31). While there have been only a few reports dealing with thermophilic bacteria that preferentially degrade collagens and their enzymes (3, 16, 30, 34), we have thus far isolated a thermophilic and collagen-degrading bacterium, Geobacillus collagenovorans MO-1, from soil around a cold spring in Kyoto, Japan, and have studied an array of collagenolytic enzymes produced by strain MO-1 (14, 19, 32). This strain produces three enzymes related to collagen breakdown: a collagenolytic protease that directly hydrolyzes collagen molecules and two distinct metallopeptidases (Pz peptidases) that hydrolyze the synthetic substrate 4-phenylazobenzyloxycarbonyl-Pro-Leu-Gly-Pro-d-Arg, which contains the collagen-specific tripeptide. The collagenolytic protease is recognized in the culture supernatant, while the two Pz peptidases are in the cytoplasm fraction of the cell. It is supposed that in strain MO-1, macromolecular collagens are first degraded by the collagenolytic protease extracellularly and that the resulting oligopeptides are then transported inside the cells to be further hydrolyzed by two Pz peptidases. From enzymological analyses, it was found that the collagenolytic protease is an extracellular enzyme that belongs to a family of serine proteases; however, the enzyme has a much larger molecular mass than do other extracellular serine proteases from bacteria (32). While the collagenolytic protease from strain MO-1 has a broad specificity for protein substrates, one of its distinct characteristics is its strong activity toward collagen. Most collagenases have larger molecular masses than ordinary proteases have (32), and they are structurally equipped with the mechanism to break down collagens by such means as hydrolysis and binding (19). In view of these facts, we proposed that the collagenolytic protease from strain MO-1 also has an advantageous apparatus for collagen degradation (19). To examine whether the collagenolytic protease from strain MO-1 is truly a collagenolytic protease specific to collagen, we cloned the gene for the collagenolytic protease and analyzed its structure. Subsequently, in this study, we investigated whether a region with unknown function binds collagen and, if so, which segment is responsible for collagen binding.
MATERIALS AND METHODS
Microorganisms and growth conditions.
G. collagenovorans MO-1 was used for this study of the collagenolytic protease and the cloning of its gene. The features of this strain have been described in previous reports (14, 19). This strain was grown in L broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl, pH 7.2) with reciprocal shaking (95 rpm) at 65°C for 8 h for chromosomal DNA isolation. For gene cloning and expression of recombinant genes, Escherichia coli DH5α and BL21(DE3), respectively, were used. All recombinant strains were aerobically cultivated in L broth appropriately supplemented with ampicillin (50 μg/ml) at 37°C.
Determination of internal amino acid sequence.
To determine the internal protein sequences, the purified collagenolytic protease sample (10 μg) was subjected to partial degradation by Staphylococcus aureus V8 protease as described in our previous report (14). Partially digested proteins were recovered on a polyvinylidene difluoride membrane by semidry electrotransfer and then applied to a PPSQ-21 protein sequencer (Shimadzu, Kyoto, Japan) using automated Edman degradation.
Cloning of the gene for collagenolytic protease.
To obtain the chromosomal DNA of G. collagenovorans MO-1, the sample prepared in the previous study was used here (14). A pair of PCR primers were designed from the N-terminal and internal amino acid sequences of the collagenolytic protease (primer F-1, CGCGGATCCATGAAATAYTCNAAAGARYTKGTS; primer R-1, CCGGAATTCTTGNARCGCGTTSACGTTCATAAA [Y equals C or T; R equals A or G; S equals C or G; N equals A, C, G, or T]) to amplify the DNA probe for colony hybridization. A 0.7-kb probe for the collagenolytic protease was prepared by PCR (30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 3 min), using rTaq polymerase (TOYOBO, Osaka, Japan). The probe was labeled using the digoxigenin labeling kit (Roche, Mannheim, Germany) according to supplier's specifications. To screen a colony containing the desired DNA fragment, colony hybridization was carried out by a method described previously (14). Both strands of the cloned DNA fragment were sequenced using PRISM dye primer sequencing kits on a 310NT DNA sequencer (Applied Biosystems, Foster City, CA).
Plasmid construction for GST fusion proteins and their purification.
To prepare the protein samples for collagen-binding analysis, the glutathione S-transferase (GST) fusion system was employed. Plasmids for fusion proteins between GST and various regions of the collagenolytic protease were constructed by using the BamHI and EcoRI sites of the vector plasmids pGEX-1λT or pGEX-4T-1 (GE Healthcare Bio-Sciences Corp., NJ). A DNA fragment coding for the catalytic region (Met165 to Pro659) of the collagenolytic protease was prepared by PCR with two primers (MO-matu, CGCGGATCCATGAAATATAGCAAAGAGCTG; Cat-R, CCGGAATTCAGGAGTAGATAGTGCCGC). Other fragments coding for the intervening region (Int) or three divided segments (Int1, Int2, and Int3) of the collagenolytic protease were prepared in a similar manner with the respective pairs of primers (for Int, CGCGGATCCCCTGTCATCGTGACGGAG and CCGGAATTCGTGCAAATCCGGGATGTTTCC; for Int1, CGCGGATCCCCTGTCATCGTGACGGAG and CCGGAATTCTGGAGCGTCCACGATTGG; for Int2, CGCGGATCCGCGTGGGATGAAAATAGC and CCGGAATTCGTCAAAGCTTGCTTCTACCG; for Int3, CGCGGATCCAAAGATACACAAACGATTTCC and CGCGGATCCTCCTTTATTATTGATGGCGG). The fusion proteins were overexpressed in E. coli BL21 at 37°C by the addition of isopropyl-β-d-galactopyranoside (final concentration, 0.2 mM) for 2 h after the cell growth reached an optical density at 660 nm of 0.5. Cells harvested from the culture (50 ml) were resuspended in 5 ml of 50 mM Tris-HCl (pH 7.5) (buffer I) and disintegrated by sonication. The cell extract obtained by centrifugation at 10,000 × g for 20 min was applied to glutathione Sepharose 4B (gel volume, 66.5 μl; GE Healthcare Bio-Sciences Corp.) equilibrated with buffer I. After being washed in 2 ml of buffer I, proteins trapped on the affinity gel were eluted with 250 μl of 10 mM glutathione in buffer I. The eluted solution was dialyzed against 250 ml of buffer I and utilized for the assay. The GST assay was carried out with 1-chloro-2,4-dinitrobenzene as a substrate according to a previously described method (5).
