Abstract
Endonuclease V, encoded by the nfi gene, initiates removal of the base analogs hypoxanthine and xanthine from DNA, acting to prevent mutagenesis from purine base deamination within the DNA. On the other hand, the RdgB nucleotide hydrolase in Escherichia coli is proposed to prevent hypoxanthine and xanthine incorporation into DNA by intercepting the noncanonical DNA precursors dITP and dXTP. Because many base analogs are mutagenic when incorporated into DNA, it is intuitive to think of RdgB as acting to prevent similar mutagenesis from deaminated purines in the DNA precursor pools. To test this idea, we used a set of Claire Cupples' strains to detect changes in spontaneous mutagenesis spectra, as well as in nitrous acid-induced mutagenesis spectra, in wild-type cells and in rdgB single, nfi single, and rdgB nfi double mutants. We found neither a significant increase in spontaneous mutagenesis in rdgB and nfi single mutants or the double mutant nor any changes in nitrous acid-induced mutagenesis for rdgB mutant strains. We conclude that incorporation of deaminated purines into DNA is nonmutagenic.
Three of the four DNA bases—cytosine, adenine, and guanine—have amino groups. Deamination of these three bases (Fig. 1) generates the base analogs uracil, hypoxanthine, and xanthine, which have pairing specificities different from the original bases. Therefore, the cell employs excision repair to prevent mutagenesis due to spontaneous DNA base deamination. For example, in Escherichia coli, uracil-DNA glycosylase encoded by the ung gene initiates excision of uracils from DNA (12, 30). The ung mutants experience a 30-fold increase in GC→AT transitions (13), owing to the deamination of cytosine to uracil in the DNA (31). Deamination of adenine to hypoxanthine in the DNA (24) is also known to cause mutations: hypoxanthine preferentially base pairs with cytosine (3, 18, 35, 39, 51), and its formation in DNA, for example, as a result of nitrous acid exposure, leads to AT→GC transitions (45). Hypoxanthine removal from DNA in E. coli is initiated by the product of the nfi gene, endonuclease V (Endo V) (53, 54), and, to a much lesser extent, by the product of the alkA gene, 3-methyladenine-DNA glycosylase (43, 44). Although nfi mutants do not show increased spontaneous mutagenesis, they are more mutable by nitrous acid (45), an agent known to promote deamination of DNA bases (55).
FIG. 1.
Structures of the canonical DNA bases, their deaminated derivatives, and HAP.
Bases can deaminate not only in DNA but in nucleotides as well, both chemically (32, 46) and enzymatically (56). Deamination of DNA precursors should lead to accumulation of noncanonical nucleoside triphosphates in the cell's DNA precursor pool. These noncanonical nucleotides, such as dITP, dXTP, and dUTP, can be incorporated into the newly synthesized DNA, albeit less efficiently than regular DNA precursors (3, 38, 42, 51). Noncanonical DNA precursors are actively intercepted and removed from the DNA precursor pool. For example, dUTP is hydrolyzed to dUMP by deoxyuridine triphosphatase (2, 16), encoded by the dut gene (19). Indirect evidence (5; B. Budke and A. Kuzminov, unpublished data) strongly suggests that dITP and dXTP are the relevant in vivo targets of the RdgB protein, which is characterized in vitro as the dITP- and XTP-pyrophosphatase of E. coli (dXTP has yet to be tested) (7, 8).
Interception of noncanonical DNA precursors can be an important strategy in preventing mutagenesis due to the presence of a particular base analog in the DNA. For example, while mutM and mutY mutants, deficient in base excision repair of, or opposite to, 8-oxoguanine in DNA, show increased GC→TA transversion mutagenesis (15), mutT mutants, deficient in the interception of the noncanonical DNA precursor 8-oxo-dGTP, show an even stronger transversion mutagenesis in the opposite direction (AT→CG) (15). In order to see whether inactivation of rdgB is mutagenic due to incorporation of deaminated purines into DNA, we determined the spectra of spontaneous and nitrous acid-induced mutations in rdgB mutants and compared them to those of wild-type cells, nfi single mutants, and rdgB nfi double mutants.
MATERIALS AND METHODS
Strains, mutations, plasmids, and growth conditions.
All strains were derivatives of E. coli K-12. The 11 strains from Claire Cupples (CC strains) have various lacZ mutations, each of which reverts by a specific base pair change or frameshift, the mutant lacZ genes being harbored on an F′ episome in a Δ(pro-lac) strain (10, 11). The CC strains were maintained on M9 plus 0.2% glucose minimal plates to select for retention of the episome (37). Otherwise, strains were routinely grown in LB broth (10 g tryptone-5 g yeast extract-5 g NaCl per liter, pH 7.2) or on LB plates (15 g agar per liter of LB broth). The rdgB61 mutation, which is a precise deletion-replacement of the rdgB gene with a kanamycin resistance marker (5), was introduced into the CC strains by P1 transduction. The nfi gene was inactivated by transducing strains with the nfi-1 mutation, which has a chloramphenicol resistance marker inserted at position 443 of the nfi gene (17). The rdgB61 and nfi-1 mutations were confirmed by PCR. For the rdgB locus, primers were YggVF4 (5′-GTCCTCGCAACCGGCAATG-3′) and YggWR2 (5′-GCATCCGGCAATCAACGTC-3′), producing a 1,819-bp product with the wild-type locus and a 2,681-bp product with the rdgB61::kan allele. For the nfi locus, primers were NfiF1 (5′-GTGATTATGGATCTCGC-3′) and NfiR1 (5′-CGCTCCAGACGCAGATG-3′), producing a 754-bp product with the wild-type locus and a 2,624-bp product with the nfi-1::cat allele. Antibiotics were used at the following concentrations: spectinomycin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 10 μg/ml.
