Abstract
MicroRNAs (miRNAs) constitute a novel, extensive class of small RNAs (~21 nucleotides), and play important gene-regulation roles during growth and development in various organisms. Here we conducted a homology search to identify homologs of previously validated miRNAs from silkworm genome. We identified 24 potential miRNA genes, and gave each of them a name according to the common criteria. Interestingly, we found that a great number of newly identified miRNAs were conserved in silkworm and Drosophila, and family alignment revealed that miRNA families might possess single nucleotide polymorphisms. miRNA gene clusters and possible functions of complement miRNA pairs are discussed.
Keywords: miRNA, Silkworm, Computational prediction
INTRODUCTION
MicroRNAs (miRNAs) represent a class of noncoding small RNAs (~22 nucleotides) identified from various organisms extending from nematodes to humans (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). They derive from pre-miRNAs extracted from primary transcripts (pri-miRNA) and can form stable hairpin structures. Pre-miRNAs are processed by RNase III enzyme Dicer, which also cleaves long, perfectly double-stranded RNA into 21~22 nucleotides (nt) siRNA duplexes (Hannon, 2002).
There are two types of mechanisms for miRNA-dependent regulation of gene expression: (1) target mRNAs which can perfectly complement miRNAs cleaved by RISC (RNA-induced silencing complex); (2) miRNAs which inhibit the expression of mRNAs that are imperfectly complementary to miRNA. Functions of several miRNAs have been reported before, although biological functions of miRNAs are not yet completely understood. The founding members of miRNAs are lin-4 and let-7 in C. elegans (Lee et al., 1993; Reinhart et al., 2000), which are also called small temporal RNAs because their inactivative mutation affects developmental timing (Banerjee and Slack, 2002). Lin-4 and let-7 exerted their functions by complementing to 3′ untranslated region (3′ UTR) of target mRNAs and inhibiting their translation processes (Lee et al., 1993; Reinhart et al., 2002). In Drosophila, the defect of miR-14 leads to reaper-dependent cell death and increases levels of triacylglycerol and diacylglycerol, which are required for fat metabolism (Xu et al., 2003); miRNA encoded in bantam locus controls the cell proliferation and regulates the proapoptotic gene hid (Brennecke et al., 2003). These findings raise the possibility that miRNAs perform as negative regulators to control cell proliferation, differentiation, and other developmental processes. Similar to animal miRNAs, plant miRNAs exhibit temporal and tissue-specific expression patterns (Llave et al., 2002; Mourelatos et al., 2002; Park et al., 2002; Reinhart and Bartel, 2002). Many target genes which perfectly complement plant miRNAs were found, however, identification of putative miRNAs targets in animals is much more complex, because imperfect complementarities between miRNAs and target genes increase the difficulties in investigating the interaction between miRNAs and target mRNAs.
Up to now, hundreds of miRNAs, usually having multiple copies in vivo, have been identified from various species. For instance, in nematodes, most identified miRNAs present very high steady-state levels—more than 1000 molecules per cell, with some exceeding 50000 molecules per cell (Lim et al., 2003). Llave et al.(2002) identified 125 small RNAs between 16 and 25 nt; only four of these met the criteria (Ambros et al., 2003) of miRNAs. Similarly, Park et al.(2002) identified 230 unique sequences, five of which appeared to be miRNAs. Nowadays, a great proportion of known miRNAs were identified by computational approaches or by the combination of biochemical methods and computational procedures. It seems that few miRNAs have not been uncovered in model organisms. On the other hand, miRNAs from non-model organisms, such as silkworm, have not yet been reported.
Recently, draft sequence of domesticated silkworm genome has been accomplished. It revealed that there are 18 510 genes in the silkworm genome, which far exceeds the official gene count of 13379 for fruitfly and that a great number of silkworm genes have homologs in fruitfly (Xia et al., 2004). These accomplishments made it possible to investigate function genes in silkworm genome by implementing computational procedures. In the present study, we conducted a homology search to select new silkworm miRNA genes that are homologous with identified miRNAs from other organisms. As a result, we identified many paralogs of known miRNAs with one or more different nucleotides with most potential miRNAs perfectly complementing with query sequences from Drosophila.
