Abstract
Translational initiation factor 2 (IF2) is a guanine nucleotide-binding protein that can bind guanosine 3′,5′-(bis) diphosphate (ppGpp), an alarmone involved in stringent response in bacteria. In cells growing under optimal conditions, the GTP concentration is very high, and that of ppGpp very low. However, under stress conditions, the GTP concentration may decline by as much as 50%, and that of ppGpp can attain levels comparable to those of GTP. Here we show that IF2 binds ppGpp at the same nucleotide-binding site and with similar affinity as GTP. Thus, GTP and the alarmone ppGpp can be considered two alternative physiologically relevant IF2 ligands. ppGpp interferes with IF2-dependent initiation complex formation, severely inhibits initiation dipeptide formation, and blocks the initiation step of translation. Our data suggest that IF2 has the properties of a cellular metabolic sensor and regulator that oscillates between an active GTP-bound form under conditions allowing active protein syntheses and an inactive ppGpp-bound form when shortage of nutrients would be detrimental, if not accompanied by slackening of this synthesis.
Keywords: fast kinetics, GTP, translation regulation, nutritional stress
Initiation factor 2 (IF2) is the only initiation factor that is ribosome-bound throughout the entire translation initiation pathway, participating initially in the formation of the 30S initiation complex (30SIC) and subsequently in the assembly of the 70S initiation complex (70SIC), a process that ultimately results in formation of the first peptide bond (initiation dipeptide) and generates the first ribosomal pretranslocation complex (for reviews, see refs. 1–5). Thus, it could be predicted that IF2 functions are accompanied/modulated by conformational changes that could be either consequence or cause of the interactions of IF2 with its various ligands (30S and 50S ribosomal subunits, fMet-tRNA, GTP, GDP·Pi, and GDP). Crystallographic (6) and NMR (7) studies have, in fact, shown that, depending upon the nature of their ligand (i.e., GTP or GDP), aIF5B, the archaeal homologue of bacterial IF2, as well as isolated IF2G2, the G domain of IF2, undergo large structural changes. On the other hand, chemical probing (8) and cryo-EM (9, 10) have clearly shown that several conformational changes of the factor occur during the early, middle, and late events of translation initiation.
Bacterial cells growing under optimal nutritional conditions contain a high (>1 mM) concentration of GTP and a vanishingly low level of GDP. Thus, IF2 is expected to exist and to bind the 30S subunit almost exclusively in the GTP form, because it displays similar affinity for GTP and GDP, both Kds being in the 10- to 100-μM range (11). The IF2·GTP was shown to have a higher affinity for the 30S ribosomal subunit than either IF2·GDP or free IF2 (11). The adjustment of fMet-tRNA in the P site (ref. 12 and refs. therein) and the release of IF2 from 70SIC (13–15) have been attributed to the IF2-dependent GTP hydrolysis, which is very rapidly triggered by the first contact between the 50S subunit and the 30S initiation complex during the transition from 30SIC to 70SIC (16). Nevertheless, an essential role for this hydrolysis for either activity has never been conclusively proven (2, 16), so that the reason for the evolutionary conservation of the guanine nucleotide-binding domain of IF2 remains an open and intriguing question.
After different types of nutritional stress, the alarmone guanosine 3′,5′-(bis) diphosphate (ppGpp) can accumulate at considerable concentrations (i.e., up to 2 mM) in the cell at the expense of GTP, whose level can drop by >50% (refs. 17–24 and M. Cashel, personal communication). Like the other GTP-binding proteins involved in translation (EF-Tu, EF-G, and RF3), IF2 contains a guanine nucleotide-binding site that can bind GTP and GDP, as well as ppGpp (25–28).
In this article, we present a study of the interaction between ppGpp and IF2 and demonstrate that ppGpp inhibits translation by selectively targeting IF2 activities, thus suggesting that the G2 domain of IF2 confers on this factor the properties expected for a metabolic sensor and a translational regulator. In turn, this property would provide an adaptive advantage that could explain the evolutionary conservation of the guanine nucleotide-binding domain of IF2.
Results
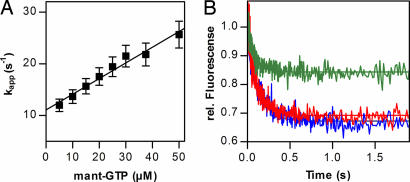
Binding of GTP to IF2 was studied essentially as in ref. 29 by using FRET from Trp in IF2 to mant-GTP, which results in an increase of mant fluorescence upon binding of the nucleotide to the factor. Time courses of binding were measured at constant concentration of IF2 and varying excess concentrations of mant-GTP. The data were analyzed by exponential fitting to determine the value of kapp for each titration point. For a binding reaction of the type IF2 + mant-GTP = IF2·mant-GTP, the value kapp = kon·[mant-GTP] + koff, where kon and koff are association and dissociation rate constants, respectively. The rate constants were calculated from the slope and y axis intercept of the linear dependence of kapp on the concentration of mant-GTP (Fig. 1A). The values were kon = (0.4 ± 0.1) × 106 M−1·s−1 and koff = 15 ± 5 s−1.
Fig. 1.
IF2 binding of GTP and ppGpp. (A) Concentration dependence of kapp of mant-GTP–IF2 interaction. (B) Time courses of mant-GTP dissociation (5 μM) from IF2 upon addition of 25 μM GTP (red), 25 μM ppGpp (blue), or buffer (green).