Endo V-recognized DNA modifications.
The 22.5-kbp plasmid pK96 (28) was introduced into each CC strain by CaCl2-facilitated colony transformation. pK96 plasmid DNA was isolated from rapidly growing LB broth cultures by alkaline-sodium dodecyl sulfate lysis (4), and 50 ng of it was treated with 0.075 U of Endo V (Trevigen) in 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.4-50 mM NaCl-2 mM MgCl2 at 37°C for 30 min, as described previously (5). The reaction was ethanol precipitated and electrophoresed on a 1.0% agarose Tris-acetate-EDTA gel for 20 h at 2.35 V/cm, after which the gel was transferred to a positively charged nylon membrane and hybridized to a pK96-specific 32P-labeled probe, generated by random primer labeling. The percentage of relaxed circular plasmid DNA in the total plasmid DNA was determined by phosphorimager as the ratio of the signal from the relaxed circular band to the sum of the signals from the relaxed and supercoiled bands of pK96.
Determination of mutation frequency.
Fresh colonies no more than 5 days old were grown on M9 plus glucose minimal medium and used to start 2-ml overnight LB cultures, with an initial cell density of ∼107 CFU per ml. The cultures were incubated with shaking at 28°C for 16 to 20 h and used directly to determine the lac reversion frequencies for each strain except CC101, CC103, and CC106 (and their derivatives). The 2-ml overnight cultures of CC101, CC103, and CC106, the strains that gave a low yield of lac revertants (less than one revertant per 1.0 ml of saturated broth culture), were diluted sixfold into a total of 12 ml of LB and outgrown to saturation before everything was plated on a single plate. Each saturated culture was titered for total viable cells carrying the F′ episome by plating serial dilutions onto M9 minimal medium plates supplemented with 0.2% glucose. The concentration of lac revertants in each culture was then determined by plating the culture directly onto M9 minimal medium plates supplemented with 0.2% lactose, incubating the culture at 30°C for 3 days, and scoring each plate for lac revertants by counting uniformly large colonies. When more than 200 μl of cell culture was to be plated, the cells were collected by centrifugation and resuspended in the residual medium and then plated on the minimal medium plus lactose. In separate experiments (not shown), we made sure that the number of Lac+ revertants per single plate increased linearly with the increasing volume of the saturated culture plated, up to at least 12 ml. The volumes of culture plated to yield 10 to 300 colonies per plate were 50 μl for CC107, CC108, and CC109; 100 μl for CC111; 1 ml for CC110; 1.5 ml for CC102, CC104, and CC105; and 12 ml for CC101, CC103, and CC106.
Nitrous acid mutagenesis.
In a procedure modified from reference 17, overnight LB cultures of the strains to be mutagenized were pelleted from the volumes listed above (except for strains CC107, CC108, and CC109, which were pelleted from 1.0 ml), resuspended in 1.0 ml of 0.1 M sodium acetate buffer, pH 4.6, and titered on M9 plus glucose minimal medium. Freshly made 0.4 M NaNO2 in 0.1 M acetate buffer, pH 4.6, was added to each cell suspension to yield a final concentration of 40 mM NaNO2, and the suspensions were incubated at 22°C for 10 min, after which the cells were collected by a 1-min centrifugation in a microcentrifuge, resuspended in 1.0 ml of M9 salts (which effectively stopped the treatment), titered on M9 glucose minimal agar for survivors, and plated directly (100 μl for strains CC107, CC108, and CC109; 1.0 ml for all other CC strains) on M9 lactose minimal agar for lac revertants. Between 10 and 50% of the cells survived the treatment.
UV mutagenesis.
Overnight LB cultures of strains to be mutagenized were subcultured 1:100 into fresh LB and grown to mid-logarithmic phase (optical density at 600 nm, 0.3), and 1.0 ml of the cell culture was pelleted and resuspended in 1.0 ml of 0.1 M MgSO4 with 0.1% Triton X-100 to facilitate the spreading of a ∼0.2-mm-thick layer of cell suspension over the rimmed top of a 9-cm plastic petri plate cover. Each culture thus spread was administered a precise dose of UV in a UV cross-linker sufficient to kill 97% of the cells in the suspension; 250 μl of the UV-irradiated suspensions were used to inoculate 5-ml LB cultures, which were shaken at 28°C overnight and then processed as described above for spontaneous mutagenesis.
MNNG mutagenesis.