MATERIALS AND METHODS
Identified mature miRNAs of model organisms (Mar. 2005, downloaded from http://166.111.30.65/micrornadb/index.php) were used as query sequences to BLAST against silkworm genome (http://www.dna.affrc.go.jp/database/silkworm.html) with default parameters and a non-stringent cutoff of E>1.8 (Lim et al., 2003). Then we picked out flanking sequences of hit sites (about ±60 nt) from silkworm genome sequences. The extracted sequences were submitted to Mfold Web server (Zuker, 2003) to predict fold-back structures. Sequences were eliminated either more than 3 internal nucleotides mismatched to query sequences or total mismatched nucleotides of over 4 nt, or the free energy of the second structures were higher than −1.05×102 kJ/mol (Lim et al., 2003).
For further validation, all identified sequences that can form stable stem-loop structures were aligned with their family members and subjected to phylogenetic analysis on the relationship between members in each family by using ClustalX and PHYLIP (download from http://evolution.genetics.washington.edu/phylip.html).
RESULTS
Identification of 24 miRNA genes from silkworm genome
In the present research, we adopted a homology search procedure to search for miRNA genes that are homologous with identified miRNAs from model organisms. As a result, 24 novel potential miRNA genes were identified from silkworm genome (Table 1). With each of them given a name according to the common criteria such as bmo-let-7 (Bombyx mori, bmo-). Twenty-two newly identified sequences were homologous with query miRNAs from Drosophila and two had C. briggsae homologs. We failed to find any sequences matching query miRNAs from the other species, excepting that seven sequences (bmo-let-7, bmo-mir-1, bmo-mir-7, bmo-mir-8, bmo-mir-9a, bmo-mir-124, and bmo-mir-263b) had homologs in several species in addition to Drosophila.
Table 1.
Candidate microRNAs in silkworm genome
| Namea | Sourceb | Match extendc | Matched sequenced | Fold-back sizee |
| bmo-let-7 | AADK01002497 (10858-10878) | 21/21 | TGAGGTAGTAGGTTGTATAGT | 96 |
| bmo-miR-1 | AADK01005296 (6087-6108) | 22/22 | TGGAATGTAAAGAAGTATGGAG | 97 |
| bmo-miR-7 | AADK01002696 (14044-14066) | 23/23 | TGGAAGACTAGTGATTTTGTTGT | 98 |
| bmo-miR-8 | AADK01023997 (815-837) | 23/23 | TAATACTGTCAGGTAAAGATGTC | 98 |
| bmo-miR-9a | Contig551684 (34-56) | 23/23 | TCTTTGGTTATCTAGCTGTATGA | 88 |
| bmo-miR-10c | AADK01012541 (8182-8201) | 17/21 | CTCTCTAGAACCGAATTTGT | 96 |
| bmo-miR-14 | AADK01001451 (17523-17543) | 21/21 | TCAGTCTTTTTCTCTCTCCTA | 96 |
| bmo-miR-31c | AADK01000344 (33951-33971) | 20/23 | GGCAAGAAGTCGGCATAGCTG | 98 |
| bmo-miR-34 | Contig567840 (1436-1457) | 22/22 | TGGCAGTGTGGTTAGCTGGTTG | 97 |
| bmo-miR-71 | AADK01011192 (8467-8485) | 19/19 | TGAAAGACATGGGTAGTGA | 64 |
| bmo-miR-124 | AADK01010415 (7041-7063) | 23/23 | TAAGGCACGCGGTGAATGCCAAG | 98 |
| bmo-miR-263b | AADK01025663 (3376-3396) | 21/21 | CTTGGCACTGGGAGAATTCAC | 96 |
| bmo-miR-263c | Contig476700 (2833-2853) | 21/24 | AATGGCACTGGAAGAATTCAC | 99 |
| bmo-miR-275 | AADK01007727 (11262-11284) | 23/23 | TCAGGTACCTGAAGTAGCGCGCG | 70 |