Nucleotide dissociation from IF2·mant-GTP was studied in the presence of excess nonfluorescent GTP or ppGpp (Fig. 1B). Ligand competition experiments indicated similar extent and rate of mant-GTP dissociation from IF2 in the presence of GTP or ppGpp, suggesting competition for the same nucleotide- binding site on IF2. In this case, the rate of GTP/ppGpp binding is limited by the rate of dissociation of mant-GTP from IF2; in the presence of a large excess of unlabeled nucleotide, rebinding of mant-GTP is negligible, whereas the binding of GTP/ppGpp is very fast. Thus, the rate by which the fluorescence decreases equals the dissociation rate constant of the mant-GTP, koff = 8 s−1. Upon dilution of IF2·mant-GTP with buffer, some mant-GTP dissociated, in agreement with the low stability of the complex, albeit not to the same extent as in the presence of nucleotide competitor. The apparent dissociation rate constant, kapp = 16 s−1, is equal to kon·[mant-GTP] + k−1, in good agreement with the values determined in mant-GTP titrations.
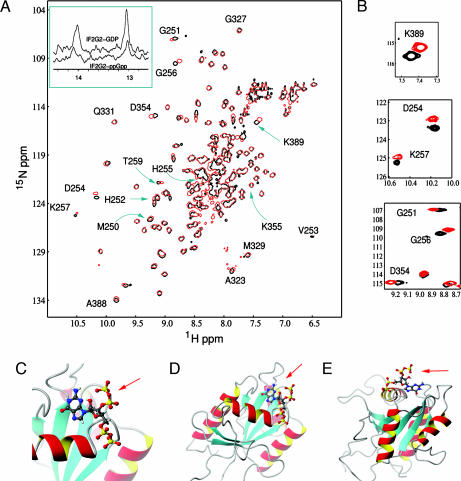
Subsequently, NMR titration experiments were carried out by using the isolated “minimal” guanine nucleotide-binding domain (G2) of Bacillus stearothermophilus IF2, whose 3D structure has recently been elucidated in free and GDP-bound forms (ref. 7 and unpublished work). After ascertaining that no ppGpp hydrolysis occurs on the time scale of NMR experiments, 15N-HSQC spectra of IF2G2 were recorded in the presence of either GDP or ppGpp. The 2D spectra are very similar, with the signals of almost all residues being superimposable (Fig. 2A). Also similar are the two 1H resonances because of the HN1 of the guanine nucleotide H-bonded to Asp354 (14 ppm) and to Hε2 atom of the P loop residue His255 (13 ppm), which are characteristic of the G2·GDP and G2·ppGpp complexes (Fig. 2A Inset) but absent in G2·GDPNP and in free G2.
Fig. 2.
Interaction of IF2G2 with GDP and ppGpp monitored by NMR spectroscopy. (A) Overlay of the 15N-HSQC spectra of IF2G2·GDP (black) and IF2G2·ppGpp (red). The assigned residues close to guanosine nucleotide are indicated. (Inset) Downfield region of the 1D 1H NMR watergate spectra of the same complexes. (B) Expansions of the 2D spectral regions displaying the most pronounced differences between IF2G2·GDP and IF2G2·ppGpp. (C–E) Views of the NMR 3D structure of IF2G2·ppGpp (7). The protein is represented by ribbons and ppGpp by “balls and sticks,” with the 3′ diphosphate indicated by red arrows.
Overall, these results indicate that GDP and ppGpp bind to the same site in G2, establishing the same network of hydrogen bonds, and that the structure of the G2 domain is very similar in both complexes. However, aside from these expected similarities, distinct spectral differences for several residues directly involved in nucleotide binding are also evident, the largest chemical-shift differences (Fig. 2B) being observed for residues G251, G256, and K257 (belonging to the P loop of IF2) and for D354 and K389 involved in guanine ring binding in IF2G2·GDP complex (ref. 7 and E.T., unpublished work). These differences, which may underlie the different functional effects that the two ligands have proven to have on the IF2 functions (see below), are likely more pronounced in intact ribosome-bound IF2, because, as seen from the modeled 3D structure of G2 in the complex with ppGpp, the 3′ diphosphate protrudes out of the IF2 domain (Fig. 2 C–E). This circumstance could explain the selective inhibition of IF2 functions by ppGpp (see below), because from its location, the 3′ diphosphate could perturb other domains of IF2 and interfere with its interaction with 50S subunits or with other ligands.
Having established in the above experiments that ppGpp binds to IF2 and that all guanine nucleotides target the same active site of the factor, the functional consequences of these IF2–ligand interactions were examined. However, because the GTP level remains fairly high even under nutritional stress, the effects of ppGpp and GDP on the initiation functions of IF2 were tested in the presence of a substantial level of GTP to mimic the conditions existing in the cell during the stringent response.
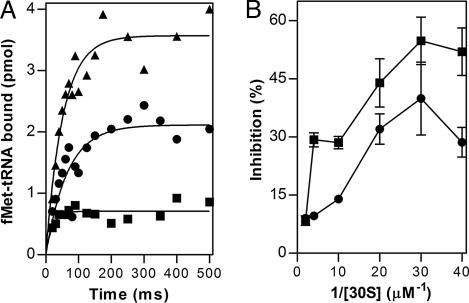
The kinetics of IF2-dependent binding of fMet-tRNA to mRNA-programmed 30S ribosomal subunits was analyzed first. Compared with the control in which only 1 mM GTP was present in the reaction mixture, initiator tRNA binding was substantially decreased in the presence of mixtures containing GTP and either GDP or ppGpp (Fig. 3A). The inhibition caused by ppGpp is much stronger than that caused by GDP and concerns the level of fMet-tRNA bound and not the rate at which it is bound, which is essentially the same regardless of the nucleotide present. A similar inhibition, albeit of lower intensity, is seen also with mixtures containing equal amounts of GTP and either GDP or ppGpp (not shown). The lower levels of fMet-tRNA bound in the presence of GDP and ppGpp compared with GTP can be explained, at least in part, by a lower affinity for the 30S subunit of IF2·GDP and IF2·ppGpp compared with IF2·GTP. This interpretation is supported by the experiment of Fig. 3B showing that the level of fMet-tRNA bound to 30S in the presence of GDP or ppGpp progressively approaches that reached in the presence of GTP by increasing concentrations of both IF2 and 30S ribosomal subunit in the reaction mixture.