The N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) mutagenesis protocol was essentially as described previously (37). The CC cultures to be mutagenized were administered a dose of MNNG sufficient to kill 50 to 90% of the cells. Log-phase cultures were washed twice in sodium citrate buffer, pH 5.5, incubated with 150 μg/ml MNNG for 15 to 30 min, washed twice in 1× M9 minimal salts, and titered for survivors. Mutagenized cultures were outgrown 1:20 into LB overnight cultures and processed for lac revertants the next day.
HAP mutagenesis.
6-Hydroxylaminopurine (HAP) (MP Biomedicals) was dissolved in 100% dimethyl sulfoxide (DMSO) on a 100°C heating block at 10 mg/ml, aliquoted into microcentrifuge tubes, and stored frozen at −20°C. HAP mutagenesis was carried out by diluting saturated 2-ml overnight cultures of the wild-type and rdgB mutant CC strains to a 10−5 dilution and using 5 μl (approximately 500 cells) to inoculate a 500-μl LB culture containing either 1% DMSO and the indicated concentration of HAP or 1% DMSO alone. The cultures were grown overnight at 30°C to saturation and then processed for Lac+ revertants according to the standard protocol; 200 μl of the overnight cultures were plated onto M9 plus 0.2% lactose minimal medium.
RESULTS
CC strains can detect increased mutagenesis.
Our goal was to examine the mutagenic potential and spectra of the rdgB61, nfi-1, and rdgB61 nfi-1 mutants of E. coli by using the set of CC strains (10, 11). Each of these 11 Lac− strains detects a specific base substitution or frameshift mutation by reverting to the Lac+ phenotype. We started by increasing the plating volumes in order to reliably detect the very rare GC→CG transversions (CC103) and AT→GC transitions (CC106) (11), which in our hands occurred about once per 1010 cells. We then confirmed that all of the strains of the set can detect increased mutagenesis when treated with DNA-modifying agents or combined with certain mutants.
All 11 CC strains except CC101, CC103, and CC106 responded to various extents to the nitrous acid-induced mutagenesis (Fig. 2). CC103 and CC106 did not respond to nitrous acid treatment (Fig. 2) but convincingly responded to MNNG (Fig. 3), in agreement with the literature (10, 11). We also tested CC101, CC103, CC105, and CC106 for mutagenesis with UV, but only CC105 showed a weak response (not shown). The only strain that failed to respond to any of these treatments, CC101 (AT→CG transversions), also failed to show increased mutagenesis when treated with 8-oxoguanine, hydrogen peroxide, or menadione. However, CC101 responded with more than a 10,000-fold increase in reversion to the introduction of a mutT mutation (Ella Rotman, personal communication). All of these tests confirm the original reports that (i) Lac− strains of the CC set revert to Lac+ with widely varying frequencies and (ii) in all strains, the reversion frequencies can be increased by specific DNA-modifying treatments (10, 11) or by specific mutations (52).
FIG. 2.
Confirmation of the ability to detect increased mutagenesis in some of the CC strains after treatment with nitrous acid (in acetate buffer). The data are medians of six to nine experiments ± first and third quartiles.
FIG. 3.
Confirmation of the ability to detect increased mutagenesis in selected CC strains with MNNG. The data are averages of two experiments run on different days.
Spontaneous mutagenesis spectra of rdgB and nfi mutants.
Next, we introduced rdgB61 and nfi-1 mutations into each of the 11 CC strains by P1 transduction and confirmed the mutants by PCR. Earlier we had shown that DNA of rdgB mutants (in an AB1157 background) accumulates modifications recognized by Endo V in vitro, but there are no such modifications in the DNA isolated from rdgB+ strains independently of their nfi gene status (5). To make sure that deaminated purines were still incorporated into the DNA of rdgB mutants in the CC background, we measured the density of Endo V-recognized DNA modifications, with the help of a large plasmid (5).
The 22.5-kbp pK96 plasmid was isolated from the original 11 CC strains, as well as from all their rdgB and nfi mutant derivatives, and treated in vitro with Endo V. The relative amount of the relaxed circular plasmid versus supercoiled plasmid was then determined by Southern hybridization. In all strains, the average level of relaxed circular pK96 in untreated samples was between 12.5% and 22%; the remainder was supercoiled plasmid (Fig. 4). When pK96 from the original CC strains, as well as from their nfi mutant derivatives, was treated with Endo V in vitro, there was no increase in the fraction of relaxed circular species compared to the untreated pK96 (Fig. 4). In contrast, when plasmid DNA isolated from the rdgB or rdgB nfi mutant derivatives of CC strains was treated with Endo V in vitro, the fraction of relaxed circular DNA was increased on average to 36 to 49%, suggesting that DNA of rdgB mutants does accumulate the expected density of Endo V-recognized DNA modifications (calculated earlier at 1 per 1.1 × 105 nucleotides [5]), presumably hypoxanthines and xanthines.
FIG. 4.
Accumulation of Endo V-recognized modifications in the DNA of rdgB mutants. A 22.5-kbp plasmid (pK96) was isolated from the 11 CC strains, as well as from their 11 ΔrdgB61, 11 nfi-1, and 11 ΔrdgB61 nfi-1 derivatives. The DNA was treated in vitro with Endo V, run on an agarose gel to separate supercoiled from relaxed circular species, transferred to a membrane, and hybridized with a pK96-specific probe. The fraction of relaxed circular DNA was then determined in the total plasmid preparation for every sample, and the averages for the 11 samples of the same genotype ± standard errors were plotted.