| bmo-miR-276b | AADK01021256 (906-927) | 22/23 | AGCGAGGTATAGAGTTCCTACG | 97 |
| bmo-miR-276a* | AADK01021256 (941-962) | 22/22 | TAGGAACTTCATACCGTGCTCT | 97 |
| bmo-miR-277 | Contig132079 (12099-12121) | 23/23 | TAAATGCACTATCTGGTACGACA | 108 |
| bmo-miR-279b | Contig467689 (714-732) | 19/22 | TGACTAGATCCACACTCAT | 87 |
| bmo-miR-282b | AADK01024756 (1421-1445) | 24/28 | ACCTAGCCTCTCCTTGGCTTTGTCTGT | 87 |
| bmo-miR-283 | AADK01009773 (8152-8172) | 21/21 | TAAATATCAGCTGGTAATTCT | 96 |
| bmo-miR-305 | AADK01007727 (11352-11374) | 23/23 | ATTGTACTTCATCAGGTGCTCTG | 63 |
| bmo-miR-307 | AADK01002680 (19093-19112) | 20/20 | TCACAACCTCCTTGAGTGAG | 95 |
| bmo-miR-iab-4 | AADK01004318 (3477-3498) | 22/22 | ACGTATACTGAATGTATCCTGA | 97 |
| bmo-miR-iab-4* | AADK01004318 (3515-3538) | 24/24 | CGGTATACCTTCAGTATACGTAAC | 97 |
Each miRNA gene is given a name according to common convention
Silkworm genome sequences that contain novel miRNA genes
The extent of the match is indicated in the format of “number of matched ncleotides/query sequence length”
Matched sequences in silkworm genome, and mismatched nucleotides are indicated as little characters
Predicted length of identified miRNAs precursors
means the miRNA is produced from antisence strand
Most identified miRNA genes perfectly matched the query sequences, while the remaining were homologous with query sequences with one or more mismatching nucleotides. Imperfectly matched miRNA genes either had different nucleotides at their 5′/3′ terminus compared to query sequences or contained diverged nucleotides in their internal regions. In total, the number of mismatched nucleotides in potential miRNA genes was less than 4 nt, providing they had the same length as their query sequences. Although these sequences extended from 19 to 27 nt, they had a much tighter distribution, centering on 21~24 nt which were coincident with known specificity of Dicer processing. Interestingly, most identified sequences had composition preferable beginning with an uridine at the 5′ terminus, which also had been observed in miRNAs from other organisms (Lee and Ambros, 2001; Lagos-Quintana et al., 2002; Reinhart et al., 2002). Twenty-four potential miRNAs can be grouped into 22 families, each comprising of 2 to 28 genes. Within these families, consensual sequences either span through the whole length of miRNAs or are predominantly at their 5′/3′ terminus.
Of the 22 families, 6 largest families contained over 10 miRNAs while 9 families only had 2 members from Drosophila and silkworm.
Homology analysis
According to the criteria of miRNA annotation, homologs of previously validated miRNAs need not meet as stringent criteria to be annotated as additional miRNA loci. Very close homologs in other species can be annotated as miRNA homologs without experimental validation, if they have phylogenic conservation and predicted fold-back precursor secondary structure (Ambros et al., 2003). In order to furher verify candidate miRNAs, pre-miRNAs from each family were subjected to family alignment process using Clustal X, including known miRNAs and novel candidates.