Collagen-binding assay.
The binding capabilities of the collagenolytic protease and GST fusion proteins containing the various regions of the collagenolytic protease to collagen were analyzed by two assay methods: (i) the batch method followed by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE) and imaging analysis and (ii) the surface plasmon resonance (SPR) method. The collagenolytic protease from strain MO-1 used here was purified in our previous study (14).
The batch method was modified from that previously reported by Matsushita et al. (13). Two hundred microliters of ice-chilled buffer I was added to 5 mg of type I collagen (bovine Achilles tendon; Nacalaitesque, Kyoto, Japan) in a microtube. Soon after the buffer was removed with a pipette, the same amount of the buffer was added to be suspended and kept at 0°C for 60 min. Then, the mixture was centrifuged at 14,000 rpm for 5 min and the supernatant was discarded. Fifty microliters of the protein solution (2.5 to 3.5 μg included) to be investigated was added and kept at 0°C for 180 min. An aliquot of the sample (18 μl) was removed just at the beginning and centrifuged as described above to save the resulting supernatant as a negative control. After 180-min incubation at 0°C, another aliquot (18 μl) was removed from the remaining mixture and centrifuged as described above. The samples were subjected to SDS-PAGE with 12.5% (wt/vol) sheet gels. After SDS-PAGE, the band patterns of the gel were compared by using an LAS-1000 Plus imaging analyzer (FUJIFILM, Tokyo, Japan), and the quantitative analyses for protein bands in the gel were densitometrically carried out by the attached program, Multi Gauge version 2.1.
The SPR method was performed at room temperature in a MultiSprinter system (TOYOBO, Osaka, Japan), based on a method described previously (7). Different amounts of acid-soluble collagen (type I; Nacalaitesque) were immobilized onto the spots of a COOH sensor chip with bifunctional cross-linkers, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, and N-hydroxysuccinimide. For the blank control, bovine serum albumin (BSA) was spotted onto the same chip. The sample protein solution (GST fusion proteins, 1 mg/ml) in buffer I was refluxed at a flow rate of 50 μl/min.
Release of the collagenolytic protease from collagen.
To search for the condition to release the collagenolytic protease from collagen, various concentrations of NaCl or detergent were added to collagen, which bound the fusion protein GST-Int in a microtube. The collagen binding the collagenolytic protease (collagen complex) was prepared by incubating for 180 min at 0°C according to the batch method, described above. The collagen complex was centrifuged at 14,000 rpm for 5 min, and the supernatant was completely removed. Fifty microliters of the releasing solution (0, 30, 145, and 1,000 mM NaCl; 1% [wt/vol] Triton X-100, 1% [wt/vol] Tween 20, or 1% [wt/vol] Tween 60 in Tris-HCl [pH 7.5]; the numbers indicate the final concentration in the mixture) was added to the sedimented collagen complex in the microtube, and incubation was carried out at 0°C for 30 or 60 min. The microtube was centrifuged as described above, and 18 μl of the sample was removed and then transferred into a new microtube. The microtube was centrifuged again, and the supernatant was subjected to SDS-PAGE. The amount of fusion protein GST-Int released from collagen into the solution was quantitatively analyzed by calculating the densitometric ratio of the collagenolytic protease on the SDS-PAGE pattern using the same system (LAS-1000 Plus/Multi Gauge) as that used for the batch analysis.
Nucleotide sequence accession number.
The nucleotide sequence data for the gene for the collagenolytic protease from strain MO-1 will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession number AB260948.
RESULTS
Gene cloning and nucleotide sequence analysis.