Fig. 3.
Effect of ppGpp and GDP on IF2-dependent 30S initiation complex formation. (A) fMet-tRNA-binding kinetics to mRNA-programmed 30S subunits measured by rapid filtration (see Materials and Methods) in the presence of 1 mM GTP (▴), 0.25 mM GTP + 0.75 mM GDP (●), and 0.25 mM GTP + 0.75 mM ppGpp (■). (B) Level of fMet-tRNA bound as a function of ligand concentration. Different ligand concentrations in mixtures containing constant concentrations of guanosine nucleotides were obtained, keeping amounts of all 30S initiation complex components (30S, mRNA, fMet-tRNA, IF1, and IF2) constant but varying the reaction volumes (from 15 to 300 μl). The actual 30S concentrations ranged from 25 to 500 nM, and the stoichiometric ratios ligand/30S were IF1, IF2, and IF3 = 1.5; fMet-tRNA and mRNA = 2.0. The reciprocal of 30S subunits concentration in the reaction mixtures is indicated in the abscissa. The levels of 30S-bound fMet-tRNA 30S subunit in the presence of GDP (0.5 mM) + GTP (0.25 mM) (●) and ppGpp (0.5 mM) + GTP (0.25 mM) (■) are expressed as % inhibition with respect to the controls containing GTP (0.75 mM) alone.
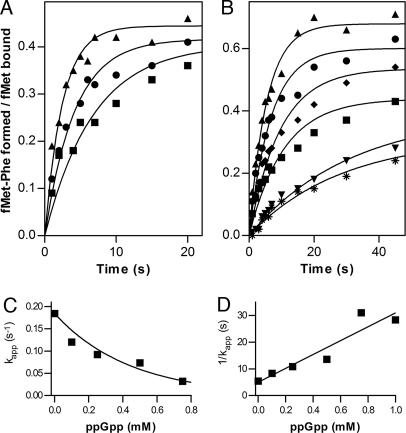
The effect of guanine nucleotides on the kinetics of IF2-dependent initiation dipeptide (fMet-Phe) formation was studied by quench-flow experiments. The rate of dipeptide formation in the presence of GTP alone was faster than in the presence of both GTP and ppGpp, whereas with a mixture of GTP and GDP, the rate is intermediate (Fig. 4A). The inhibition of initiation dipeptide formation depends on ppGpp concentration, because its apparent rate is progressively reduced (up to ≥6-fold) when, in the presence of a constant level of GTP, the concentration of ppGpp is increased from 0 to 0.75 mM (Fig. 4 B and C). Furthermore, because the reaction rate was reduced by 50% when GTP and ppGpp concentrations are about equal, and because the rates are determined from curves normalized for the actual amount of 30S IC formed in each case, it can be surmised that IF2 has similar affinities for the two nucleotides; the value Km/Ki = 1.2 calculated from the slope of the Dixon plot (Fig. 4D) seems compatible with this premise.
Fig. 4.
Effect of GDP and ppGpp on the kinetics of IF2-dependent initiation dipeptide formation. IF2-dependent formation of initiation dipeptide (fMet-Phe) in the presence of 1 mM GTP (▴); GTP + GDP, 0.5 mM each (●); and GTP + ppGpp (■), 0.5 mM each (A), and a constant concentration (0.25 mM) of GTP and increasing concentrations of ppGpp (0 mM, ▴; 0.10 mM, ●; 0.25 mM, ♦; 0.50 mM, ■; 0.75 mM, ▾; and 1.0 mM, ∗) (B). The amount of initiation dipeptide formed is expressed as fMet-Phe formed/fMet-tRNA bound in the 30SIC. (C) Variation of the kapp of dipeptide formation as a function of [ppGpp]. (D) Dixon plot showing the linear dependence of 1/kapp as a function of increasing [ppGpp]. The slope is equal to Km/(Vmax·[S]·ki). Initiation dipeptide formation was analyzed by quench-flow experiments at 37°C, as described in Materials and Methods.