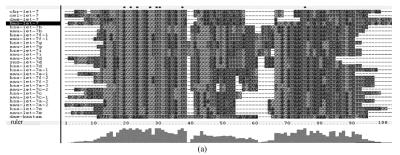
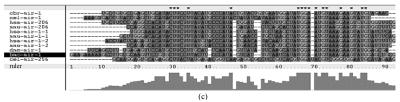
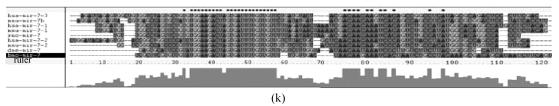
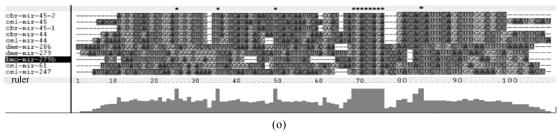
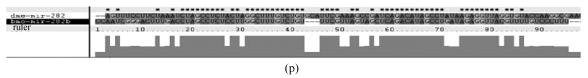
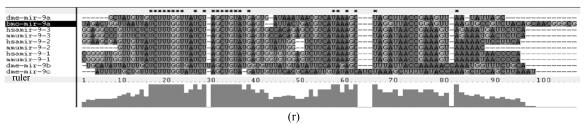
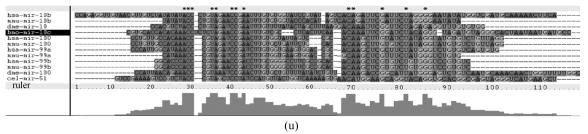
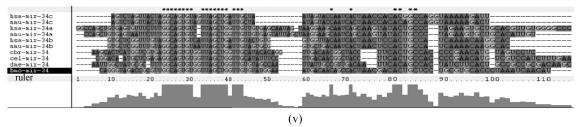
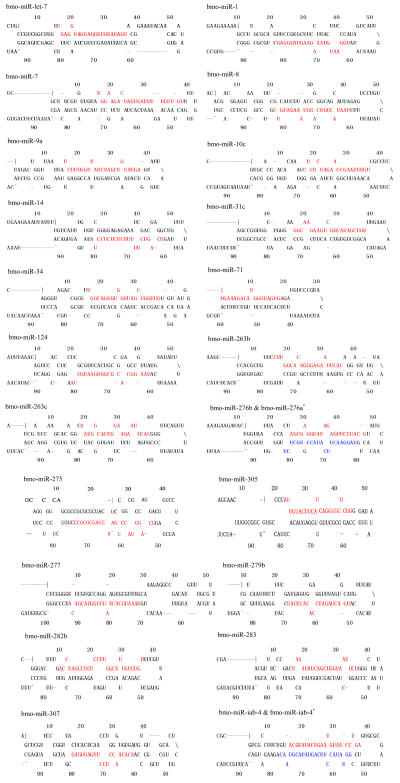
Although let-7 family contained more than 30 members, extending from worm through human, both miRNA-encoding regions and complementary sequences showed high conservation (Fig.1). Mir-124 family, mir-276 family and mir-71 family showed dramatically high conservation that over 20 continuous nucleotides were identical. Four families (mir-307, mir-7, mir-263b and mir-8) contained over 10 continuously identical nucleotides within miRNA-coding regions. However, both flanking regions and loops are variable (Fig.1). Interestingly, alignment results showed that most families might have characteristics of single nucleotide polymorphism.
Fig. 1.