Prior to the probe preparation, the internal amino acid sequence of the collagenolytic protease from strain MO-1 was determined to be Asn-Ser-Phe-Met-Asp-Val-Asp-Ala-Leu-Gln. Based on this result and the N-terminal sequence reported previously (19), two primers, F-1 and R-1, were designed to synthesize the DNA probe by PCR. The obtained DNA probe (0.8 kbp) was subject to DNA sequencing and was confirmed to contain the DNA sequences coding for the determined N-terminal and internal amino acid sequences at both edges. One hybrid plasmid containing the gene for the collagenolytic protease was screened from a library of the EcoRI-digested G. collagenovorans chromosomes through colony hybridization using the DNA probe. The DNA fragment contained in the hybrid plasmid was 6.0 kbp in length and was found to lack the region encoding the C-terminal segment of the collagenolytic protease. Therefore, other hybrid plasmids having the missing region were subsequently screened from two libraries of HindIII- or EcoRI-digested chromosomes. As a result, two hybrid plasmids containing the expected region were obtained and found to include a HindIII-HindIII fragment (0.7 kbp) or an EcoRI-EcoRI fragment (4 kbp). The DNA fragments, excluding the overlapping regions, were reconstructed together in one plasmid named pGC-9, which covers the entire gene of the collagenolytic protease. The DNA fragment derived from strain MO-1 in pGC-9 was 9.8 kbp in length.
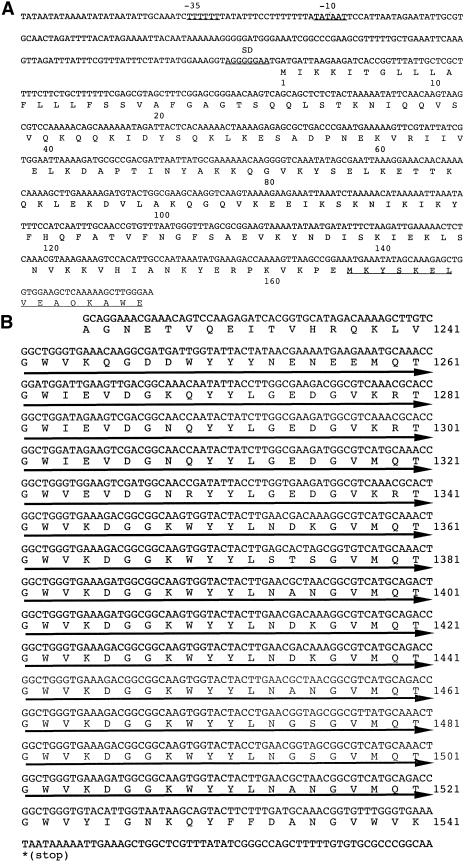
From DNA sequence analysis, two open reading frames appeared in a 9.8-kbp DNA fragment, one corresponding to the homologue of UDP-N-acetylglucosamine-2-epimerase and the other corresponding to the collagenolytic protease containing the known N-terminal and internal amino acid sequences. The open reading frame (4,623 bp) for the collagenolytic protease is capable of coding for a protein of 1,541 amino acids with a putative molecular mass of 170,962 Da. This capability is by far greater than that estimated for the purified collagenolytic protease (105 kDa) in our previous report (19). The N-terminal sequence of the purified protein occurred at amino acid position 165, indicating that the open reading frame includes the pre- and prosequences for the secretion and maturation of the collagenolytic protease. To obtain evidence supporting this idea, we investigated the motif of the region between the putative start codon and the N-terminal region experimentally determined (amino acid position 1 to 180) (Fig. 1A) by free Internet software for a motif search (http://www.cbs.dtu.dk/services/SignalP/). The result indicated that the N-terminal segment between amino acid positions 1 and 22 most probably functions as a signal sequence for secretion (data not shown). Furthermore, combinatorial homology searches using SSEARCH, FASTA, and BLAST programs revealed that the collagenolytic protease from strain MO-1 resembled lactocepins (lactococcal cell envelope-associated proteinases) from Lactobacillus paracasei subsp. paracasei NCDO 151 (SwissProt accession number Q02470) and Oceanobacillus iheyensis HTE831 (Swissprot accession number Q8CVB2) (6, 23). L. paracasei lactocepin has been confirmed to have a prosequence following the signal sequence, which shows a significant homology with the segment between amino acids positions 23 and 164 of the collagenolytic protease. The molecular mass of the segment including the pre- and prosequences of the collagenolytic protease is 18,718 Da. Judging from the molecular masses of the processed segment at the N terminus, the open reading frame, and the purified protein, it is likely that there is another processed site at the C terminus. From the putative calculation of the molecular mass, the processed site is presumably located near amino acid position 1130. Another characteristic is that there are 14 direct repeats of 20 amino acids between amino acid positions 1242 and 1521 in the C-terminal segment (Fig. 1B) showing a high homology with choline-binding protein PspC from Streptococcus pneumoniae (10, 24). In addition, the collagenolytic protease was purified from the culture supernatant of strain MO-1, not from cellular fractions. It is definite that the enzyme is secreted into the culture without being displayed on the cell surface.
FIG.1.
(A) Nucleotide and amino acid sequences of the 5′-flanking region near the translational start codon of the collagenolytic protease from strain MO-1 and the N terminus of the mature enzyme. A set of putative promoters (−35 and −10 regions) are labeled −35 and −10, and the ribosome-binding site is labeled SD. The amino acid sequence obtained from protein sequencing with the purified collagenolytic protease is underlined. (B) Nucleotide and amino acid sequences of the C-terminal region of the collagenolytic protease. Fourteen direct repeats of 20 amino acids are indicated by thick arrows under the sequence.
