Abstract
Mutation of Bruton's tyrosine kinase (Btk) causes human X-linked agammaglobulinemia and murine X-linked immunodeficiency syndrome (xid). Quantitative aspects of B lymphocyte development and function have been demonstrated to depend on Btk level in vivo by using a murine transgenic model system. A sensitive intracellular immunofluorescent assay was developed to measure Btk protein on a per cell basis to test the hypothesis that its dosage is dynamically regulated during B cell development or functional responses. Marrow-derived hematopoietic stem cells, common lymphoid progenitor cells, and developing B and myeloid lineages expressed Btk protein at comparable levels. Resting peripheral B lineage cells had a significantly lower amount of Btk than marrow-derived cells in both wild-type and xid mice. Activation of the B cell antigen receptor up-regulated Btk protein level 10-fold within several hours by a phosphatidylinositol 3-kinase-dependent, posttranscriptional mechanism. In contrast, the protein level of Btk R28C in activated B lymphocytes from xid mice remained low. Bypass of the antigen receptor signaling pathways by treatment of cells with phorbol myristic acid and ionomycin rescued up-regulation of Btk protein in xid splenic B cells. These combined results suggest that certain receptor signals mediated by Btk regulate the level of expression of Btk protein in responding B lymphocytes to potentiate signal transduction. Dynamic regulation of Btk protein dosage is an additional mechanism to modulate B lymphocyte immune functions.
Regulation of the level of expression of key proteins, such as transcription factors, protooncogenes, cell surface receptors, and intracellular signal transducers, plays a major role in the careful orchestration of lymphocyte development and immune function (1–3). For example, maturation of lymphocytes is marked by stage-specific expression of recombination activating genes (Rag-1 and Rag-2), which are required for continued development of these cells (4, 5). The rapid, developmentally programmed change in level of critical protein mediators of B lymphocyte function can be accomplished through several types of molecular mechanisms, including gene transcription, RNA stability, protein synthesis, or degradation (1–3).
Bruton's tyrosine kinase (Btk) is a signal transducing protein expressed in all hematopoietic lineages, except T cells (6–10). Btk has a particularly important role in B lymphocytes, where it functions in multiple receptor pathways, including the B cell antigen receptor (BCR), interleukin 5 and 10 receptors, CD19, CD38, and CD40 (11). Btk belongs to the structurally homologous Btk/Tec family of intracellular tyrosine kinases that have similar roles in receptor signal transduction pathways but distinct patterns of cell expression (12). Certain mechanisms regulating activation of Btk/Tec kinases, such as the influence of upstream signaling proteins phosphatidylinositol (PI) 3-kinase and Src family kinases, also appear to be conserved amongst most family members (12). These upstream regulators influence Btk signaling by modulating the phosphorylation of specific tyrosine residues, its intrinsic kinase activity, and association of Btk with the plasma membrane (12, 13). Cell culture models confirm that Btk/Tec kinases function interchangeably in certain situations, like the mediation of receptor-dependent increases in intracellular calcium level (14–16). The natural pattern of coexpression of Btk/Tec kinases creates redundant signaling capacity in certain cell types, such as platelets (17), T cells (18), and mast cells (19). The T lymphocyte defect observed in Itk−/−/Rlk−/− mice is severe compared with the milder phenotype of either single knockout (18). Thus, the total cellular amount of Btk/Tec family kinases is another potential site for regulation of receptor signaling pathways critical for lymphocyte function.
Naturally occurring genetic syndromes and transgenic model systems reveal that normal B cell development and function depends on the total amount of functional Btk protein expressed. A spontaneous murine Btk point mutation (Btk R28C) (20, 21), or the targeted deletion of the Btk gene (22), results in X-linked immunodeficiency (xid) syndrome. This phenotype is characterized by a partial block in B cell development and defective responses to certain types of immune stimulation (23). Humans with a Btk mutation usually display a more severe phenotype, X-linked agammaglobulinemia, resulting from a near-total B lineage developmental block and absent humoral immune responses (11, 24). Intracellular detection of the relative level of Btk protein expression in B lineage cells has been proposed as a diagnostic tool to evaluate human patients with symptoms of X-linked agammaglobulinemia (25). The xid and X-linked agammaglobulinemia phenotypes reveal that loss of Btk function is not compensated by coexpression of other Btk/Tec family members in B cells.
Btk's rate-limiting function is confirmed by the dosage-dependent reconstitution of B cell development and immune responses using a transgenic expression system. Btk transgene expressed at 25 or 50% of the level of the endogenous allele yielded a graded recovery of Btk-dependent lymphocyte functions (26). Importantly, Btk overexpression (150% of normal level) did not result in increased B cell functions beyond wild-type responses and, in some assays, actually diminished responses. This result demonstrates that the level of endogenous Btk expression in wild-type B cells directs an optimal immune response. Transgenic expression of an activated Btk allele (Btk E41K) produces a more profound phenotype of immunodeficiency than xid, resulting from a nearly total developmental block at the transition from preB to immature B cell stage, and demonstrates the exquisite sensitivity of B cells to the level of Btk activity (27, 28).
A sensitive and specific intracellular immunofluorescent flow cytometric assay was established to detect the amount of Btk protein expressed on a per cell basis to test the hypothesis that Btk expression is dynamically regulated. Btk was detected in all developing hematopoietic lineages from bone marrow except T cells. The high Btk level consistently observed in all marrow-derived B lineage cells contrasted with the variable levels of expression of other regulatory proteins, such as Rag2, Bcl2, and Tdt. Btk level was significantly lower in B cells from peripheral sites, such as spleen, compared with marrow. Stimulation of splenic B cells through crosslinking of the BCR led to a rapid increase in Btk expression through a posttranscriptional regulatory mechanism. Because Btk function is rate-limiting for certain B cell immune responses, regulation of its protein expression may be a distinct mechanism to control the flow of information through certain receptor signaling pathways.