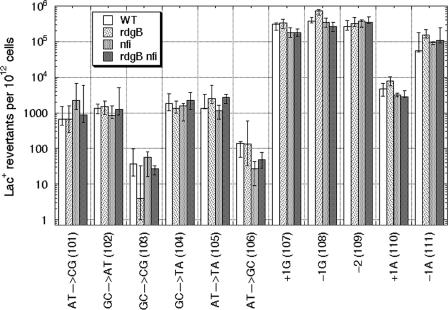
We then determined the frequencies of spontaneous reversions in the 11 CC strains, as well as in their rdgB, nfi, and rdgB nfi mutant derivatives. The reversion frequencies obtained from each experiment did not follow a normal distribution, owing to jackpots and experiments in which strains with low reversion frequencies did not yield any revertants. Therefore, the median was used to describe the reversion frequency of each strain. Overall, we found no substantial change in the frequency or spectrum of spontaneous mutations in either the rdgB61 mutant or nfi-1 mutant CC strains compared to the original rdgB+ nfi+ CC strains (Fig. 5). The double mutants also showed essentially the same spectrum (Fig. 5). We conclude that neither rdgB nor nfi defects, alone or in combination with each other, increase spontaneous mutagenesis.
FIG. 5.
Spontaneous mutagenesis in the 11 CC strains, as well as in their rdgB, nfi, and rdgB nfi mutant derivatives. The data are medians of eight or nine experiments ± first and third quartiles.
Nitrous acid-induced mutagenesis.
There was a possibility that the noncanonical DNA precursors intercepted by RdgB were only weakly mutagenic and that their low levels of incorporation in untreated cells (about 40 modifications per genome equivalent [5]) were not enough to cause increased mutagenesis. Acute deamination of the DNA precursors in the cell might have elevated this mutagenesis above the detection limit. On the other hand, our failure to detect mutagenic consequences of deaminated purines in the DNA precursor pools might have been due to their rapid repair in the DNA of CC strains, including nfi-1 mutants, which were supposed to be defective in this repair. To induce acute deamination of bases in the DNA and DNA precursor pools, as well as to verify the normal kinetics of repair of deaminated DNA bases in Nfi+ strains and the repair defect of nfi-1 derivatives, we determined the mutation spectrum induced by a short treatment with high concentrations of nitrous acid, the only DNA-damaging agent reported to reveal the nfi defect (45).
Nitrous acid deaminates purine and pyrimidine bases and is known to induce GC→AT transitions due to cytosine deamination (55). In our hands, treatment with nitrous acid yielded a large increase in GC→AT transitions (2,171), GC→TA transversions (60.8), and −1 frameshifts (120.9 and 651.9) and a modest increase in AT→TA transversions (9.3) and +1 frameshifts (17.2 and 29.7) in wild-type CC strains (Fig. 6A). No significant increase in either frameshifts or base substitutions over the wild-type level was found in rdgB61 strains mutagenized with nitrous acid, with the exception of +1 frameshifts in CC107 (Fig. 6). The nfi and rdgB nfi mutants treated with nitrous acid gave the expected (although weak in our hands) increase over the wild-type and rdgB single-mutant levels in GC→AT and AT→GC transitions (Fig. 6), corroborating the results of Schouten and Weiss (45). We conclude that the kinetics of deaminated base excision repair is normal in Nfi+ derivatives of CC strains, while transition mutagenesis is elevated in nfi mutant derivatives of the CC set in response to nitrous acid treatment.
FIG. 6.
Nitrous acid-induced mutagenesis. The data are medians of six experiments ± first and third quartiles. (A) Wild-type (WT) cells versus nfi mutants. (B) rdgB single mutants versus rdgB nfi double mutants.
HAP-induced mutagenesis.
Besides its “natural” substrates ITP, XTP, and dITP, RdgB is proposed to hydrolyze dHAP-TP (6), a deoxynucleoside triphosphate of the base analog HAP, which can be considered an intermediate between adenine and hypoxanthine (Fig. 1). HAP is a universal mutagen (reviewed in references 1 and 27), which is explained by the fact that in its coding capacity, HAP acts as an analog of either adenine or guanine (1, 41). Strains of the yeast Saccharomyces cerevisiae that are deficient in the Ham1 protein, which is an RdgB homolog, are hypermutable in the presence of HAP (27, 40). In contrast, E. coli controls detoxification of HAP with unknown molybdoproteins (26). In E. coli moa mutant strains, which are unable to synthesize the molybdenum cofactor and are thus hypersensitive to HAP, the rdgB defect decreases survival and increases mutagenesis after HAP treatment, suggesting that RdgB of E. coli also intercepts the DNA precursor form of HAP, dHAP-TP (6). However, the effects of rdgB and nfi mutations on HAP mutagenesis in otherwise wild-type cells have not been evaluated.