In the region between amino acid positions 165 and 654, there is close homology with the catalytic regions of lactobacillus lactocepins introduced above. Lactocepins belong to a subtilisin family (S8) of serine peptidases, and their typical active site residues have been established through genetic studies (22). In the collagenolytic protease, Asp194, His260, and Ser590 are presumed to be the active residues, since their equivalent residues work as a catalytic triad located in the N-terminal region of the lactocepin molecules (25). This feature on the active site suggests that the hydrolyzing mechanism of the collagenolytic protease is not specifically restricted to collagen. In addition to the characteristics described above, there is a long region (amino acid positions 655 to 1130) intervening between the catalytic region and 14 repeats of 20 amino acids. The intervening region shares no significant homology with lactocepins. Five types of secondary structure predictions were carried out for the region by using the systems SSTHREAD (http://www.ddbj.nig.ac.jp/search/ssthread.html), PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/), Chou-Fasman (http://mbs.cbrc.jp/papia-cgi/ssp_menuJ.pl), Non predictor (http://cubic.bioc.columbia.edu/services/NORSp/submit.html), and PREDATOR (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html). Their combined results revealed that β-strands exclusively occupy the region and, therefore, that the region most likely constitutes β-sheets (data not shown). Since the function of the intervening region was unclear, the collagenolytic protease containing the region was subjected to collagen-binding analysis.
Collagen-binding analysis of the purified collagenolytic protease.
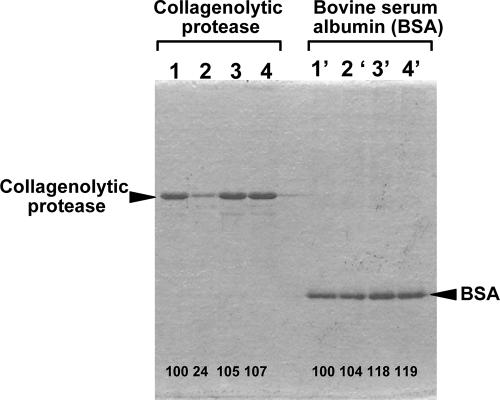
The collagenolytic protease purified from strain MO-1 was investigated to determine whether it could bind insoluble collagen and other insoluble proteins. Among insoluble proteins, elastin and feather keratin, in addition to type I collagen, were chosen for the binding analysis. Since the collagenolytic protease can degrade collagen molecules along with temperature elevation and, in addition, the background on the SDS-PAGE was drastically increased along with the elevation of temperature, mainly due to hydration of collagen, the incubation of the collagenolytic protease with insoluble proteins was performed at 0°C on ice to prevent, as much as possible, collagen breakdown. As shown in Fig. 2, the collagenolytic protease significantly bound with collagen but not with the other insoluble proteins, keratin and elastin, during 180-min incubation. In the binding assays performed by using the patterns of SDS-PAGE, most of the collagenolytic protease binding with collagen was precipitated with collagen, while the slight amount left in the supernatant appeared as a thinner band on SDS-PAGE. On the other hand, bovine serum albumin used instead of the collagenolytic protease, as a negative control protein, showed no interaction with the three insoluble proteins, including collagen, and was mostly recovered in the supernatant after centrifugation following incubation with the respective insoluble proteins. Bands for bovine serum albumin of unchanged thicknesses at the same positions were observed. These results demonstrated that the collagenolytic protease purified from strain MO-1 has the potency to specifically bind with collagen but not with other insoluble proteins.
FIG. 2.
Collagen binding of the collagenolytic protease from strain MO-1 and bovine serum albumin. The binding assays were carried out according to the method described in Materials and Methods. The collagenolytic protease (2.5 μg) was incubated without any insoluble protein (lane 1) or with collagen (5 mg; lane 2), keratin (5 mg; lane 3), or elastin (5 mg; lane 4) at 0°C for 180 min. Bovine serum albumin (2.5 μg) was incubated in variations corresponding to those of the collagenolytic protease (lanes 1′ to 4′). Numbers at the bottom of the gel are the densitometric ratios of each band compared with that of the control lane.
Collagen-binding analysis of GST fusion proteins.
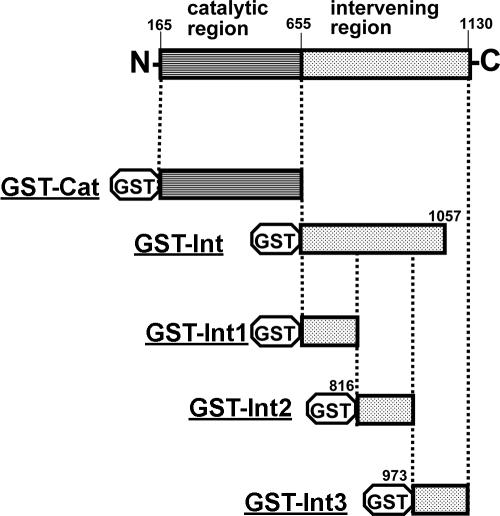
Fusion proteins of GST and various regions of the collagenolytic protease were constructed (Fig. 3), overexpressed in E. coli BL21, and purified to homogeneity. The catalytic region and the region intervening between the catalytic region and 14 repeats of 20 amino acids were subjected to binding analysis. The result showed that GST-Int had a binding capability comparable to that of the purified collagenolytic protease. In contrast, GST itself and the fusion protein including the catalytic region (GST-Cat) were kept on the supernatant without attaching on collagen. GST-Cat was found to be enzymatically active at elevated temperatures; therefore, the fusion protein was correctly folded in E. coli cells. This result indicated that the intervening region is responsible for collagen binding in the collagenolytic protease. A further investigation to find the segment for binding collagen in the intervening region was carried out. The intervening region was divided into three segments of approximately the same size (about 160 amino acids; see Fig. 3 for the positions). Calculated molecular masses were as follows: Int1, 17.4 kDa; Int2, 17.7 kDa; and Int3, 17.5 kDa. These proteins were fused to the C terminus of GST in the same manner as the fusion proteins constructed above (Fig. 3). After GST fusion proteins were purified to homogeneity with glutathione-immobilized affinity gel chromatography, their collagen-binding abilities were analyzed under the same condition in triplicate. The GST fusion proteins GST-Int2 and GST-Int3 showed significant collagen-binding capability (two-thirds and one-third of that of the mature collagenolytic protease, respectively), whereas the fusion protein GST-Int1 hardly bound collagen. In addition, the sum of the binding capabilities of the fusion proteins GST-Int2 and GST-Int3 was comparable to that of the mature collagenolytic protease and the fusion protein GST-Int. Thus, the segments (amino acid positions 816 to 1130) included in the fusion proteins GST-Int2 and GST-Int3 were found to be responsible for the collagen-binding capability of the collagenolytic protease from strain MO-1. From the secondary structure prediction of the segment, the segments are composed only of β-strands that possibly constitute β-sheets.