Materials and Methods
Materials.
Colonies of BALB/c, Balb/xid, Btk-knockout (22), and Btk-knockout mice heterozygous or homozygous for a Btk transgenic allele (26) were maintained in filter-top cages according to university guidelines for care of animals. Commercially available [Jackson ImmunoResearch, PharMingen, and Supertechs (Bethesda, MD)] antibodies were used to activate or immunostain cells for analysis, flow sorting, or immunomagnetic affinity selection, including goat anti-mouse IgM, FITC-anti-CD80, FITC-anti-mouse IgD, FITC-anti-mouse IgM, FITC-anti-mouse BP-1, FITC-anti-mouse CD43, FITC-anti-mouse HSA (CD24), rat anti-TER119, FITC-anti-Gr-1, FITC-anti-Thy1.2, FITC-anti-MAC-1, allophycocyanin-anti-c-kit, biotinylated anti-IL-7R, Texas Red-anti-Sca-1, hamster anti-Bcl-2, anti-Rag-2, rabbit anti-terminal deoxytransferase (TdT), phycoerythrin (PE)-donkey anti-rabbit IgG, and PE-donkey anti-mouse IgG. Immunomagnetic affinity selection was performed by using anti-FITC-coupled magnetic beads (Miltenyi Biotec, Auburn, CA) and MACS immunomagnetic columns (Miltenyi). Antigen affinity-purified rabbit polyclonal antibody was prepared from serum of animals immunized with GST-Btk 1-197 (27, 29) and was used for intracellular detection of Btk (30). Reverse transcription (RT)–PCR was performed by using Superscript II reverse transcriptase (GIBCO/BRL) and Taq polymerase (GIBCO/BRL).
Methods.
Single cell suspensions were prepared from bone marrow or spleen as described (30, 31) and were maintained on ice during the staining and selection procedures. Erythrocytes were lysed by incubation of cells in 0.15 M NH4Cl, 0.1M KCl, and 0.1 mM EDTA (30, 31). Cells were incubated with anti-CD16/CD32 (PharMingen) to decrease binding to the Fc receptor. Designation of B lineage developmental stages was based on the nomenclature of Hardy et al. (32). Subpopulations of cells from bone marrow or peripheral tissues were purified by sorting with either fluorescence-activated cell sorter (FACS) or immunomagnetic affinity columns. In the latter case, cells were incubated with anti-B220 immunomagnetic beads, or the primary antibody followed by addition of anti-FITC immunomagnetic beads, then were passed through the MACS immunomagnetic affinity column and were eluted according to the manufacturer's protocol. Flow cytometry analysis of cell populations selected by immunomagnetic affinity columns demonstrated purity of >90% for the desired staining characteristics.
Purified cells were fixed with 4% paraformaldehyde in PBS and were permeabilized and immunostained, and the fluorescence signals were analyzed by using a FACScan (Becton Dickinson) or a FACSVantage (Becton Dickinson). In certain experiments, erythrocyte-depleted murine splenocytes were stimulated by addition of goat anti-mouse IgM (20 μg/ml) and were incubated at 37°C in RPMI containing 2% fetal bovine serum for the indicated periods of time. After stimulation, the splenocytes were fixed in paraformaldehyde, and B cells were recovered by positive selection using anti-B220 immunomagnetic affinity columns. The B cells were then permeabilized and stained as described (30). Fluorescence data plots were generated by using a lymphocyte gate as defined by forward and side scatter characteristics and simultaneously detecting FITC, PE, propidium iodide, Texas Red, or allophycocyanin fluorescence in the FL1-FL5 windows.
Results and Discussion
Btk Expression in Developing Hematopoietic Cells.
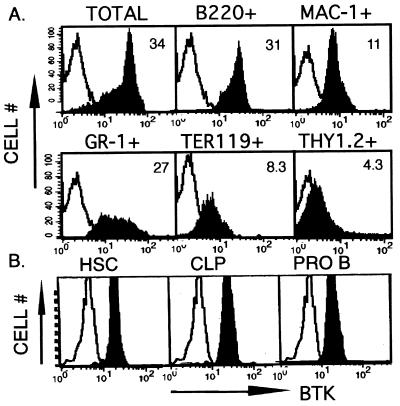
To evaluate whether expression of the endogenous Btk allele is dynamically regulated during the development of B lymphocytes, we used a quantitative, intracellular immunofluorescent assay. Analysis of murine bone marrow cells demonstrates the presence of several subpopulations with differing levels of Btk expression per cell (Fig. 1A). Fractionation of wild-type (BALB/c) bone marrow lineages based on cell surface markers revealed uniform or bimodal distributions of Btk expression in most of the hematopoietic lineages. In contrast, wild-type T lineage cells, or hematopoietic cells derived from Btk knockout animals, stained similar to controls using nonspecific rabbit Ig (Fig. 1A; data not shown).
Figure 1.