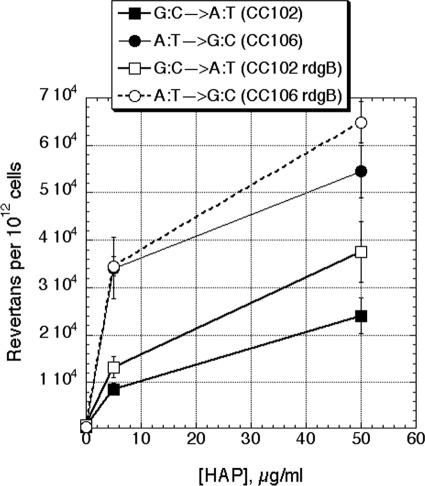
Although we demonstrated that the rdgB defect has no mutagenic consequence by itself, we predicted that there would be such a consequence in cells grown in the presence of HAP. To test this prediction, we determined the magnitude of HAP-induced mutagenesis in the two transition indicator strains of the CC set (CC102, GC→AT; CC106, AT→GC) under both RdgB+ and rdgB mutant conditions. We found that treatment with 5 μg/ml and 50 μg/ml of HAP, respectively, stimulates mutagenesis 16- to 60-fold and 45- to 95-fold in RdgB+ cells (Fig. 7). Surprisingly, we did not find a significant increase in HAP-induced mutagenesis in rdgB mutants over the RdgB+ levels (Fig. 7). At face value, this result suggests that RdgB recognizes dHAP-TP in vivo inefficiently.
FIG. 7.
HAP-induced mutagenesis. The data points are averages of four independent measurements ± standard errors.
DISCUSSION
Although formation of deaminated purines in DNA is mutagenic, we demonstrate that formation of deaminated purines in the DNA precursor pools and subsequent incorporation of these base analogs into DNA is not. We failed to detect any change in mutation spectra due to the rdgB defect, which blocks interception of dITP and dXTP in the DNA precursor pools, or due to the double rdgB nfi defect, which additionally prevents the incorporated deaminated purines from being excised from the DNA. Control experiments show that the strains of the CC set that we used (i) exhibit the expected mutagenic response to nitrous acid, confirming the normal repair kinetics of the deaminated bases in the DNA of nfi+ cells and the lack of alternative repair pathways in the nfi mutant cells; (ii) accumulate Endo V-recognized DNA modifications under rdgB mutant conditions; and (iii) are mutable by HAP, but independently of their rdgB status. There should be an explanation of why the incorporated deaminated purines are so different in their mutagenic properties from purines deaminated within DNA. Since the two deaminated purines, hypoxanthine and xanthine, are quite different in their pairing characteristics while RdgB is proposed to intercept both corresponding DNA precursors, dITP and dXTP, there is likely to be more than one explanation.
It is thoroughly established that hypoxanthine, which forms in DNA due to deamination of adenine, is highly mutagenic (Fig. 8). Since hypoxanthine has a strong pairing preference for cytosine (3, 18, 39, 51), changing adenines to hypoxanthines in DNA leads to AT→GC transitions. With this fact in mind, it is intuitively apparent that incorporation of hypoxanthine in the DNA of rdgB mutants should be also mutagenic and that, in addition, this is probably why the cell is trying to keep hypoxanthine out of its DNA. Hypoxanthine in its DNA precursor form, dITP, is readily incorporated into DNA by isolated nuclei (38) or by purified DNA polymerases (3, 42, 51). Moreover, there is at least one report that dITP can be mildly mutagenic under PCR conditions in which one of the canonical deoxynucleoside triphosphates is limiting (49). On the other hand, neither dUTP nor dITP is mutagenic under standard replication conditions in vitro (47) or when being “transformed” at high concentrations into live E. coli cells (20). We would like to argue that, exactly because of its strong pairing preference for cytosine, incorporated hypoxanthine cannot be mutagenic, because it acts strictly as a guanine analog (Fig. 8). Generally speaking, only base analogs capable of alternative pairing should be mutagenic when incorporated into DNA, while base analogs with strict pairing preferences cannot be mutagenic as DNA precursors. For example, uracil, which pairs only with adenine, is not mutagenic when incorporated but is highly mutagenic when formed in DNA by cytosine deamination. This peculiar point was independently raised previously (48).
FIG. 8.
Overall scheme of deaminated purine contamination and decontamination in DNA metabolism. dITP, dITP in DNA precursor pools; H, hypoxanthine in DNA; ???, lack of knowledge about excision repair of hypoxanthines downstream of Endo V nicking. Hypoxanthine in DNA may produce double-strand breaks in three ways: (i) by replication forks running into Endo V-induced nicks (shown), (ii) by direct cleavage by Endo V in single-stranded regions of the replication fork (because its hypoxanthine-specific endonuclease activity attacks both single- and double-stranded DNA [53]), and (iii) by two simultaneous Endo V nicks in the opposite strands of the same DNA duplex. We believe that the two latter mechanisms are less likely than the first one because of the very low density of Endo V-recognized DNA modifications, even in rdgB mutants (5).