FIG. 3.
Physical maps for GST-collagenolytic protease fusion proteins. Numbers are amino acid positions in the collagenolytic protease (on the basis of the open reading frame).
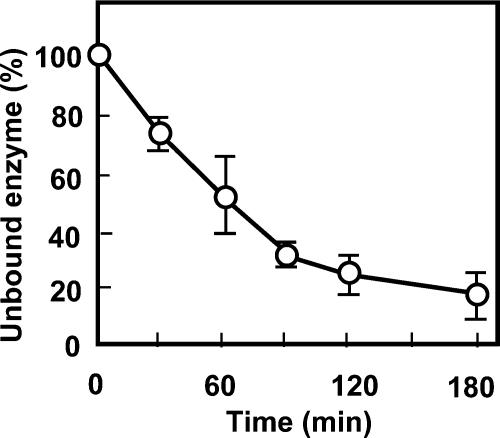
A time course analysis of the intervening region in collagen binding was carried out by use of the GST fusion protein GST-Int (Fig. 4), which gradually increased the ratio of bound collagen to total collagen, which reached the midpoint in 60 min, when 50% of the GST-Int molecules bound collagen. The binding of the collagenolytic protease with collagen was completed in about 90 min.
FIG. 4.
Time course of collagen binding of the collagenolytic protease. The assay was done for the purified fusion protein GST-Int by following the method described in Materials and Methods. Every 30 min, an aliquot of the sample was removed and subjected to SDS-PAGE with a 12.5% sheet gel. After the gel was destained, bands for the fusion protein GST-Int at each time were compared by use of quantitative analysis software in the imaging analyzer LAS-1000 Plus. The band for the collagenolytic protease added at the beginning was set as 100%.
The collagen binding of GST-Int was confirmed by SPR using a MultiSprinter system. Two proteins, acid-soluble collagen and BSA, were immobilized on a COOH tip by use of bifunctional cross-linkers, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and N-hydroxysuccinimide. Examination of the sensorgrams for collagen and BSA indicated that the two proteins exhibited different patterns of signal to immobilized type I collagen. The increase in SPR signals by the flow of the fusion protein GST-Int was detected only in the spot of collagen but not in BSA (data not shown). Since the increase was rather slow, the result supported the idea of slow binding of the collagenolytic protease to collagen. Furthermore, when the buffer (50 mM Tris-HCl [pH 7.5]) flowed to dissociate the fusion protein GST-Int binding on collagen, the signal was so slowly decreased that the dissociation rate could not be successfully obtained. However, this finding suggested that the binding of the fusion protein GST-Int is too tight to be released reversibly from collagen.
Condition for the collagenolytic protease to release from collagen.
Since the running buffer in SPR failed to detach the collagenolytic protease from collagen, the condition to release the collagenolytic protease from collagen was investigated through the addition of a salt solution (NaCl) or a nonionic detergent. The buffer (50 mM Tris-HCl [pH 7.5]) did not allow the collagenolytic protease to dissociate from collagen, which was in good accordance with the result obtained through SPR. On the other hand, the more the NaCl concentration was increased, the more collagenolytic protease was released from collagen into the solution in 60 min. However, three nonionic detergents, Triton X-100, Tween 20, and Tween 60, were ineffective for the release of the collagenolytic protease from collagen, even at 60 min. These results indicated that the collagenolytic protease binds collagen through hydrogen bonding between collagen and the intervening region and can be released in a highly ionic environment. Furthermore, the hydrophobic interaction is not critical for the binding of collagen and collagenolytic protease.