(A) Btk expression in multiple murine bone marrow-derived hematopoietic lineages. Bone marrow cells were immunostained as indicated with FITC-conjugated antibodies for one of the following lineage markers: B (B220+), T, and natural killer (Thy 1.2+) monocyte (Mac1+), granulocyte and mast (Gr1 +), or erythroid (Ter119+). After incubation with anti-FITC-conjugated magnetic beads, marker-positive cells were purified by retention on and elution from a magnetic column. Purified cells were fixed and immunostained with a nonspecific antibody (open histograms) or anti-Btk (filled histograms) followed by PE-anti-rabbit. The number within each graph represents the mean fluorescence intensity of cells in the anti-Btk-immunostained population. (B) Expression of Btk in hematopoietic stem cells and common lymphoid progenitors. Bone marrow cells were immunostained as indicated with antibodies for cell surface proteins defining totipotent hematopoietic stem cells (c-kithi, Sca-1hi, IL-7R−, Lin−), common lymphoid progenitors (c-kitlo, Sca-1lo, IL-7R+, Lin−), or early B lineage cells (B220+, CD43+, IgM−). Cells were purified by FACS, were permeabilized with saponin 0.03% (in Hank's balanced salt solution, 2% FCS, 0.02% sodium azide), and were immunostained with a nonspecific antibody (open histograms) or anti-Btk (filled histograms) followed by FITC-anti-rabbit.
Detection of Btk protein in multiple marrow-derived hematopoietic lineages suggests that initiation of Btk expression is a common developmental feature in these cell types or that Btk expression is initiated in a shared cellular precursor and is maintained during subsequent development. To distinguish between these alternative hypotheses, hematopoietic stem cells, common lymphoid progenitors, and progenitor B cells were purified by FACS from whole murine bone marrow on the basis of positive and negative cell surface marker criteria (33), then were stained with either control Ig or a Btk-specific antibody. Btk protein was detected in both hematopoietic stem cells and common lymphoid progenitors at a level similar to that observed in the progenitor B cells (Fig. 1B), indicating that its expression originates at the earliest point in hematopoietic development and is maintained.
Sustained Btk Expression in Developing B Lymphocytes.
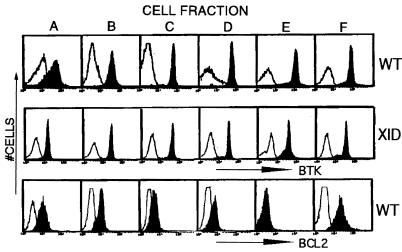
Bone marrow B lineage cells progress through a tightly regulated set of developmental stages leading to the recombination of Ig loci, formation of antigen receptor complexes, negative selection of cells recognizing autoantigens, and generation of immunocompetent B cells (1, 32, 34–36). In addition to progressive genetic changes at the Ig loci, there are selective changes in expression of many proteins that influence these developmental processes (1–3). To test for a developmental influence upon Btk protein level, six distinct B cell subpopulations of increasing maturity were purified using a modification of the Hardy fractionation protocol (32). Bone marrow B lineage cells (37), including prepro-B (fraction “A”), pro-B (“B”), pro-B plus large preB (“C”), small preB (“D”), immature B (“E”), and mature B cells (“F”), express a similar amount of Btk per cell (Fig. 2 Top). The earliest developmental stage (“A”) had a broader range of Btk expression than observed in cells of later stages in this experiment, but a narrower distribution was found in other experiments. A partial block in late B cell development is observed in the murine phenotype of Btk loss of function (xid or knockout alleles) (22, 38). However, marrow-derived B lineage cells from xid mice have sustained expression of the mutant form of Btk protein quite similar to wild-type cells (Fig. 2 Middle). The maintenance of Btk protein level in xid B lineage cells suggests that Btk expression during development is independent of its signal transducing function. In contrast with the consistent level of Btk expression, several other proteins (Rag2, Bcl-2, Tdt) were observed to have B cell developmental stage-specific patterns of expression consistent with previous reports (Fig. 2 Bottom; data not shown) (37, 39–42).
Figure 2.
Btk or Bcl-2 expression during B lineage development. Bone marrow cells from wild-type (Top and Bottom) or xid (Middle) mice were sequentially stained with FITC-conjugated antibodies recognizing B lineage markers (32), and then were fractionated by using either anti-B220 or anti-FITC-conjugated immunomagnetic bead columns. In each step, the marker-positive cells are retained in the magnetic column, and the marker-negative cells are recovered in the flow through fraction. After marrow harvest, erythrocytes were lysed as described in the Methods, and monocytic cells were depleted by using FITC-anti-Mac-1 followed by anti-FITC immunomagnetic bead columns. The first purification step was performed by staining Mac-1− marrow cells with FITC-anti-IgD to obtain fraction “F” (IgD+ cells). The IgD− cell population was then stained with FITC-anti-IgM to yield fraction “E” (IgM+). The IgD−IgM− cell population was immunostained with FITC-anti-BP-1. BP-1+ cells were next stained with FITC-anti-CD43 whereas BP-1− cells were stained with FITC-anti-HSA. A positive selection with anti-B220 immunomagnetic beads was used to recover B lineage cells from these latter steps to yield the final four cell fractions: BP-1−, HSA−, B220+ (“A”); BP-1−, HSA+, B220+ (“B”); BP-1+, CD43+, B220+ (“C”); BP-1+, CD43−, B220+ (“D”). Purified cells were fixed and immunostained with a nonspecific antibody (open histograms), anti-Btk [filled histograms (Top and Middle)], or anti-Bcl-2 [filled histograms (Lower)] followed by the appropriate fluorescent-tagged secondary antibody.
Decreased Btk Expression in Peripheral B Lymphocytes.