Sequence alignments of pre-miRNAs in each miRNA family. (a) let-7 family; (b) mir-124 family; (c) mir-1 family; (d) mir-14 family; (e) mir-275 family; (f) mir-305 family; (g) mir-276 family; (h) mir-263b family; (i) mir-283 family; (j) mir-307 family; (k) mir-7 family; (l) mir-8 family; (m) mir-iab-4 family; (n) mir-31c family; (o) mir-279 family; (p) mir-282 family; (q) mir-263a family; (r) mir-9a family; (s) mir-277 family; (t) mir-71 family; (u) mir-10 family; (v) mir-34 family
Members of each miRNA family that contain novel silkworm miRNAs are subjected to sequence alignment; Precursors of silkworm miRNAs are indicated by black ground. The length of pre-miRNAs consistent with that of foldback predicted sequence. Dark dot in the first line of pictures indicates constant nucleotide
Phylogeny analysis
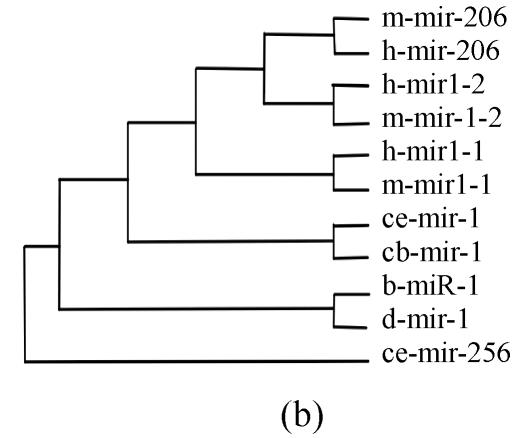
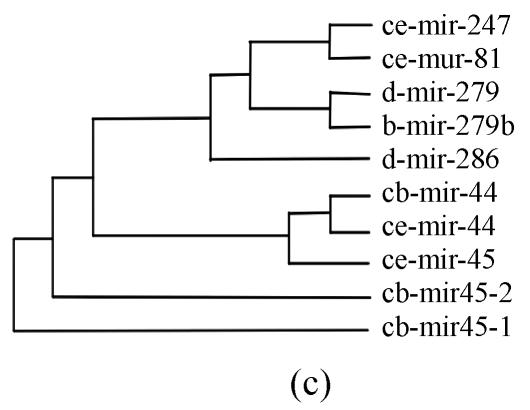
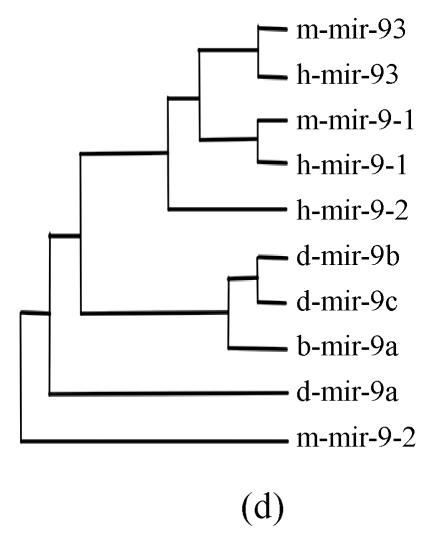
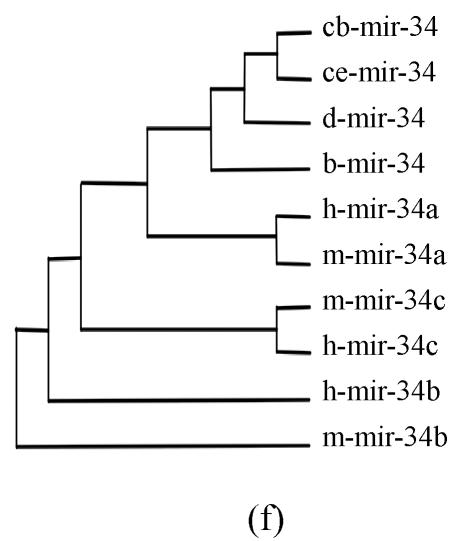
Pre-miRNAs from six largest families were subjected to phylogeny analysis using dnapairs program in PHYLIP package for analyzing revolution relationship of members from each family. Most silkworm miRNAs were located together in branches with related Drosophila miRNAs, with the exception of bmo-mir-10c, which was relatively less related to any Drosophila microRNA (Fig.2). Commonly, human miRNAs had close revolution relationship with mouse miRNAs (Fig.2). It might attribute to the fact that a number of miRNAs in the two species share identical sequences. Let-7 family contained more than 30 sequences from divergent species, with most of them from mammalians (Fig.1). Interestingly, both m-let-7c-1 and h-let-7c located at shortest branches of let-7 family tree (Fig.2), indicating they may be the earliest members of let-7 family.