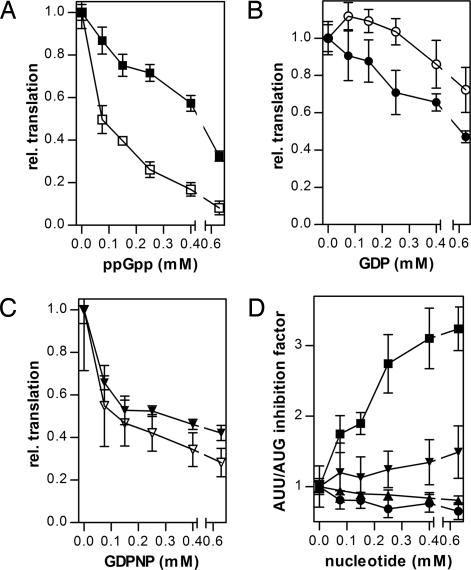
The experiments described above have demonstrated that ppGpp inhibits IF2 activity at both early (30SIC formation) and late (formation of the first peptide bond) steps of the translation initiation pathway. In the following experiments, we investigated whether ppGpp inhibition of these partial reactions results in an overall reduction of mRNA translation. For this purpose, a method to evaluate specifically the effect of ppGpp on the initiation functions of IF2 had to be devised, because ppGpp can also bind to elongation factors EF-Tu and EF-G and inhibit their functions (28, 30). We took advantage of the fact that translation of mRNAs initiating with the noncanonical AUU triplet is substantially more IF2-dependent than that of mRNAs, beginning with a canonical AUG triplet (31, 32), and compared the effect of ppGpp and of other guanine nucleotides on the translation of two model mRNAs (027AUGmRNA and 027AUUmRNA), identical in their coding sequence but bearing different initiation codons (i.e., AUG vs. AUU). Although increasing concentrations of ppGpp cause progressive translational inhibition with both mRNAs, 027AUUmRNA translation is clearly more inhibited than 027AUGmRNA translation (Fig. 5A). On the other hand, increasing concentrations of GDP (Fig. 5B) and of the nonhydrolyzable GTP analog GDPNP (Fig. 5C) inhibit translation of the two model mRNAs to approximately the same extent. The differential sensitivity of mRNA translation to inhibition by ppGpp, GDP, and GDPNP as a function of the nature of the mRNA initiation codons is illustrated by the different inhibition ratios (027AUUmRNA/027AUGmRNA). As seen from Fig. 5D, this ratio remains ≅1 at all concentrations of GDP or GDPNP but increases with increasing ppGpp, becoming ≥3 when the alarmone concentration reaches 0.4 mM. These results not only confirm the premise that ppGpp is capable of inhibiting mRNA translation (33) but also demonstrate that it does so by targeting primarily the initiation function of IF2. The selectivity by which ppGpp blocks the initiation functions of IF2 compared with the elongation functions of EF-Tu and EF-G is even greater than superficially surmised from the selective inhibition of 027AUUmRNA translation. In fact, the 027mRNAs encode a 36-aa oligopeptide plus the N-terminal formyl-methionine (31), and the inhibition of EF-Tu and EF-G functions could be reiterated several times, whereas that of IF2, taking part only once in the translational process, has only one chance to be inhibited.
Fig. 5.
Inhibition of mRNA translation by various guanosine nucleotides. Translation in cell-free systems containing a constant concentration of GTP (0.25 mM) and programmed with 027AUGmRNA (closed symbols) and 027AUUmRNA (open symbols) is plotted as a function of the indicated concentrations of ppGpp (A), GDP (B), and GDPNP (C). The levels of translation indicated on the ordinate are normalized for the level of translation (taken as 1) obtained in the absence of added guanine nucleotide inhibitors. (D) Translation inhibition ratios (027AUUmRNA/027AUGmRNA) plotted as a function of the indicated concentrations of ppGpp (■), GDP (●), GDPNP (▾), and GTP (▴).
Discussion
Several physiological conditions and nutritional downshifts, such as amino acid starvation in a relA+ strain (19), nitrogen (21), and carbon (20) limitation or reduction of glucose availability (24), which severely slow down protein synthesis and cell growth, induce a rapid accumulation of ppGpp whose cellular level may increase up to millimolar concentrations, whereas that of GTP decreases (up to ≅50%). These changes determine the inhibition of rRNA and tRNA transcription and affect other cellular activities such as DNA replication, transport, and metabolic stability of different molecules (34, 35). Translation is also affected by ppGpp (ref. 33 and refs. therein) with a correlation between the abundance of ppGpp and the extent of translation inhibition that is relieved only when ppGpp returns to its very low basal level. So far, this inhibition has been primarily attributed to an interference of the alarmone with the activity of EF-Tu and EF-G (28). Finally, ppGpp was also reported to control translation in the stationary phase by facilitating the formation of inactive ribosome (100S) dimers (36). The results presented in this article suggest that, even in the presence of a substantial level of GTP, ppGpp concentrations normally attained in vivo under nutritional stress inhibit translation by preferentially targeting the initiation function of IF2. From this point of view, ppGpp differs from both GDP and GDPNP, which inhibit protein synthesis without such a preference (Fig. 5 A–C).
The crystallographic structures of the IF2 homologue aIF5B (6), the NMR structures of free IF2G2 and in complex with GDP (7), and the cryoEM reconstructions of complexes at different stages of initiation (10) clearly indicate that the 3D structures of IF2·GTP and IF2·GDP are substantially different from each other. However, on the basis of NMR studies, the structures of IF2G2 in complex with ppGpp and GDP are likely to be very similar. Thus, even though important functional consequences could arise from subtle structural differences of the factor, the different effect of ppGpp and GDP (or GDPNP) on the IF2 functions is most likely because of the 3′ diphosphate of the alarmone ppGpp. In fact, as seen in Fig. 2 C–E, this negatively charged diphosphate protrudes from the G domain of IF2. From this position, the negatively charged 3′ diphosphate likely interferes with other domains of the factor and/or with its interaction with one or more of its ligands (the ribosomal subunits fMet-tRNA).