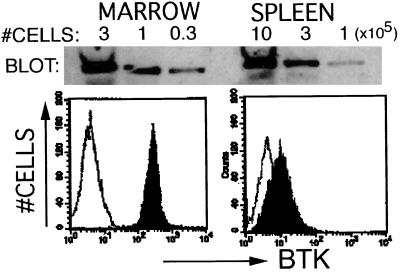
B lineage cells expressing intact BCR exit the bone marrow to populate the blood, spleen, lymph nodes, and other peripheral sites. Antigen binding to the BCR activates B cells in peripheral tissues, leading to further cell maturation, with Ig gene rearrangement, clonal expansion, and antibody secretion (1, 34–36). Bone marrow and peripheral populations also include B cell populations that have undergone antigen-provoked maturation (1, 34–36). Btk level was compared in B cells from marrow and peripheral lymphoid tissues to test whether Btk expression level varied in cells derived from distinct anatomic locations. Immunofluorescent staining revealed a dramatic decrease in Btk expression in the peripheral B cells relative to marrow cells, as demonstrated by comparison of IgD+ cells from marrow and spleen (Fig. 3). Btk immunoblot analysis of these cell fractions confirms this observation (Fig. 3). Similar low Btk levels were observed in B cell populations from lymph nodes, spleen, and peritoneum as opposed to the consistent high levels in marrow-derived populations (Fig. 2; data not shown). Reduced expression did not require intact Btk signaling function because a decrease in Btk protein is observed in splenic B cells from xid cells (data not shown).
Figure 3.
Decreased Btk in splenic B cells compared with bone marrow. Total bone marrow cells and splenic lymphocytes were stained with anti-IgD. The IgD+ B cells from spleen and marrow were purified by immunomagnetic affinity column and immunostained (lower panels) with a nonspecific antibody (open histograms) or anti-Btk (filled histograms) followed by PE-anti-rabbit. Alternatively, FACS-purified IgD+ B cells from spleen and marrow were lysed with Laemmli sample buffer for immunoblot analysis (upper panels). Serial dilutions of each lysate representing decreasing numbers of cell equivalents (×105 cells/sample) were loaded as indicated. After resolution by gel electrophoresis, protein was transferred to nitrocellulose and was immunoblotted with anti-Btk. The Btk protein was visualized by using horseradish peroxidase-coupled anti-rabbit secondary followed by enzyme chemiluminescent substrate.
Increased Btk Dosage in Activated Splenic B Cells.
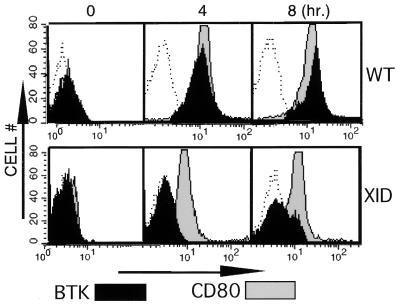
Antigen-binding to IgM+ murine B cells rapidly induces complex biochemical signaling events that ultimately can provoke increased IgM secretion, cell maturation, proliferation, or cell death (43–45). Because Btk dosage is a genetically defined parameter mediating BCR function (26), Btk expression in splenic B cells was examined before and after BCR crosslinking to test the hypothesis that activation of this signaling pathway modulates Btk level. Activation of the BCR maximally increased (≈7- to 10-fold higher mean fluorescence intensity) Btk expression within 4 hours, and this higher level was maintained in the B cells for at least 4 hours more (Fig. 4 Upper). Btk expression decreased to unstimulated control levels within 24 hours after stimulation (data not shown). In contrast, the level of the Btk enzyme in xid cells fails to increase after BCR crosslinking (Fig. 4 Lower). However, loss of Btk function did not confer a global deficit in BCR signaling because cells from both normal and xid mice responded to stimulation by increasing the level of the surface marker CD80 (46, 47) (Fig. 4 Upper and Lower). Comparison of the relative level of protein expression of the xid and wild-type alleles (Figs. 2 and 4) suggests that different mechanisms regulate Btk dosage during B cell development and BCR-mediated splenocyte stimulation. Because Btk is rate-limiting for certain BCR-mediated immune functions, this dosage regulation could be a sensitive mechanism to influence the intensity of cellular responses to Btk-dependent signal transduction.
Figure 4.
BCR crosslinking increases Btk protein dosage in murine splenic B lymphocytes. Wild-type (Upper) or xid (Lower) splenic B lymphocytes were stimulated by cross-linking BCR with anti-IgM (20 μg/ml) for increasing periods of time (0, 4, 8 hours) as indicated. Cells were fixed and immunostained with a nonspecific antibody (open histograms), anti-Btk (filled histograms), or anti-CD80 (gray histograms) followed by the appropriate fluorescent-tagged secondary antibody.
To test whether up-regulation of Btk in BCR-crosslinked splenic B cells in culture reflects in vivo immune responses, wild-type and xid mice were immunized with a single dose of either T cell-dependent or -independent antigen, and then splenic B cells were purified and tested for Btk expression. For both classes of immunogen, increased Btk protein was found in a subpopulation (≈10–15%) of B cells from wild-type mice (data not shown). Previous analysis of splenic lymphocyte responses to specific antigens suggests that a much smaller percentage of cells (<1%) is directly activated through antigen–receptor interactions (48). Thus, in vivo Btk up-regulation in T-dependent and -independent immune responses may reflect both direct and indirect effects of splenic lymphocyte activation. Interestingly, Btk signaling function was necessary for up-regulation in vivo, as the xid protein was not increased for either class of antigen.
Posttranscriptional Up-Regulation of Btk.