Interestingly, according to this logic, the other deaminated purine, xanthine, which can pair with either thymine or cytosine (14, 23), should be highly mutagenic if incorporated into DNA. In the case of xanthine, the explanation for the lack of mutagenesis has to be entirely different. Since xanthine pairs equally poorly with both T and C, we suggest that xanthine cannot have a significant influence on mutagenesis because of the poor utilization of dXTP by DNA polymerases (3, 21, 34, 42, 50). Therefore, the combination of a strict pairing in the case of readily incorporatable hypoxanthine and an inefficient incorporation in the case of alternatively pairing xanthine leads to a somewhat counterintuitive conclusion, although fully backed by our results, that there should be no elevation of mutagenesis in rdgB single mutants or even in rdgB nfi double mutants.
As a test of this logic, we grew the two transition indicator strains in the presence of HAP, a purine base analog readily convertible into a noncanonical DNA precursor, dHAP-TP (6, 27), which incorporates into the DNA, causing transitional mutagenesis due to its approximately equal pairing with T or C (1, 41). Since it was proposed that RdgB can hydrolyze dHAP-TP in addition to dITP and dXTP (6), we expected that HAP-induced mutagenesis would be further and substantially increased in rdgB mutants. We indeed found a significant transitional mutagenesis in cultures grown in the presence of HAP, demonstrating the importance of intercepting the noncanonical DNA precursor dHAP-TP. However, the additional increase in mutagenesis in rdgB mutants exposed to HAP turned out to be statistically insignificant, suggesting that dHAP-TP is not a good substrate for RdgB in vivo.
Another test of this logic was the mutagenicity of nitrous acid, which deaminates bases in DNA as well as in DNA precursors. Nitrous acid increased the frequency of GC→AT and AT→GC transitions and GC→TA and AT→TA transversions, in addition to frameshift mutations. However, according to the above logic, nitrous acid should not induce additional mutagenesis in rdgB mutants, and it did not. On the other hand, the nfi-1 mutants yielded the expected weak increase in transitional mutagenesis (45). Furthermore, we found that the rdgB61 nfi-1 double mutants had no more nitrous acid-induced mutations than the nfi-1 single mutants did. These results show that RdgB does not protect the cell from mutations induced by nitrous acid, further corroborating our general point that incorporation of deaminated nucleotides is nonmutagenic.
When RdgB homologs from various organisms were first implicated in sanitizing the DNA precursor pool, one of the assumptions was that these enzymes act primarily to prevent mutagenesis (7, 8, 22, 29), which is often a consequence of incorporation of base analogs into DNA, 8-oxoguanine and HAP providing good examples (27, 36). To the contrary, our findings allow us to conclude that the primary function of RdgB in E. coli does not include prevention of mutagenesis, despite its clear role in sanitization of the DNA precursor pools. Rather, the main function of RdgB seems to be to prevent Endo V-triggered chromosomal fragmentation (Fig. 8), as suggested by (i) the unviability of rdgB recA, rdgB recBC, and rdgB ruv double mutants (5, 9, 33); (ii) the significant chromosomal fragmentation in rdgB recBC double mutants (5, 25); and (iii) the suppression of both unviability and chromosomal fragmentation by inactivation of Endo V (5; Budke and Kuzminov, unpublished data). It remains to be tested whether the rdgB defect actually increases the amount of dITP and dXTP in the DNA precursor pools and whether the Endo V-recognized modifications in the DNA of rdgB mutants are indeed hypoxanthines and xanthines.
Acknowledgments
We thank Claire Cupples (Concordia University, Montreal, Quebec), Mary Berlyn (E. coli Genetic Stock Center), and Jeffrey Miller (University of California, Los Angeles) for the sets of CC strains.
This work was supported by grant RSG-05-135-01-GMC from the American Cancer Society and by startup funds from the University of Illinois.