Our results show that ppGpp inhibits two IF2-dependent partial reactions, one occurring on the 30S subunit, the other on the 70S ribosome, and that ppGpp affects in different ways these IF2 activities. In fact, the lower level of fMet-tRNA bound in the presence of IF2·ppGpp (or IF2·GDP) compared with IF2·GTP is caused by a lower affinity of the first two complexes for the 30S subunit, whereas the reduced rate of initiation dipeptide formation is because of a defect in placing fMet-tRNA in the P site of the 50S. The rate reduction of initiation dipeptide formation is much more pronounced with ppGpp than with GDP and is not caused by an interference with the A site function, because also the rate of fMet-puromycin formation is drastically reduced by ppGpp (data not shown).
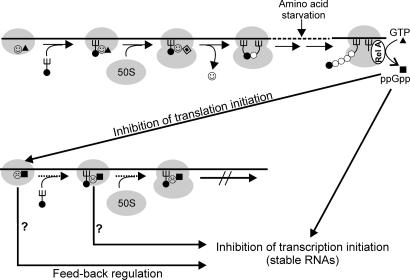
Synthesis of ppGpp is due primarily to the activation of RelA by a signal generated during translation elongation (19–21, 37). It is reasonable to imagine that such a signal would be transmitted to the translation initiation apparatus to block or slow down new translational events (Fig. 6). During exponential growth, the GTP concentration largely exceeds that of GDP and is >1 order of magnitude higher than that of ppGpp (refs. 17 and 18 and M. Cashel, personal communication), whereas after stress, the level of the latter can even exceed that of GTP (19). Thus, we can imagine a scenario in which IF2 oscillates between an active GTP form and an inactive (or only partially active) ppGpp form. If the metabolic conditions of the cell are favorable for active translation and growth, almost all IF2 molecules are GTP-bound, the small amount of IF2-GDP physiologically formed upon GTP hydrolysis undergoing rapid exchange with GTP. However, under the unfavorable metabolic conditions that generate ppGpp, this alarmone would at least partially replace GTP to generate an inactive IF2–ppGpp complex, which would slow down translation and possibly transmit a feedback signal to the transcriptional machinery (Fig. 6). Furthermore, idle IF2 can also contribute to increasing the cellular level of ppGpp, because we have evidence that it can bind and hydrolyze pppGpp, generating ppGpp (unpublished results). As pointed out in an earlier publication (43), a central role of IF2 in gearing translational activity to the metabolic state of the cell can be surmised, considering that the IF2—fMet–tRNA interaction, the fundamental raison d'être of the factor, depends on the availability of methionine and tetrahydrofolate, two molecules playing a major metabolic role as C1 donors.
Fig. 6.
Schematic representation of the postulated regulatory circuits involving IF2 and ppGpp. (Upper) The essential steps of translation initiation: formation of 30SIC in the presence of IF2-GTP ( ▴), association of 50S subunit and formation of 70SIC, GTP (▴) hydrolysis to GDP (
▴), association of 50S subunit and formation of 70SIC, GTP (▴) hydrolysis to GDP ( ), IF2 dissociation (
), IF2 dissociation ( ), and initiation dipeptide formation. Amino acid starvation during elongation triggers RelA-dependent synthesis of ppGpp (■), which inhibits stable RNA transcription (Lower) and, as shown in this study, the initiation functions of IF2 (30SIC formation and initiation dipeptide formation). A possible function of translation initiation intermediates containing IF2-ppGpp (
), and initiation dipeptide formation. Amino acid starvation during elongation triggers RelA-dependent synthesis of ppGpp (■), which inhibits stable RNA transcription (Lower) and, as shown in this study, the initiation functions of IF2 (30SIC formation and initiation dipeptide formation). A possible function of translation initiation intermediates containing IF2-ppGpp ( ) in feedback inhibition of stable RNA transcription is suggested by earlier reports that components of the translation initiation apparatus play an active part in this regulation. In particular, IF2, fMet-tRNA, and ppGpp were found to interact with the RNA polymerase and influence its activity at stable RNA promoters (38, 39), whereas IF2 (40), IF3 (41), and initiation-competent 30S subunits (42) were shown to be required for feedback repression of stable RNA transcription.
) in feedback inhibition of stable RNA transcription is suggested by earlier reports that components of the translation initiation apparatus play an active part in this regulation. In particular, IF2, fMet-tRNA, and ppGpp were found to interact with the RNA polymerase and influence its activity at stable RNA promoters (38, 39), whereas IF2 (40), IF3 (41), and initiation-competent 30S subunits (42) were shown to be required for feedback repression of stable RNA transcription.
Thus, the present data indicate that, in addition to performing an essential role in translation, IF2 might also function as a regulatory switch, depending upon the nature of its ligand (GTP or ppGpp). In turn, the adaptive advantage offered by this function could represent at least one of the reasons for the evolutionary conservation of the G2 module of IF2. The intrinsic capacity of G2 to act as a hinge during the transition from 30S to 70S initiation complex could be another reason for the conservation of this module. This premise is suggested by both genetic and biochemical data (2, 44) and by the evidence that GTP hydrolysis is not required for dipeptide formation and IF2 dissociation (16). A conformational transition about this hinge, driven by subunit association, would allow the dissociation of the IF2-fMet-tRNA interaction, which simultaneously loosens the IF2–ribosome interaction and allows the tRNA acceptor end to become a substrate for the peptidyl transferase reaction. In this model, GTP hydrolysis is seen not as a necessary source of chemical energy but instead as a means to get rid of an inhibitor (e.g., nonhydrolyzed GTP) of this essential conformational change (2, 44).