Fig. 2.
Phylogeny analysis of six large families. (a) let-7 family; (b) mir-1 family; (c) mir-279 family; (d) mir-9 family; (e) mir-10 family; (f) mir-34 family
Members from six large families, which contain novel miRNAs in the present study, were submitted to construct phylogeny trees in order to analysis the relationship between family members from different species. Abbreviations are indicated as follows: h: human; r: Rattus norvegicus; m: Mus musculus; d: Drosophila melanogaster; b: Bombyx mori; cb: Caenorhabditis briggsae; ce: Caenorhabditis elegans
Clusters and complement miRNA pairs
Mapping miRNA genes into silkworm genome, we discovered bmo-miR-275 and bmo-miR-305 were hosted in single sequence as their homology from Drosophila. However, the distance between these two silkworm miRNA genes was farther. As similar phenomenon had been reported before, we predicted that there might be more clusters existing in silkworm. Indeed, applying similar strategy, we found another two clusters (bmo-miR-iab-4/bmo-miR-iab-4* and bmo-miR-276b/bmo-miR-276a*). Moreover, bmo-miR-iab-4/bmo-miR-iab-4* and bmo-miR-276b/bmo-miR-276a* can complement with each other, respectively (Fig.3). Fold-back prediction showed these four miRNAs came from two miRNA precursors.
Fig. 3.
Predictions of secondary structures of novel miRNAs from silkworm genome
Each of novel miRNAs is submitted to Mfold Web server to verify whether they can form fold back structure and whether their secondary structure is stable or not. Mature miRNA indicated by red or blue. miRNAs produced from antisense strand of pre-miRNA are indicated as blue characters
DISCUSSION
Silkworm is a model organism of lepidoptera that is widely used in industry, especially in textile production. Up to now, hundreds of miRNAs have been identified from various species; although, miRNAs from silkworm have not yet been reported. In the present study, by applying homologous alignment procedure, we found 24 potential miRNA genes located in various sites of the silkworm genome. Most of the novel miRNA genes were highly homologous to query miRNAs from other species and the foldback prediction showed each of the potential miRNAs located in either arm of hairpin structure, which is consistent with query miRNAs. Moreover, sequences extracted from flanking regions of hit loci can form stable stem-loop structure (dG≤−1.05×102 kJ/mol). Based on these observations and the annotation criteria for homology of previously validated miRNAs (Ambros et al., 2003), we proposed that these loci could be annotated as miRNA genes. Of course, we cannot exclude the possibility that they are pseudo-miRNAs due to lack of direct experimental evidence.
Phylogeny analysis revealed that within six largest miRNA families in the present research, human miRNAs usually had closer revolution relationship with mouse miRNAs than the other species (Fig.2). This phenomenon might be due to the miRNAs characteristic of high conservation, as organisms that have close phylogeny relationship usually share a number of miRNAs, although their pre-miRNAs might be dramatically different.
Besides, single nucleotide polymorphism (SNP) patterns were obvious for most of the miRNA families, especially in the conservation regions that produce matured miRNAs. In mammalians, SNPs are important for discriminating different species because they usually have different SNP contribution patterns. So we proposed that SNPs might also represent the differentiation of miRNAs from various species. As most SNPs reside in conserved regions encoding mature miRNAs, changes in these sites may influence the interaction between Dicer and miRNA precursors, and then causing asymmetric or symmetric cleavage. In animals and plants, the mechanisms of miRNA-dependent regulation were different; changes in some sites of mature miRNAs might also direct the way of gene regulation. These hypotheses need further experiments to verify, as characteristics of SNP in miRNAs have not yet been reported.