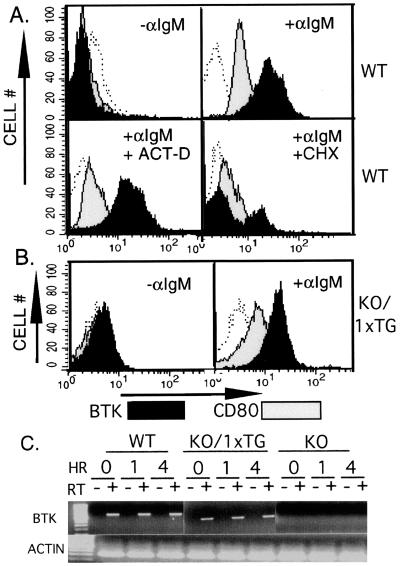
Alteration in protein level could be mediated at several levels, such as transcription, translation, or protein half-life. The maximal increase in Btk requires approximately 4 hours, compatible with any of these mechanisms. To distinguish among the possible mechanisms, analysis of Btk up-regulation was performed in the presence of inhibitors of mRNA or protein synthesis. Actinomycin D or cycloheximide abolished BCR-mediated CD80 up-regulation, but only cycloheximide inhibited the increase in Btk protein level (Fig. 5A). These results are consistent with the conclusion that up-regulation of Btk is posttranscriptional whereas CD80 requires increased transcription. This mechanism is further supported by a comparison of Btk expressed from two distinct genetic loci. BCR-induced Btk up-regulation was compared in splenocytes derived from wild-type mice or ones expressing a Btk transgene (having a knockout allele at the endogenous locus) (26). This Btk transgene lacks most of the untranslated 5′ region of Btk mRNA, and the endogenous promoter or enhancer elements have been replaced by the IgM promoter region (49). In lymphocytes expressing the Btk transgene (KO/1 × TG), BCR crosslinking increased Btk protein level with similar kinetics as in wild-type cells but to a slightly lower maximal level at 4 hours (Fig. 5B; data not shown).
Figure 5.
(A) Up-regulation of Btk by a posttranscriptional mechanism. Wild-type splenic B lymphocytes were stimulated by cross-linking BCR with anti-IgM (20 μg/ml) for 4 hours in the presence of cycloheximide (100 μM), actinomycin D (100 μM), or carrier (DMSO 1%) alone as indicated. Cells were fixed and immunostained with a nonspecific antibody (open histograms), anti-Btk (filled histograms), or anti-CD80 (gray histograms) followed by the appropriate fluorescent-tagged secondary antibody. (B) Up-regulation of transgenic Btk protein after BCR crosslinking. Splenic B lymphocytes expressing a single copy of the Btk transgene (KO/1 × TG) were stimulated by cross-linking BCR with anti-IgM (20 μg/ml) for 4 hours as indicated. Cells were fixed and immunostained with a nonspecific antibody (open histograms), anti-Btk (filled histograms), or anti-CD80 (gray histogram) followed by the appropriate fluorescent-tagged secondary antibody. (C) Semiquantitative assay of Btk mRNA. Splenic B lymphocytes expressing endogenous Btk (WT), a single copy of the Btk transgene (KO/1 × TG), or no detectable Btk protein (KO) were stimulated by cross-linking BCR with anti-IgM (20 μg/ml) for 0, 1, or 4 hours as indicated. Total RNA was purified from the cells and was tested (0.8–3.2 μg/sample) for Btk mRNA expression before (−) and after (+) reverse transcription (RT) of the RNA sample. RT was performed by using oligo(dT) primer. PCR was then performed for 35 cycles using Btk-specific sense (bp 781–799, gctgaaaaaggtcgtggc) and antisense (bp 1,517–1,500, cttcgaccacgtcaacat) primers. The results shown were obtained by using 3.2 μg of total RNA per sample. Actin RNA transcripts were amplified (+ RT condition only) to demonstrate the integrity of the RNA samples (Lower).
Semiquantitative RT-PCR was performed to test directly whether the level of Btk mRNA varied after receptor stimulation in wild-type or transgenic animals. No receptor stimulation-dependent change in Btk mRNA was observed in wild-type or transgene-expressing splenocytes (Fig. 5C; data not shown) using an assay capable of distinguishing 2-fold differences in Btk mRNA. Btk mRNA was undetectable with this assay in control or activated samples derived from Btk knockout B lymphocytes, a finding compatible with the markedly reduced abundance of mutant Btk mRNA transcripts demonstrated by Northern blot analysis (22). Simultaneous evaluation of serially diluted samples indicated that Btk mRNA in B splenocytes from Btk transgenic animals (KO/1 × TG) was slightly lower (≈50% less) than in the wild type (Fig. 5C; data not shown). Thus, three separate experimental approaches (inhibitor studies, transgene alleles, and RT-PCR) support the conclusion that BCR-induced up-regulation occurs through a posttranscriptional mechanism.
Btk Up-Regulation Depends on a Signal Transmitted by PI 3-Kinase.