That IF2 might also function as a regulatory switch cannot be considered entirely unorthodox. In fact, it should be borne in mind that IF2 is a member of the large superfamily of guanine-binding proteins, which often act as molecular switches in all kingdoms of life where they regulate different types of cellular processes. Indeed, bacteria contain 11 universally conserved GTPases, and many of them seem to have properties compatible with the performance of similar roles (45, 46). Most of these proteins bind RNA and/or ribosome and regulate different aspects of ribosome function and/or transmit to downstream effector pathways signals generated on the ribosome, often at the onset of stress conditions. Of particular interest are the members of the Era and Obg protein families, which, like IF2, interact with ribosomes and are involved in stress response. IF2 is in fact one of the cold-shock proteins (32), whereas Era is involved in regulating both carbon and nitrogen metabolism and in the control of chromosome segregation and cell cycle. Ribosome-bound Era would activate cell division, whereas the free form would participate in terminating cell division (47). Finally, Obg is implicated in chromosome partitioning, initiation of replication, and sporulation. It is remarkable that the recently solved 3D structure of Obg has revealed the presence of a ppGpp molecule in its GTP-binding site, a finding that has led to the proposal that Obg could control Bacillus subtilis sporulation by sensing directly the GTP/ppGpp ratio in the cell (48).
Materials and Methods
Ribosomes, Factors, tRNAs, and mRNA.
Escherichia coli MRE600 ribosomal subunits, tRNAPhe and tRNAfMet, were provided by Y. P. Semenkov and S. V. Kirillov (Petersburg Nuclear Physics Institute, Gatchina, Russia). IF1, IF3, and IF2 were prepared from overproducing strains constructed in our laboratory and purified to electrophoretic homogeneity essentially as described (49). EF-Tu and [14C]Phe-tRNAPhe and f[35S]Met-tRNAfMet and fMet-tRNAfMet were prepared essentially as described in Rodnina et al. (50, 51). The sequence 027AUGmRNA and 027AUUmRNA (52) and the method to transcribe and purify them have been described (31).
Binding of mant-GTP to IF2.
Fluorescence stopped-flow experiments were carried out at 20°C essentially as described here and in ref. 29 by using a SX-18MV spectrometer (Applied Photophysics, Surrey, U.K.). Two appropriate 60-μl samples in buffer A (50 mM Tris·HCl, pH 7.5/70 mM NH4Cl/30 mM KCl/7 mM Mg acetate/1 mM DTT) were rapidly mixed; 6–10 individual transients were averaged; and the data were evaluated by fitting to a single exponential function with a characteristic time constant (kapp), amplitude (A), and another variable for the final signal (F∞) according to the equation F = F∞+ A exp(−kapp·t), where F is the fluorescence at time t. Calculations were performed by using GraphPad Prism (GraphPad, San Diego, CA).
Synthesis and Purification of ppGpp.
E. coli MG1655 (53) carrying relA on the pALS10 plasmid (33) was grown at 37°C in LB broth supplemented with ampicillin (100 μg/ml). When the cells reached A600 ≅ 1.35, RelA expression was induced by addition of 1 mM isopropyl β-d-thiogalactoside. After 1 h of incubation, the cells were harvested, frozen at −80°C, and used as a source of low-salt-washed ribosomes for the synthesis of ppGpp. Synthesis and purification of ppGpp were carried out essentially as described (54). The purity of ppGpp was assessed by HPLC on a CC 125/4 Nucleosil PEI 4000-7 column (Macherey & Nagel, Düren, Germany) on an Amersham Äktabasic (Amersham Pharmacia, Uppsala, Sweden) apparatus.
Initiation Dipeptide Formation.
This was obtained by rapid mixing in a quench-flow KinTek (Austin, TX) apparatus, preformed 30S initiation complexes, and a mixture containing 50S ribosomal subunits and EF-Tu-GTP-[14C]Phe-tRNAPhe. The 30S initiation complexes were prepared in buffer A containing 1 μM 30S subunits; 1.5 μM each IF1, IF2, and IF3; 2 μM f[35S]Met-tRNAfMet; 2 μM mRNA; and guanosine nucleotides, as indicated in Fig. 4. After a 20-min incubation at 37°C, the amount of complex formed was determined by nitrocellulose filtration. The EF-Tu-GTP-[14C]Phe-tRNAPhe ternary complex was prepared (50) by mixing 1,100 pmol of EF-Tu and 900 pmol of [14C]Phe-tRNAPhe in 100 μl of buffer A containing 0.5 mM GTP and purified by size-exclusion chromatography (NAP Columns; Amersham). After mixing 14-μl aliquots of the content of syringe A, corresponding to 15 pmol of 30S subunits (>90% of which engaged in the 30S initiation complex) with 14-μl aliquots of the content of syringe B containing 20 pmol of 50S subunits and ≅20 pmol of the purified EF-Tu-GTP-[14C]Phe-tRNAPhe ternary complex, the reaction mixtures were aged at 37°C for the indicated times, quenched with 0.8 M KOH, incubated for 20 min at 37°C, and finally neutralized with acetic acid. The fMet-[14C]Phe initiation dipeptide formed was quantified by HPLC as described (50).
Kinetics of 30S Initiation Complex Formation.
Binding of fMet-tRNA to 30S was performed by rapid filtration on nitrocellulose by using a modified SFM-400 Biologic (Grenoble, France) instrument. Equal volumes (40 μl) of mixture A containing 1 μM 30S; 1.5 μM IF1, IF2, and IF3 in buffer A; and mixture B, containing 2 μM [35S]fMet-tRNA and 2 μM mRNA in buffer A, were rapidly mixed and aged at 37°C for the times indicated in Fig. 3A, and the reaction stopped by rapid dilution with buffer A (first 720 μl, then 3 ml) followed by filtration on BA-85 [Schleicher & Schuell (Whatman), Florham Park, NJ] nitrocellulose filters. The amount of fMet-tRNA bound was determined by radioactivity liquid scintillation counting.