DISCUSSION
We have studied the degradation of collagens by a thermophilic bacterium, Geobacillus collagenovorans MO-1. In this study, we revealed the primary structure of the collagenolytic protease from strain MO-1 by gene cloning. Two features of the structure of the enzyme were confirmed by study of its open reading frame: (i) active site residues for a subtilisin family (S8) of serine peptidases and (ii) a large molecular mass. These characteristics correspond well to those obtained from the enzymological analysis in our previous report (19). Although the fact that the collagenolytic protease from strain MO-1 is conspicuously sensitive to the inhibitors of serine peptidases is in good agreement with the active site residue results, the broad substrate specificity of other enzymes belonging to the family is insufficient to ensure the specificity of the collagenolytic protease from strain MO-1 in collagen degradation. The majority of known collagenolytic proteases belong to a family of metallopeptidases containing a zinc ion at their active sites, where collagens are specifically recognized and hydrolyzed (12, 28). The greater molecular mass of the collagenolytic protease from strain MO-1, calculated from the open reading frame, also supports the idea that collagenolytic proteases tend to have a greater molecular mass than those of other proteases. However, the calculated molecular mass of collagenolytic protease (170 kDa) is much greater than that of the purified protein (105 kDa) (19, 32). The reasons are stated below.
In contrast, two unpredictable features were clarified in this study: (i) the signal sequence for secretion and the prosequence at the N terminus of the collagenolytic protease and (ii) the unusual 14 repeats of 20 amino acids at the C terminus. Met, the N-terminal amino acid empirically determined from the mature collagenolytic protease from strain MO-1 (19), negated the idea that the enzyme would be equipped with the signal and prosequences. The unusual repeats of 20 amino acids showed a significant homology with choline-binding protein PspC from S. pneumoniae, functioning as a cell-surface anchor (2), but Western blotting analysis using a polyclonal antibody against the collagenolytic protease from strain MO-1 followed by cell fractionation of strain MO-1 failed to provide evidence that the collagenolytic protease is displayed on the cell surface (data not shown). Another characteristic sequence of a cell surface anchor, the specific amino acid sequence LPXTG, is well known (1, 11). This LPXTG motif was not observed in the vicinity of the unusual repeats of the collagenolytic protease from strain MO-1. Therefore, the characteristic region containing the repeats is most likely secreted from the cell and cleaved in the process of maturation of the collagenolytic protease, judging from the apparent molecular mass (105 kDa) obtained by use of the purified protein (19).
We found, in addition to these features, a long region (53 kDa, calculated on the basis of the gene) intervening between the catalytic region and the unusual repeats. The intervening region shared no significant homology with any other proteins in the primary sequence; thus, its function remains unknown. In the previous report, we pointed out the abnormally large molecular mass of this collagenolytic protease and thus expected the existence of the characteristic structure giving priority over the decomposition of collagens in such a large molecule of the enzyme. In the case of Clostridium histolyticum ColG, there is a definite collagen-binding segment (14.1 kDa) in a molecular structure with a comparable molecular mass of 116 kDa (12). Together with the fact that subtilisin can fully function in a compact area of 30 kDa (21, 26), these findings allow us to presume that the intervening region of the collagenolytic protease from strain MO-1 plays a role in collagen binding to facilitate collagen degradation. Thus, we next investigated the ability of the intervening region to bind collagen. We found that the mature collagenolytic protease from strain MO-1 containing the intervening region at the C terminus bound collagen but not the other insoluble proteins, elastin and keratin (Fig. 2). In contrast, bovine serum albumin could bind none of the three insoluble proteins. Furthermore, GST-Int showed a collagen-binding ability comparable to that of the mature collagenolytic protease, whereas the GST itself and the fusion protein GST-Cat, containing the catalytic region, showed no binding ability. The ability of the intervening region to bind to collagen was also supported by SPR analysis. SPR analysis also demonstrated the binding of the collagenolytic protease to collagen is slow but tight, since the reversible release of the enzyme protein from collagen with the running buffer was unsuccessful. From further truncation analysis, the binding capability was attributed to two-thirds of the intervening region (Int2 and Int3), which is rich in β-strands and is approximately 35 kDa in molecular mass. The molecular sizes of the two segments seemed rather large for bacterial collagen-binding segments, since bacterial collagen-binding domains are much smaller than eukaryotic collagen-binding domains, as seen, for example, with ∼200 amino acid residues for hemopexine-like domains in matrix metalloproteases and ∼100 amino acids for clostridial collagen-binding domains (33). However, these results indicate that the collagenolytic protease from strain MO-1 has a region which binds collagen molecules at the C-terminal part of the matured form, and these segments may contribute to the degradation of collagens. It is plausible that the binding of the collagenolytic protease on collagen through the intervening region mediates and contributes to the cleavage of collagen at the active site of the enzyme.
Bacterial collagen-binding proteins of which the structures and the collagen-binding mechanism have been studied in detail are Clostridium histolyticum ColG and Staphylococcus aureus collagen adhesin (CNA) (13, 22, 27). The common features of the two are (i) a lack of homology in sequence and structure with eukaryotic collagen-binding domains and (ii) a characteristic β-sheet structure composed of numerous β-strands. We found that the collagenolytic protease from strain MO-1 shares these features with the two other bacterial proteins. However, strictly speaking, there is a difference in that ColG and CNA share neither the same fold nor any common residues with each other or with the collagenolytic protease from strain MO-1. Furthermore, while ColG has a central hot spot for binding on the interface of collagen and protein (33), CNA binds collagen on the residues spreading over the surface of the β-sheet groove of the CNA protein (27). In addition, while hydrogen bonds between the hot-spot residue (Tyr994) of ColG and the backbone of collagen are thought to be critical for binding (33), the hydrophobic interaction between the concave surface of CNA and collagen is apparently important (27). In the collagenolytic protease from strain MO-1, hydrogen bonds most likely predominate over the hydrophobic interaction for collagen binding, since a higher concentration of NaCl could release collagen from the enzyme surface but a nonionic detergent could not. In this respect, the collagenolytic protease from strain MO-1 shows a resemblance with ColG; however, the region which is responsible for collagen binding, encompassing the segments Int2 and Int3, extends over a larger region than that of ColG. This fact implies that the collagenolytic protease from strain MO-1 holds collagen on the surface of the larger region containing segments Int2 and Int3 with moderate binding ability, which is different from the mechanism proposed for ColG (13).