The molecular basis of the genetic defect observed in xid appears to result from the deceased capacity of the Btk R28C allele to bind to the second messenger membrane phospholipid phosphatidylinositol 3,4,5-PO4 (14, 50, 51). BCR crosslinking was performed before or after treatment of cells with a membrane permeant inhibitor (52–54) of PI 3-kinase activity to test the hypothesis that PI 3-kinase activity is required for up-regulation of Btk in wild-type splenocytes. Inhibition of PI 3-kinase in wild-type cells before BCR crosslinking decreased Btk up-regulation, similar to the phenotype seen in Btk xid (Table 1). The result does not distinguish whether PI 3-kinase activity up-regulates Btk by increasing the rate of protein synthesis, by stabilizing Btk protein, or a combination of these mechanisms. However, LY294002 addition 1 hour after BCR crosslinking blocked up-regulation (measured 4 hours after crosslinking) as effectively as pretreatment (Table 1), demonstrating that the mechanism(s) up-regulating Btk protein level requires prolonged signaling through PI 3-kinase. Up-regulation was not rapidly reversed by PI 3-kinase inhibition because addition of LY294002 at 3 hours after crosslinking had little effect on Btk level measured at 4 hours.
Table 1.
Pharmacologic agents influence Btk protein level
| Experiment | Btk | Treatment | Anti-IgM | Fluorescence* |
|---|---|---|---|---|
| 1 | WT | None | (−) | 4 |
| WT | None | (+) | 42 | |
| WT | LY294002 (−0.5 hour) | (+) | 8 | |
| WT | LY294002 (+1 hour) | (+) | 8 | |
| WT | LY294002 (+3 hours) | (+) | 36 | |
| 2a | WT | None | (−) | 9 |
| WT | None | (+) | 28 | |
| WT | PMA + ionomycin | (−) | 28 | |
| 2b | xid | None | (−) | 5 |
| xid | None | (+) | 7 | |
| xid | PMA + ionomycin | (−) | 18 |
Influence of activators or inhibitors of intracellular signaling pathways on Btk protein expression. Experiment 1: Balb/c splenic lymphocytes were stimulated by cross-linking BCR with anti-IgM (20 μg/ml) for 4 hours at 37°C. LY294002 (20 μM) or carrier (DMSO 1%) alone was added 0.5 hours before, 1 hour after, or 3 hours after receptor crosslinking. Experiment 2: Balb/c (a) or Balb/xid (b) splenic lymphocytes were stimulated by cross-linking BCR with anti-IgM (20 μg/ml), or by treatment with phorbol myristic acid (10 nM) and ionomycin (100 nM) or carrier (DMSO 1%) alone, and then were incubated for 4 hours at 37°C. Cells were fixed and immunostained with a nonspecific antibody or anti-Btk followed by PE-anti-rabbit. The mean channel fluorescence intensity for each sample was rounded to the nearest whole number. PMA, phorbol myristic acid; WT, wild type.
*Mean channel fluorescence intensity.
Receptor-Independent Btk Up-Regulation.
Because Btk functions at a crossroads of many important intracellular signaling pathways (13, 16), pharmacological activation of pathways distal to surface receptors may also influence Btk level. Splenic B lymphocytes can be activated by treatment with a combination of phorbol myristic acid and ionomycin, leading to activation of protein kinase C and increased intracellular free calcium levels (14–16, 55, 56). Direct activation of these two signaling pathways increased the level of both Btk wild-type and xid proteins (Table 1).
Distinct mechanisms appear to regulate endogenous Btk protein level in bone marrow B lineage cells in comparison to antigen receptor-activated splenic B cells. Up-regulation of Btk protein expression in cell culture assays suggests that in vivo activation of B cells in antigen responses could modulate the signaling function of this rate-limiting component. One example would be if the antigen-receptor complex binding affinity directly correlated with the increase in Btk. Because multiple cell surface receptor pathways require Btk signaling function in vivo, these pathways could cooperate or antagonize in the Btk up-regulation to help select individual cells in an immune response. An intriguing possibility is that up-regulation of Btk by one receptor stimulus could sensitize the cells to signaling through a second pathway.
The molecular structure of the PH domain indicates that Btk R28 residue has an important binding interaction with phosphatidylinositol 3,4,5-PO4. The xid mutation (R28C) renders this protein incapable of responding to the upstream regulatory influence of PI 3-kinase, a phenotype recapitulated by the treatment of wild-type B cells with PI 3-kinase inhibitor. The pharmacologic and genetic experiments defining PI 3-kinase's role in Btk up-regulation cannot distinguish whether this influence occurs by decreasing Btk association with the activated receptor signaling complex or is caused by the loss in signaling to effectors downstream of Btk. Direct activation of two signaling pathways that function downstream of Btk is sufficient to up-regulate Btk R28C in xid splenic B cells. This result strongly suggests that Btk signal transducing capacity is a mediator of the up-regulation of its own protein level. Dynamic changes in Btk protein level may modulate B cell immune responses in conjunction with the regulatory influences of tyrosine phosphorylation and plasma membrane localization (6–10, 14–16, 27, 50, 51). The mechanism for up-regulation of Btk protein may include an increase in protein half-life, an increase in protein synthesis, or a combination of both.
Acknowledgments
We thank J. Johnson for technical assistance and J. C. White for help in preparation of the manuscript. S.N. is an Associate of the Howard Hughes Medical Institute. M.I.W. is supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (Fellowship DRG-086). O.N.W. is an Investigator of the Howard Hughes Medical Institute. K.A. is supported by a Jose Carreras International Leukemia Society grant (1997). I.L.W. is supported by National Institutes of Health Grant CA42551.