In Vitro Translation.
Translation assays were carried out essentially as described (31, 55).
NMR Spectroscopy.
Expression and purification of B. stearothermophilus IF2G2 (residues 241–414) and [15N]-labeled IF2G2 were carried out as described (7). A series of 1D 1H WATERGATE 1D 31P and 2D 15N-FHSQC spectra of [15N]-IF2G2 with different amounts of GDP or ppGpp was acquired on a Bruker (Billerica, MA) AVANCE 600-MHz spectrometer equipped with an inverse BBI probe with z gradients. NMR experiments were performed at 315 K with samples containing 0.4–0.9 mM IF2G2 in 20 mM K phosphate (pH 6.5) buffer in 90% H2O/10% D2O containing 150 mM KCl, 5 mM MgCl2, 1 mM DTT, and 0.6–1.2 mM GDP or ppGpp. Trace amounts of an EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN) and 0.01% NaN3 were also added to the NMR samples. Further details are given in ref. 7.
Acknowledgments
We are grateful to Dr. M. Cashel (National Institutes of Health, Bethesda, MD) for providing the bacterial strain and the protocol for the production and purification of ppGpp and to Dr. P. L. Ipata (University of Pisa, Pisa, Italy) for invaluable suggestions. This work was supported in part by Ministero dell'Università e della Ricerca PRIN 2005 grants (to C.O.G., C.L.P., and A.L.T.), the German-Italian DAAD/Vigoni bilateral exchange program (C.O.G. and M.V.R.), the Alfried Krupp von Bohlen und Halbach Stiftung, and the Fonds der Chemischen Industrie (M.V.R.).
Abbreviations
- IF2
initiation factor 2
- ppGpp
guanosine 3′,5′-(bis) diphosphate
- 30SIC
30S initiation complex
- 70SIC
70S initiation complex
- IF2G2
G domain of IF2.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gualerzi CO, Brandi L, Caserta E, La Teana A, Spurio R, Tomšic J, Pon CL. In: The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. Garrett RA, Douthwaite SR, Liljas A, Matheson AT, Moore PB, Noller HF, editors. Washington, DC: ASM Press; 2000. pp. 477–494. [Google Scholar]
- 2.Gualerzi CO, Brandi L, Caserta E, Garofalo C, Lammi M, La Teana A, Petrelli D, Spurio R, Tomšic J, Pon CL. Cold Spring Harbor Symp Quant Biol. 2001;66:363–376. doi: 10.1101/sqb.2001.66.363. [DOI] [PubMed] [Google Scholar]
- 3.Boelens R, Gualerzi CO. Curr Protein Pept Sci. 2002;3:107–119. doi: 10.2174/1389203023380765. [DOI] [PubMed] [Google Scholar]
- 4.Ramakrishnan V. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 5.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roll-Mecak A, Cao C, Dever TE, Burley SK. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 7.Tishchenko E. PhD thesis. Utrecht, The Netherlands: Univ of Utrecht; 2005. [Google Scholar]
- 8.Marzi S, Knight W, Brandi L, Caserta E, Soboleva N, Hill WE, Gualerzi CO, Lodmell JS. RNA. 2003;9:958–969. doi: 10.1261/rna.2116303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP. Nat Struct Mol Biol. 2005;12:1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- 11.Pon CL, Paci M, Pawlik RT, Gualerzi CO. J Biol Chem. 1985;260:8918–8924. [PubMed] [Google Scholar]
- 12.La Teana A, Pon CL, Gualerzi CO. J Mol Biol. 1996;256:667–675. doi: 10.1006/jmbi.1996.0116. [DOI] [PubMed] [Google Scholar]
- 13.Hershey JWB. In: Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. Neidhardt FC, Ingraham JL, Brooks Low K, Magasanik B, Schaechter M, Umbarger HE, editors. Washington, DC: Am Soc Microbiol; 1987. pp. 613–647. [Google Scholar]
- 14.Luchin S, Putzer H, Hershey JW, Cenatiempo Y, Grunberg-Manago M, Laalami S. J Biol Chem. 1999;274:6074–6079. doi: 10.1074/jbc.274.10.6074. [DOI] [PubMed] [Google Scholar]
- 15.Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. EMBO J. 2003;22:5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomšic J, Vitali L. A., Daviter T, Savelsbergh A, Spurio R, Striebeck P, Wintermeyer W, Rodnina MV, Gualerzi CO. EMBO J. 2000;19:2127–2136. doi: 10.1093/emboj/19.9.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salser W, Janin J, Levinthal C. J Mol Biol. 1968;31:237–266. doi: 10.1016/0022-2836(68)90442-7. [DOI] [PubMed] [Google Scholar]
- 18.Cashel M. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 19.Cashel M, Gallant J. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 20.Lazzarini RA, Cashel M, Gallant J. J Biol Chem. 1971;246:4381–4385. [PubMed] [Google Scholar]
- 21.Irr JD. J Bacteriol. 1972;110:554–561. doi: 10.1128/jb.110.2.554-561.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cashel M. Annu Rev Microbiol. 1975;75:301–318. doi: 10.1146/annurev.mi.29.100175.001505. [DOI] [PubMed] [Google Scholar]