Since strain MO-1 is not a pathogen, the collagen-binding ability of the collagenolytic protease is not related to pathogenicity, which is completely different from the cases of Bacillus anthracis MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) (15, 35), C. histolyticum ColG (13), and Staphylococcus aureus CNA (27) but is advantageous for the collagenolytic protease from strain MO-1. Studies of collagen-binding domains or proteins from pathogenic strains are directly linked to prophylactic and therapeutic developments. It has been indicated that the collagen-binding domain of C. histolyticum ColG binds to all kinds of collagen fibrils in vivo and in vitro, and its collagen-binding ability implies the potential for applications to a drug delivery system (18, 29). Although the main function of collagenolytic protease for strain MO-1 is to degrade collagens in collaboration with two Pz peptidases as a nutrient source for cells (14), it is possible to apply the collagen-binding segment to such a drug delivery system by taking advantage of the nonpathogenicity of strain MO-1. In addition, we are searching for specific oligopeptide transporters in strain MO-1 that transport collagen peptides cleaved by the extracellular collagenolytic protease for two cytosolic Pz peptidases from extracellular space to the inside of a cell. It has been reported that in gram-positive bacteria such as lactobacilli, macromolecular proteins are degraded by extracellular proteases and the resulting oligopeptides are often transported by oligopeptide-binding proteins into the cells to be further hydrolyzed by various peptidases for optimum growth (4, 8). Our focus is on the establishment of the cascade of collagen degradation from the collagenolytic protease to Pz peptidases through the oligopeptide transporter(s) in strain MO-1. To the best of our knowledge, this is the first report of a thermophilic collagenolytic protease containing the collagen-binding segment.
Acknowledgments
We thank Naohiro Tomari and Yoshihiro Yamamoto, Kyoto Municipal Industrial Research Institute, for allowing the use of an SPR system.
REFERENCES
- 1.Boekhorst, J., M. W. de Been, M. Kleerebezem, and R. J. Siezen. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J. Bacteriol. 187:4928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman, S. A., K. Peek, M. Prescott, and R. Daniel. 1993. Characterization of a chelator-resistant proteinase from Thermus strain Rt4A2. Biochem. J. 295:463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garault, P., D. Le Bars, C. Besset, and V. Monnet. 2002. Three oligopeptide-binding proteins are involved in the oligopeptide transport of Streptococcus thermophilus. J. Biol. Chem. 277:32-39. [DOI] [PubMed] [Google Scholar]
- 5.Habig, W. H., M. J. Pabst, and W. B. Jakoby. 1974. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249:7130-7139. [PubMed] [Google Scholar]
- 6.Holck, A., and H. Naes. 1992. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO 151. J. Gen. Microbiol. 138:1353-1364. [DOI] [PubMed] [Google Scholar]
- 7.Kyo, M., T. Yamamoto, H. Motohashi, T. Kamiya, T. Kuroita, T. Tanaka, J. D. Engel, B. Kawakami, and M. Yamamoto. 2004. Evaluation of MafG interaction with Maf recognition element arrays by surface plasmon resonance imaging technique. Genes Cells 9:153-164. [DOI] [PubMed] [Google Scholar]
- 8.Levdikov, V. M., E. V. Blagova, J. A. Brannigan, L. Wright, A. A. Vagin, and A. J. Wilkinson. 2005. The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J. Mol. Biol. 345:879-892. [DOI] [PubMed] [Google Scholar]
- 9.Linsenmayer, T. F. 1991. Collagen, p. 7-44. In E. D. Hay (ed.), Cell biology of extracellular matrix. Plenum Press, New York, N.Y.