Abbreviations
- BCR
B cell antigen receptor
- Btk
Bruton's tyrosine kinase
- FACS
fluorescence-activated cell sorter
- PE
phycoerythrin
- PI 3-kinase
phosphatidylinositol 3-kinase
- RT
reverse transcription
- xid
X-linked immunodeficiency
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050583597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050583597
References
- 1.Osmond D G. Immunol Rev. 1986;93:103–124. doi: 10.1111/j.1600-065x.1986.tb01504.x. [DOI] [PubMed] [Google Scholar]
- 2.Fitzsimmons D, Hagman J. Curr Opin Immunol. 1996;8:166–174. doi: 10.1016/s0952-7915(96)80054-9. [DOI] [PubMed] [Google Scholar]
- 3.Osmond D G, Rolink A, Melchers F. Immunol Today. 1998;19:65–68. doi: 10.1016/s0167-5699(97)01203-6. [DOI] [PubMed] [Google Scholar]
- 4.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 6.Tsukada S, Saffran D C, Rawlings D J, Parolini O, Allen R C, Klisak I, Sparkes R S, Kubagawa H, Mohandas T, Quan S, et al. Cell. 1993;72:279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 7.Vetrie D, Vorechovsky I, Sideras P, Holland J, Davies A, Flinter F, Hammarstrom L, Kinnon C, Levinsky R, Bobrow M, et al. Nature (London) 1993;361:226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 8.Yamada N, Kawakami Y, Kimura H, Fukamachi H, Baier G, Altman A, Kato T, Inagaki Y, Kawakami T. Biochem Biophys Res Commun. 1993;192:231–240. doi: 10.1006/bbrc.1993.1404. [DOI] [PubMed] [Google Scholar]
- 9.Smith C I, Baskin B, Humire-Greiff P, Zhou J N, Olsson P G, Maniar H S, Kjellen P, Lambris J D, Christensson B, Hammarstrom L, et al. J Immunol. 1994;152:557–565. [PubMed] [Google Scholar]
- 10.de Weers M, Verschuren M C, Kraakman M E, Mensink R G, Schuurman R K, van Dongen J J, Hendriks R W. Eur J Immunol. 1993;23:3109–3114. doi: 10.1002/eji.1830231210. [DOI] [PubMed] [Google Scholar]
- 11.Satterthwaite A B, Witte O N. Annu Rev Immunol. 1996;14:131–154. doi: 10.1146/annurev.immunol.14.1.131. [DOI] [PubMed] [Google Scholar]
- 12.Rawlings D J, Witte O N. Semin Immunol. 1995;7:237–246. doi: 10.1006/smim.1995.0028. [DOI] [PubMed] [Google Scholar]
- 13.Satterthwaite A B, Li Z, Witte O N. Semin Immunol. 1998;10:309–316. doi: 10.1006/smim.1998.0123. [DOI] [PubMed] [Google Scholar]
- 14.Scharenberg A M, El-Hillal O, Fruman D A, Beitz L O, Li Z, Lin S, Gout I, Cantley L C, Rawlings D J, Kinet J P. EMBO J. 1998;17:1961–1972. doi: 10.1093/emboj/17.7.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fluckiger A C, Li Z, Kato R M, Wahl M I, Ochs H D, Longnecker R, Kinet J P, Witte O N, Scharenberg A M, Rawlings D J. EMBO J. 1998;17:1973–1985. doi: 10.1093/emboj/17.7.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurosaki T. Annu Rev Immunol. 1999;17:555–592. doi: 10.1146/annurev.immunol.17.1.555. [DOI] [PubMed] [Google Scholar]
- 17.Oda, A., Ikeda, Y., Ochs, H. D., Druker, B. J., Ozaki, K., Handa, M., Ariga, T., Sakiyama, Y., Witte, O. N. & Wahl, M. I. (2000) Blood, in press. [PubMed]
- 18.Schaeffer E M, Debnath J, Yap G, McVicar D, Liao X C, Littman D R, Sher A, Varmus H E, Lenardo M J, Schwartzberg P L. Science. 1999;284:638–641. doi: 10.1126/science.284.5414.638. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami Y, Kitaura J, Hata D, Yao L, Kawakami T. J Leukocyte Biol. 1999;65:286–290. doi: 10.1002/jlb.65.3.286. [DOI] [PubMed] [Google Scholar]
- 20.Thomas J D, Sideras P, Smith C I E, Vorechovsky I, Chapman V, Paul W E. Science. 1993;261:355–358. doi: 10.1126/science.8332900. [DOI] [PubMed] [Google Scholar]
- 21.Rawlings D J, Saffran D C, Tsukada S, Largaespada D A, Grimaldi J C, Cohen L, Mohr R N, Bazan J F, Howard M, Copeland N G, et al. Science. 1993;261:358–361. doi: 10.1126/science.8332901. [DOI] [PubMed] [Google Scholar]
- 22.Khan W N, Alt F W, Gerstein R M, Malynn B A, Larsson I, Rathbun G, Davidson L, Muller S, Kantor A B, Herzenberg L A, et al. Immunity. 1995;3:283–299. doi: 10.1016/1074-7613(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 23.Conley M E, Cooper M D. Curr Opin Immunol. 1998;10:399–406. doi: 10.1016/s0952-7915(98)80112-x. [DOI] [PubMed] [Google Scholar]
- 24.Satterthwaite A B, Witte O N. Curr Opin Immunol. 1996;8:454–458. doi: 10.1016/s0952-7915(96)80029-x. [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto S, Tsukada S, Matsushita M, Miyawaki T, Niida Y, Yachie A, Kobayashi S, Iwata T, Hayakawa H, Matsuoka H, et al. Blood. 1996;88:561–573. [PubMed] [Google Scholar]