REFERENCES
- 1.Abdul-Masih, M. T., and M. J. Bessman. 1986. Biochemical studies on the mutagen, 6-N-hydroxylaminopurine. Synthesis of the deoxynucleoside triphosphate and its incorporation into DNA in vitro. J. Biol. Chem. 261:2020-2026. [PubMed] [Google Scholar]
- 2.Bertani, L. E., A. Haggmark, and P. Reichard. 1963. Enzymatic synthesis of deoxyribonucleotides, II. Formation and interconversion of deoxyuridine phosphates. J. Biol. Chem. 238:3407-3413. [PubMed] [Google Scholar]
- 3.Bessman, M. J., I. R. Lehman, J. Adler, S. B. Zimmerman, E. S. Simms, and A. Kornberg. 1958. Enzymatic synthesis of deoxyribonucleic acid. III. The incorporation of pyrimidine and purine analogues into deoxyribonucleic acid. Proc. Natl. Acad. Sci. USA 44:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 5.Bradshaw, J. S., and A. Kuzminov. 2003. RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol. Microbiol. 48:1711-1725. [DOI] [PubMed] [Google Scholar]
- 6.Burgis, N. E., J. J. Brucker, and R. P. Cunningham. 2003. Repair system for noncanonical purines in Escherichia coli. J. Bacteriol. 185:3101-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, J. H., J. H. Back, Y. I. Park, and Y. S. Han. 2001. Biochemical characterization of a novel hypoxanthine/xanthine dNTP pyrophosphatase from Methanococcus jannaschii. Nucleic Acids Res. 29:3099-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, J. H., H. Y. Park, J. H. Lee, and Y. Jang. 2002. Identification of the dITP- and XTP-hydrolyzing protein from Escherichia coli. J. Biochem. Mol. Biol. 35:403-408. [DOI] [PubMed] [Google Scholar]
- 9.Clyman, J., and R. P. Cunningham. 1987. Escherichia coli K-12 mutants in which viability is dependent on recA function. J. Bacteriol. 169:4203-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cupples, C. G., M. Cabrera, C. Cruz, and J. H. Miller. 1990. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 125:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan, B. K., P. A. Rockstroh, and H. R. Warner. 1978. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J. Bacteriol. 134:1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan, B. K., and B. Weiss. 1982. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J. Bacteriol. 151:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eritja, R., D. M. Horowitz, P. A. Walker, J. P. Ziehler-Martin, M. S. Boosalis, M. F. Goodman, K. Itakura, and B. E. Kaplan. 1986. Synthesis and properties of oligonucleotides containing 2′-deoxynebularine and 2′-deoxyxanthosine. Nucleic Acids Res. 14:8135-8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fowler, R. G., S. J. White, C. Koyama, S. C. Moore, R. L. Dunn, and R. M. Schaaper. 2003. Interactions among the Escherichia coli mutT, mutM, and mutY damage prevention pathways. DNA Repair 2:159-173. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg, G. R., and R. L. Somerville. 1962. Deoxyuridylate kinase activity and deoxyuridinetriphosphatase in Escherichia coli. Proc. Natl. Acad. Sci. USA 48:247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, G., and B. Weiss. 1998. Endonuclease V (nfi) mutant of Escherichia coli K-12. J. Bacteriol. 180:46-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill-Perkins, M., M. D. Jones, and P. Karran. 1986. Site-specific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat. Res. 162:153-163. [DOI] [PubMed] [Google Scholar]
- 19.Hochhauser, S. J., and B. Weiss. 1978. Escherichia coli mutants deficient in deoxyuridine triphosphatase. J. Bacteriol. 134:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hori, M., C. Ishiguro, H. Harashima, and H. Kamiya. 2005. In vivo mutagenicities of damaged nucleotides produced by nitric oxide and ionizing radiation. Biol. Pharm. Bull. 28:520-522. [DOI] [PubMed] [Google Scholar]
- 21.Horlacher, J., M. Tottiger, V. N. Podust, U. Hübscher, and S. A. Benner. 1995. Recognition by viral and cellular DNA polymerases of nucleosides bearing bases with nonstandard hydrogen bonding patterns. Proc. Natl. Acad. Sci. USA 92:6329-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang, K. Y., J. H. Chung, S.-H. Kim, Y. S. Han, and Y. Cho. 1999. Structure-based identification of a novel NTPase from Methanococcus jannaschii. Nat. Struct. Biol. 6:691-696. [DOI] [PubMed] [Google Scholar]
- 23.Kamiya, H., M. Shimizu, M. Suzuki, H. Inoue, and E. Ohtsuka. 1992. Mutation induced by deoxyxanthosine in codon 12 of a synthetic c-Ha-ras gene. Nucleosides Nucleotides 11:247-260. [PubMed] [Google Scholar]
- 24.Karran, P., and T. Lindahl. 1980. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry 19:6005-6011. [DOI] [PubMed] [Google Scholar]
- 25.Kouzminova, E. A., E. Rotman, L. Macomber, J. Zhang, and A. Kuzminov. 2004. RecA-dependent mutants in E. coli reveal strategies to avoid replication fork failure. Proc. Natl. Acad. Sci. USA 101:16262-16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozmin, S. G., Y. I. Pavlov, R. L. Dunn, and R. M. Schaaper. 2000. Hypersensitivity of Escherichia coli Δ(uvrB-bio) mutants to 6-hydroxylaminopurine and other base analogs is due to a defect in molybdenum cofactor biosynthesis. J. Bacteriol. 182:3361-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozmin, S. G., R. M. Schaaper, P. V. Shcherbakova, V. N. Kulikov, V. N. Noskov, M. L. Guetsova, V. V. Alenin, I. B. Rogozin, K. S. Makarova, and Y. I. Pavlov. 1998. Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat. Res. 402:41-50. [DOI] [PubMed] [Google Scholar]