- 10.Luo, R., B. Mann, W. S. Lewis, A. Rowe, R. Heath, M. L. Stewart, A. E. Hamburger, S. Sivakolundu, E. R. Lacy, P. J. Bjorkman, E. Tuomanen, and R. W. Kriwacki. 2005. Solution structure of choline binding protein A, the major adhesin of Streptococcus pneumoniae. EMBO J. 24:34-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushita, O., T. Koide, R. Kobayashi, K. Nagata, and A. Okabe. 2001. Substrate recognition by the collagen-binding domain of Clostridium histolyticum class I collagenase. J. Biol. Chem. 276:8761-8770. [DOI] [PubMed] [Google Scholar]
- 13.Matsushita, O., C. M. Jung, J. Minami, S. Katayama, N. Nishi, and A. Okabe. 1998. A study of the collagen-binding domain of a 116-kDa Clostridium histolyticum collagenase. J. Biol. Chem. 273:3643-3648. [DOI] [PubMed] [Google Scholar]
- 14.Miyake, R., Y. Shigeri, Y. Tatsu, N. Yumoto, M. Umekawa, Y. Tsujimoto, H. Matsui, and K. Watanabe. 2005. Two thimet oligopeptidase-like Pz peptidases produced by a collagen-degrading thermophile, Geobacillus collagenovorans MO-1. J. Bacteriol. 187:4140-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 16.Murai, A., Y. Tsujimoto, H. Matsui, and K. Watanabe. 2004. An Aneurinibacillus sp. strain AM-1 produces a proline-specific aminopeptidase useful for collagen degradation. J. Appl. Microbiol. 96:810-818. [DOI] [PubMed] [Google Scholar]
- 17.Nimni, M. E. 1983. Collagen: structure, function, and metabolism in normal and fibrotic tissues. Semin. Arthritis Rheum. 13:1-86. [DOI] [PubMed] [Google Scholar]
- 18.Nishi, N., O. Matsushita, K. Yuube, H. Miyanaka, A. Okabe, and F. Wada. 1998. Collagen-binding growth factors: production and characterization of functional fusion proteins having a collagen-binding domain. Proc. Natl. Acad. Sci. USA 95:7018-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto, M., Y. Yonejima, Y. Tsujimoto, Y. Suzuki, and K. Watanabe. 2001. A thermostable collagenolytic protease with a very large molecular mass produced by thermophilic Bacillus sp. strain MO-1. Appl. Microbiol. Biotechnol. 57:103-108. [DOI] [PubMed] [Google Scholar]
- 20.Özbek, S., O. Pertz, M. Schwager, A. Lustig, T. Holstein, and J. Engel. 2002. Structure/function relationships in the minicollagen of Hydra nematocysts. J. Biol. Chem. 277:49200-49204. [DOI] [PubMed] [Google Scholar]
- 21.Rawlings, N. D., and A. J. Barrett. 1994. Families of serine peptidases. Methods Enzymol. 244:19-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid, R. J., and T. Coolbear. 2004. Lactocepin: the cell envelope-associated endopeptidase of lactococci, p. 1803-1808. In A. J. Barrett, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes, 2nd ed. Elsevier, Oxford, United Kingdom.
- 23.Rhem, M. N., E. M. Lech, J. M. Patti, D. McDevitt, M. Hook, D. B. Jones, and K. R. Wilhelmus. 2000. The collagen-binding adhesin is a virulence factor in Staphylococcus aureus keratitis. Infect. Immun. 68:3776-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Beato, A. R., R. Lopez, and J. L. Garcia. 1998. Molecular characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae. FEMS Microbiol. Lett. 164:207-214. [DOI] [PubMed] [Google Scholar]
- 25.Siezen, R. J., P. G. Bruinenberg, P. Vos, I. van Alen-Boerrigter, M. Nijhuis, A. C. Alting, F. A. Exterkate, and W. M. de Vos. 1993. Engineering of the substrate-binding region of the subtilisin-like, cell-envelope proteinase of Lactococcus lactis. Protein Eng. 6:927-937. [DOI] [PubMed] [Google Scholar]
- 26.Siezen, R. J., and J. A. Leunissen. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6:501-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Symersky, J., J. M. Patti, M. Carson, K. House-Pompeo, M. Teale, D. Moore, L. Jin, A. Schneider, L. J. DeLucas, M. Hook, and S. V. Narayana. 1997. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat. Struct. Biol. 4:833-838. [DOI] [PubMed] [Google Scholar]
- 28.Tam, E. M., T. R. Moore, G. S. Butler, and C. M. Overall. 2004. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): the differential roles of the MMP hemopexin c domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J. Biol. Chem. 279:43336-43344. [DOI] [PubMed] [Google Scholar]
- 29.Toyoshima, T., O. Matsushita, J. Minami, N. Nishi, A. Okabe, and T. Itano. 2001. Collagen-binding domain of a Clostridium histolyticum collagenase exhibits a broad substrate spectrum both in vitro and in vivo. Connect. Tissue Res. 42:281-290. [DOI] [PubMed] [Google Scholar]
- 30.Tsuruoka, N., T. Nakayama, M. Ashida, H. Hemmi, M. Nakao, H. Minakata, H. Oyama, K. Oda, and T. Nishino. 2003. Collagenolytic serine-carboxyl proteinase from Alicyclobacillus sendaiensis strain NTAP-1: purification, characterization, gene cloning, and heterologous expression. Appl. Environ. Microbiol. 69:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel, W. F. 2001. Collagen-receptor signaling in health and disease. Eur. J. Dermatol. 11:506-514. [PubMed] [Google Scholar]
- 32.Watanabe, K. 2004. Collagenolytic proteases from bacteria. Appl. Microbiol. Biotechnol. 63:520-526. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, J. J., O. Matsushita, A. Okabe, and J. Sakon. 2003. A bacterial collagen-binding domain with novel calcium-binding motif controls domain orientation. EMBO J. 22:1743-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wlodawer, A., M. Li, A. Gustchina, N. Tsuruoka, M. Ashida, H. Minakata, H. Oyama, K. Oda, T. Nishino, and T. Nakayama. 2004. Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity. J. Biol. Chem. 279:21500-21510. [DOI] [PubMed] [Google Scholar]
- 35.Xu, Y., X. Liang, Y. Chen, T. M. Koehler, and M. Hook. 2004. Identification and biochemical characterization of two novel collagen binding MSCRAMMs of Bacillus anthracis. J. Biol. Chem. 279:51760-51768. [DOI] [PubMed] [Google Scholar]