- 26.Satterthwaite A B, Cheroutre H, Khan W N, Sideras P, Witte O N. Proc Natl Acad Sci USA. 1997;94:13152–13157. doi: 10.1073/pnas.94.24.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Tsukada S, Satterthwaite A, Havlik M H, Park H, Takatsu K, Witte O N. Immunity. 1995;2:451–460. doi: 10.1016/1074-7613(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 28.Dingjan G M, Maas A, Nawijn M C, Smit L, Voerman J S, Grosveld F, Hendriks R W. EMBO J. 1998;17:5309–5320. doi: 10.1093/emboj/17.18.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahl M I, Fluckiger A C, Kato R M, Park H, Witte O N, Rawlings D J. Proc Natl Acad Sci USA. 1997;94:11526–11533. doi: 10.1073/pnas.94.21.11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nisitani S, Kato R M, Rawlings D J, Witte O N, Wahl M I. Proc Natl Acad Sci USA. 1999;96:2221–2226. doi: 10.1073/pnas.96.5.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nisitani S, Tsubata T, Murakami M, Okamoto M, Honjo T. J Exp Med. 1993;178:1247–1254. doi: 10.1084/jem.178.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo M, Weissman I L, Akashi K. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 34.Rolink A, Melchers F. Cell. 1991;66:1081–1094. doi: 10.1016/0092-8674(91)90032-t. [DOI] [PubMed] [Google Scholar]
- 35.Melchers F, Haasner D, Grawunder U, Kalberer C, Karasuyama H, Winkler T, Rolink A G. Annu Rev Immunol. 1994;12:209–225. doi: 10.1146/annurev.iy.12.040194.001233. [DOI] [PubMed] [Google Scholar]
- 36.Nishikawa S I, Era T, Ogawa M, Nishikawa S, Ohno N, Hayashi S I, Kunisada T. Curr Top Microbiol Immunol. 1992;182:27–35. doi: 10.1007/978-3-642-77633-5_4. [DOI] [PubMed] [Google Scholar]
- 37.Li Y S, Hayakawa K, Hardy R R. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerner J D, Appleby M W, Mohr R N, Chien S, Rawlings D J, Maliszewski C R, Witte O N, Perlmutter R M. Immunity. 1995;3:301–312. doi: 10.1016/1074-7613(95)90115-9. [DOI] [PubMed] [Google Scholar]
- 39.Merino R, Ding L, Veis D J, Korsmeyer S J, Nunez G. EMBO J. 1994;13:683–691. doi: 10.1002/j.1460-2075.1994.tb06307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grillot D A, Merino R, Pena J C, Fanslow W C, Finkelman F D, Thompson C B, Nunez G. J Exp Med. 1996;183:381–391. doi: 10.1084/jem.183.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wasserman R, Li Y S, Hardy R R. J Immunol. 1997;158:1133–1138. [PubMed] [Google Scholar]
- 42.Grawunder U, Leu T M, Schatz D G, Werner A, Rolink A G, Melchers F, Winkler T H. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 43.Healy J I, Goodnow C C. Annu Rev Immunol. 1998;16:645–670. doi: 10.1146/annurev.immunol.16.1.645. [DOI] [PubMed] [Google Scholar]
- 44.Melamed D, Benschop R J, Cambier J C, Nemazee D. Cell. 1998;92:173–182. doi: 10.1016/s0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- 45.Nussenzweig M C. Cell. 1998;95:875–878. doi: 10.1016/s0092-8674(00)81711-0. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein M D, Debenedette M A, Hollenbaugh D, Watts T H. Mol Immunol. 1996;33:541–552. doi: 10.1016/0161-5890(96)00005-3. [DOI] [PubMed] [Google Scholar]
- 47.Freeman G J, Freedman A S, Segil J M, Lee G, Whitman J F, Nadler L M. J Immunol. 1989;143:2714–2722. [PubMed] [Google Scholar]
- 48.Garcia de Vinuesa C, O'Leary P, Sze D M, Toellner K M, MacLennan I C. Eur J Immunol. 1999;29:1314–1323. doi: 10.1002/(SICI)1521-4141(199904)29:04<1314::AID-IMMU1314>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 49.Faust E A, Rawlings D J, Saffran D C, Witte O N. Curr Top Microbiol Immunol. 1995;194:363–370. doi: 10.1007/978-3-642-79275-5_42. [DOI] [PubMed] [Google Scholar]
- 50.Isakoff S J, Cardozo T, Andreev J, Li Z, Ferguson K M, Abagyan R, Lemmon M A, Aronheim A, Skolnik E Y. EMBO J. 1998;17:5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z, Wahl M I, Eguinoa A, Stephens L R, Hawkins P T, Witte O N. Proc Natl Acad Sci USA. 1997;94:13820–13825. doi: 10.1073/pnas.94.25.13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell K S. Curr Opin Immunol. 1999;11:256–264. doi: 10.1016/s0952-7915(99)80042-9. [DOI] [PubMed] [Google Scholar]
- 53.Reth M, Wienands J. Annu Rev Immunol. 1997;15:453–479. doi: 10.1146/annurev.immunol.15.1.453. [DOI] [PubMed] [Google Scholar]
- 54.Tsubata T. Curr Opin Immunol. 1999;11:249–255. doi: 10.1016/s0952-7915(99)80041-7. [DOI] [PubMed] [Google Scholar]
- 55.Ransom J T, Cambier J C. J Immunol. 1986;136:66–72. [PubMed] [Google Scholar]
- 56.Klaus G G, O'Garra A, Bijsterbosch M K, Holman M. Eur J Immunol. 1986;16:92–97. doi: 10.1002/eji.1830160118. [DOI] [PubMed] [Google Scholar]