Mapping miRNA genes onto genome, revealed that each of three miRNAs pairs (bmo-mir-275/bmo-mir-305, bmo-miR-iab-4/bmo-miR-iab-4* and bmo-miR-276b/bmo-miR-276a*) is clustered into single sequence as their query miRNAs, although internal length between two silkworm miRNAs is longer. Phenomenon like such cases had already been reported from various species (Lagos-Quintana et al., 2001; Lau et al., 2001; Mourelatos et al., 2002). In fact, the clustering propensity of miRNA genes was noticed from the early days of the massive direct cloning of short ncRNA molecules (Lagos-Quintana et al., 2001; Lau et al., 2001; Mourelatos et al., 2002; Aravin et al., 2003). Usually, there are two or three miRNA genes in a cluster, although larger clusters were also identified, such as h-miR-17 cluster composed of six members (Lagos-Quintana et al., 2001; Mourelatos et al., 2002), which is also conserved in other mammals (Tanzer and Stadler, 2004), or the Drosophila melanogaster cluster of eight miRNA genes (Aravin et al., 2003). Recently, Seitz et al.(2004) predicted a huge cluster of 40 miRNA genes located in the ~1 Mb human imprinted 14q32 domain, several of which were shown to be expressed. Clustered miRNA genes might show high similarity in nucleotide composition, but sometimes they could also differ (Aravin et al., 2003; Bartel, 2004). The expression profiles of clustered genes are highly similar, raising the possibility that transcription of such miRNAs is controlled by common regulatory elements (Lee et al., 2002). It was supposed that a single promoter caused transcription of the clustered miRNAs genes (Lee et al., 2002). If so, these clustered genes might simultaneously participate in a common regulation pathway.
Recent research showed that pre-miRNAs are transported to the cytoplasm, where they are recognized by the RISC containing Dicer-TRBP-Ago2. The RNase III Dicer cleaves ~22 nt from the Drosha cleavage site, thus generating ~22 nt duplex miRNA (2 nt 3′ overhangs), which remained associated with RISC as a ribonuleoprotein complex. The complex identified the guide strand of the RNA duplex, perhaps through TRBP’s and Dicer’s recognizing thermodynamic asymmetry between the two ends of the duplex. Then miRNAs* are degraded, leaving miRNA which still resided in RISC (Gregory et al., 2005). At a dramatically low rate, both arms of the hairpins could give rise to miRNAs (Lim et al., 2003) exemplified by bmo-miR-iab-4:bmo-miR-iab-4* and bmo-miR-276b:bmo-miR-276a* (Fig.3). miRNA* recovered from duplex RNA may be due to the duplex RNA lack of structural features enforcing asymmetric RISC assembly (Schwarz et al., 2003). We noticed the possibility that antisense miRNAs play important roles in negatively regulating miRNAs function, and even regulating gene expression independently as exemplified by miR-131 (miR-9*), which is a vertebrate Brd-box family miRNA and potential homolog of miR-79 (Lai et al., 2004). In all, as more details on the function of miRNAs are being revealed, we will understand better these complementary miRNA pairs.
Acknowledgments
We gratefully thank Zheng Qiu-yan (School of law, Zhejiang university), Wang Jun-feng (Institute of neuroscience, Chinsese academy of sciences) for providing useful materials and meaningful discussion of details.
Footnotes
Project supported by the Program for New Century Excellent Talents in University (No. NCET-04-0531) and the National Basic Research Program (973) of China (No. 2005CB12100)
References
- 1.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. Rna. 2003;9(3):277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5(2):337–350. doi: 10.1016/S1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24(2):119–129. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell. 2003;113(1):25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 6.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Hannon G. RNA interference. Nature. 2002;418(6894):244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 10.Lai EC, Wiel C, Rubin GM. Complementary miRNA pairs suggest a regulatory role for miRNA: miRNA duplexes. Rna. 2004;10(2):171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans . Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14(7):1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16(6):720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA me tabolism in Arabidopsis thaliana . Curr Biol. 2002;12(17):1484–1495. doi: 10.1016/S0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297(5588):1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 20.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 21.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 23.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14(9):1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339(2):327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 25.Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306(5703):1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 26.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/S0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 27.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]