- 23.Cashel M, Gentry DR, Hernandez VJ, Vinella D. In: Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. Neidhardt FC, Ingraham JL, Brooks Low K, Magasanik B, Schaechter M, Umbarger HE, editors. Washington, DC: Am Soc Microbiol; 1996. pp. 1458–1496. [Google Scholar]
- 24.Murray HD, Schneider DA, Gourse RL. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 25.Severini MT, Choli, La Teana A, Gualerzi CO. FEBS Lett. 1990;276:14–16. doi: 10.1016/0014-5793(90)80495-5. [DOI] [PubMed] [Google Scholar]
- 26.Legault L, Jeantet C, Gros F. FEBS Lett. 1972;27:71–75. doi: 10.1016/0014-5793(72)80412-5. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida M, Travers A, Clark BFC. FEBS Lett. 1972;23:163–166. doi: 10.1016/0014-5793(72)80331-4. [DOI] [PubMed] [Google Scholar]
- 28.Hamel E, Cashel M. Arch Biochem Biophys. 1974;162:293–300. doi: 10.1016/0003-9861(74)90128-3. [DOI] [PubMed] [Google Scholar]
- 29.Gromadski KB, Wieden HJ, Rodnina MV. Biochemistry. 2002;41:162–169. doi: 10.1021/bi015712w. [DOI] [PubMed] [Google Scholar]
- 30.Miller DL, Cashel M, Weissbach H. Arch Biochem Biophys. 1973;154:675–682. doi: 10.1016/0003-9861(73)90022-2. [DOI] [PubMed] [Google Scholar]
- 31.La Teana A, Pon CL, Gualerzi CO. Proc Natl Acad Sci USA. 1993;90:4161–4165. doi: 10.1073/pnas.90.9.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giuliodori AM, Brandi A, Gualerzi CO, Pon CL. RNA. 2004;10:265–276. doi: 10.1261/rna.5164904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svitil AL, Cashel M, Zyskind JW. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 34.Gallant J, Harada B. J Biol Chem. 1969;244:3125–3132. [PubMed] [Google Scholar]
- 35.Josaitis CA, Gaal T, Gourse RL. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura T, Fujita N, Ishihama A. Nucleic Acids Res. 1986;14:6857–6870. doi: 10.1093/nar/14.17.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 38.Debenham PG, Pongs O, Travers AA. Proc Natl Acad Sci USA. 1980;77:870–874. doi: 10.1073/pnas.77.2.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travers AA, Debenham PG, Pongs O. Biochemistry. 1980;19:1651–1656. doi: 10.1021/bi00549a020. [DOI] [PubMed] [Google Scholar]
- 40.Cole JR, Olsson CL, Hershey JW, Grunberg-Manago M, Nomura M. J Mol Biol. 1987;198:383–392. doi: 10.1016/0022-2836(87)90288-9. [DOI] [PubMed] [Google Scholar]
- 41.Olsson CL, Graffe M, Springer M, Hershey JWB. Mol Gen Genet. 1996;250:705–714. doi: 10.1007/BF02172982. [DOI] [PubMed] [Google Scholar]
- 42.Yamagishi M, de Boer HA, Nomura M. J Mol Biol. 1987;198:547–550. doi: 10.1016/0022-2836(87)90299-3. [DOI] [PubMed] [Google Scholar]
- 43.Gold L. Annu Rev Biochem. 1988;57:199–223. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 44.Caserta E, Tomšic J, Spurio R, La Teana A, Pon CL, Gualerzi CO. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.07.043. in press. [DOI] [PubMed] [Google Scholar]
- 45.Caldon CE, Yoong P, March PE. Mol Microbiol. 2001;41:289–297. doi: 10.1046/j.1365-2958.2001.02536.x. [DOI] [PubMed] [Google Scholar]
- 46.Caldon CE, March PE. Curr Opin Microbiol. 2003;6:135–139. doi: 10.1016/s1369-5274(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 47.Hang JQ, Meier TI, Zhao G. Eur J Biochem. 2001;268:5570–5577. doi: 10.1046/j.1432-1033.2001.02493.x. [DOI] [PubMed] [Google Scholar]
- 48.Buglino J, Shen V, Hakimian P, Lima CD. Structure (Cambridge, UK) 2002;10:1561–1592. doi: 10.1016/s0969-2126(02)00882-1. [DOI] [PubMed] [Google Scholar]
- 49.Gualerzi CO, Severini M, Spurio R, La Teana A, Pon CL. J Biol Chem. 1991;266:16356–16362. [PubMed] [Google Scholar]
- 50.Rodnina MV, Fricke R, Wintermeyer W. Biochemistry. 1994;33:12267–12275. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
- 51.Rodnina MV, Semenkov YP, Wintermeyer W. Anal Biochem. 1994;219:380–381. doi: 10.1006/abio.1994.1282. [DOI] [PubMed] [Google Scholar]
- 52.Brandi L, Lazzarini A, Cavaletti L, Abbondi M, Corti E, Ciciliato I, Gastaldo L, Marazzi A, Feroggio M, Fabbretti A, et al. Biochemistry. 2006;45:3692–3702. doi: 10.1021/bi052540k. [DOI] [PubMed] [Google Scholar]
- 53.Guyer MS, Reed RR, Steitz JA, Low KB. Cold Spring Harbor Symp Quant Biol. 1981;45:135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 54.Cashel M. Anal Biochem. 1974;57:100–107. doi: 10.1016/0003-2697(74)90056-6. [DOI] [PubMed] [Google Scholar]
- 55.Brandi A, Pietroni P, Gualerzi CO, Pon CL. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]