- 28.Kuzminov, A., and F. W. Stahl. 1997. Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J. Bacteriol. 179:880-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, S., A. G. McLennan, K. Ying, Z. Wang, S. Gu, H. Jin, C. Wu, W. Liu, Y. Yuan, R. Tang, Y. Xie, and Y. Mao. 2001. Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the ITPA gene. J. Biol. Chem. 276:18695-18701. [DOI] [PubMed] [Google Scholar]
- 30.Lindahl, T. 1974. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. USA 71:3649-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindahl, T., and B. Nyberg. 1974. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 13:3405-3410. [DOI] [PubMed] [Google Scholar]
- 32.Lucas, L. T., D. Gatehouse, and D. E. Shuker. 1999. Efficient nitroso group transfer from N-nitrosoindoles to nucleotides and 2′-deoxyguanosine at physiological pH. A new pathway for N-nitrosocompounds to exert genotoxicity. J. Biol. Chem. 274:18319-18326. [DOI] [PubMed] [Google Scholar]
- 33.Lukas, L., and A. Kuzminov. 2006. Chromosomal fragmentation is the major consequence of the rdgB defect in Escherichia coli. Genetics 172:1359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lutz, M. J., J. Horlacher, and S. A. Benner. 1998. Recognition of a non-standard base pair by thermostable DNA polymerases. Bioorg. Med. Chem. Lett. 8:1149-1152. [DOI] [PubMed] [Google Scholar]
- 35.Martin, F. H., M. M. Castro, F. Aboul-ela, and I. J. Tinoco. 1985. Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 13:8927-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Myrnes, B., P. H. Guddal, and H. Krokan. 1982. Metabolism of dITP in HeLa cell extracts, incorporation into DNA by isolated nuclei and release of hypoxanthine from DNA by a hypoxanthine-DNA glycosylase activity. Nucleic Acids Res. 10:3693-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordmann, P. L., J. C. Makris, and W. S. Reznikoff. 1988. Inosine induced mutations. Mol. Gen. Genet. 214:62-67. [DOI] [PubMed] [Google Scholar]
- 40.Pavlov, Y. I. 1986. Saccharomyces cerevisiae mutants highly sensitive to the mutagenic action of 6-N-hydroxylaminopurine. Soviet Genet. 22:1099-1107. [Google Scholar]
- 41.Pavlov, Y. I., V. V. Suslov, P. V. Shcherbakova, T. A. Kunkel, A. Ono, A. Matsuda, and R. M. Schaaper. 1996. Base analog N6-hydroxylaminopurine mutagenesis in Escherichia coli: genetic control and molecular specificity. Mutat. Res. 357:1-15. [DOI] [PubMed] [Google Scholar]
- 42.Piccirilli, J. A., T. Krauch, S. E. Moroney, and S. A. Benner. 1990. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature 343:33-37. [DOI] [PubMed] [Google Scholar]
- 43.Saparbaev, M., and J. Laval. 1994. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl. Acad. Sci. USA 91:5873-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saparbaev, M., J.-C. Mani, and J. Laval. 2000. Interactions of the human, rat, Saccharomyces cerevisiae and Escherichia coli 3-methyladenine-DNA glycosylases with DNA containing dIMP residues. Nucleic Acids Res. 28:1332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schouten, K. A., and B. Weiss. 1999. Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res. 435:245-254. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro, R., and S. H. Pohl. 1968. The reaction of ribonucleosides with nitrous acid. Side products and kinetics. Biochemistry 7:448-455. [DOI] [PubMed] [Google Scholar]
- 47.Sinha, N. K., and M. D. Haimes. 1981. Molecular mechanisms of substitution mutagenesis. An experimental test of the Watson-Crick and Topal-Fresco models of base mispairings. J. Biol. Chem. 256:10671-10683. [PubMed] [Google Scholar]
- 48.Snow, E. T., and S. Mitra. 1988. Role of carcinogen-modified deoxynucleotide precursors in mutagenesis. Mutat. Res. 200:157-164. [DOI] [PubMed] [Google Scholar]
- 49.Spee, J. H., W. M. de Vos, and O. P. Kuipers. 1993. Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res. 21:777-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki, T., M. Yoshida, M. Yamada, H. Ide, M. Kobayashi, K. Kanaori, K. Tajima, and K. Makino. 1998. Misincorporation of 2′-deoxyoxanosine 5′-triphosphate by DNA polymerases and its implication for mutagenesis. Biochemistry 37:11592-11598. [DOI] [PubMed] [Google Scholar]
- 51.Thomas, K. R., P. Manlapaz-Ramos, R. Lundquist, and B. M. Olivera. 1979. Formation of Okazaki pieces at the Escherichia coli replication fork in vitro. Cold Spring Harbor Symp. Quant. Biol. 43:231-237. [DOI] [PubMed] [Google Scholar]
- 52.Vidmar, J. J., and C. G. Cupples. 1993. MutY repair is mutagenic in mutT− strains of Escherichia coli. Can. J. Microbiol. 39:892-894. [DOI] [PubMed] [Google Scholar]
- 53.Yao, M., Z. Hatahet, R. J. Melamede, and Y. W. Kow. 1994. Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem. 269:16260-16268. [PubMed] [Google Scholar]
- 54.Yao, M., and Y. W. Kow. 1995. Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem. 270:28609-28616. [DOI] [PubMed] [Google Scholar]
- 55.Zimmermann, F. K. 1977. Genetic effects of nitrous acid. Mutat. Res. 39:127-148. [DOI] [PubMed] [Google Scholar]
- 56.Zoref-Shani, E., A. Shainberg, and O. Sperling. 1987. Pathways of adenine nucleotide catabolism in primary rat muscle cultures. Biochim. Biophys. Acta 926:287-295. [DOI] [PubMed] [Google Scholar]