Abstract
Infections caused by the bacteria Chlamydia trachomatis contribute to diverse pathologies in a variety of human populations. We previously used a systemic model of C. trachomatis infection in mice to map three quantitative trait loci that influence in vivo susceptibility differences between the C57BL/6J and C3H/HeJ inbred strains of mouse. One of these quantitative trait loci, Ctrq-3, influences an IFN-γ-dependent susceptibility difference in primary embryonic fibroblasts isolated from these strains. Here we use fine structure mapping in congenic fibroblasts carrying DNA from the susceptible parent to localize the effect of Ctrq-3 to a 1.2-megabase interval of genomic DNA that contains Irgb10 and Igtp, two members of the IFN-γ-inducible p47 family of GTPases. This class of proteins has been widely implicated in resistance to intracellular pathogens in mice. We analyzed expression of Irgb10 and Igtp in parental and congenic embryonic fibroblasts treated with IFN-γ and found that relatively resistant fibroblasts express more Irgb10 than relatively susceptible fibroblasts. However, we also found that abolishing the expression of either Irgb10 or Igtp increases susceptibility of embryonic fibroblasts to C. trachomatis. Thus, we conclude that, although a difference in Irgb10 expression is likely responsible for the effect of Ctrq-3 on susceptibility to C. trachomatis, both genes play a role in intracellular resistance to C. trachomatis.
Keywords: genetic, infection, mouse, immunity, interferon
Chlamydia trachomatis is an obligate intracellular bacterial pathogen that is a major cause of disease in a variety of different human populations (1). Infections caused by C. trachomatis are the most common bacterial source of sexually transmitted disease worldwide, with an annual incidence estimated at >90 million cases (2). C. trachomatis is also causative of the ocular infection trachoma, a leading cause of preventable blindness, particularly in developing countries where C. trachomatis is endemic (3). Although treatment and diagnostics continue to improve, there is still no vaccine for Chlamydia.
Outside of the host cell, chlamydiae exist as infectious, metabolically quiescent elementary bodies. Upon internalization, these elementary bodies undergo a rapid developmental shift to larger, metabolically active reticulate bodies. These reticulate bodies subsequently grow and divide within a parasitophorous vacuole termed an inclusion, a unique compartment whose development is largely orchestrated by the Chlamydia itself (4, 5). Critical to inclusion development is early bacterial transcription followed by targeting of the nascent inclusion to the perinuclear region, where it intercepts sphingomyelin-containing exocytic vesicles from the Golgi apparatus (6, 7). Here the reticulate bodies divide until converting back into elementary bodies that are then released to begin another round of infection. This entire cycle (entry to exit) takes ≈36–72 h to complete, depending on the particular host–serovar combination.
The involvement of the host cell in the Chlamydia developmental cycle is not completely understood. De novo host cell protein synthesis is not required at any point during the developmental cycle, and, in fact, replication of C. trachomatis is enhanced in cells treated with inhibitors of eukaryotic translation (8). These facts suggest that normal host processes might be more restrictive than facultative for Chlamydia growth and that the host actively mounts intracellular defenses against the Chlamydia. These intracellular resistance pathways, particularly those enacted in response to cytokines released during the immune response to Chlamydia, are currently a matter of intense investigation (9, 10).
We have been working with a systemic model of C. trachomatis infection in mice in an effort to identify novel host factors involved in resistance to Chlamydia. In genetic crosses involving inbred mice that are relatively resistant [C57BL/6J (B6)] and susceptible [C3H/HeJ (C3H)] to Chlamydia spp. (11–14), we recently identified three quantitative trait loci (QTL) that segregate with the splenic bacterial load in F2 progeny shortly after i.v. delivery of C. trachomatis L2 (14). These QTL map to chromosomes 2, 3, and 11 and have been called Ctrq-1, Ctrq-2, and Ctrq-3, respectively. Interestingly, primary mouse embryonic fibroblasts (MEFs) that carry the susceptible allele of Ctrq-3 are also rendered more susceptible to C. trachomatis L2, suggesting a strong cell-autonomous component to the relative resistance of B6 mice in vivo. Notably, the effect of this congenic interval in vitro is observed only upon pretreatment of the cells with IFN-γ, a cytokine that is central to the host response to Chlamydia in vivo and that restricts Chlamydia replication in vitro (15).
Our genetic model of susceptibility to C. trachomatis is well suited for identifying genes involved in host pathways critical for intracellular defense against Chlamydia infection, particularly because we have an in vitro phenotype that is amenable to manipulation and analysis. Thus, in the current study we sought to define the genetic basis for the natural variation in C. trachomatis susceptibility that we observe between MEFs from different genetic backgrounds. In this article, we describe the fine mapping of Ctrq-3 to a genomic interval on chromosome 11 containing 18 annotated genes, including two members of the p47 family of IFN-γ-inducible GTPases that play a critical role in resistance to intracellular pathogens in mice. Our data suggest that, although both p47 GTPases in our critical genetic interval, Igtp and Irgb10, are involved in the host response to C. trachomatis infection, altered expression of Irgb10 is likely responsible for the effect of Ctrq-3 on susceptibility to C. trachomatis in our model.
Results
Ctrq-3 Maps to 1.2 Megabases of DNA on Chromosome 11.
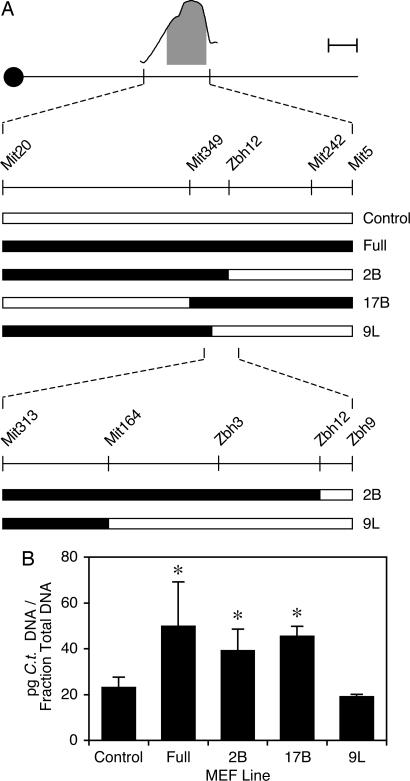
We previously showed that congenic MEFs carrying >30 megabases of chromosome 11 around Ctrq-3 from C3H on a B6 background are more susceptible to C. trachomatis L2 than noncongenic B6 MEFs; this effect is observed only upon pretreatment of the MEFs with IFN-γ (14). To further localize this in vitro effect, we derived three subcongenic lines from the large-interval B6.C3H congenic. The breakpoints for these three recombinant lines fall within the ±1.5 logarithm of odds support interval for Ctrq-3 (Fig. 1A). We prepared heterozygous MEFs from each of these lines, as well as from the original congenic and from noncarrier controls, and tested the C. trachomatis susceptibility phenotypes of each of these lines in vitro after pretreatment with IFN-γ. Two of these lines (2B and 17B) were relatively susceptible, similar to the original congenic (full); the third (9L) was relatively resistant, similar to noncarrier controls. The phenotypes of the 2B and 9L lines map the susceptibility phenotype to a maximum genetic interval of 1.2 megabases of C3H DNA between markers D11Mit164 and D11Zbh12 (Fig. 1).
Fig. 1.
Fine mapping of Ctrq-3. (A) A schematic showing breakpoints for various B6.C3H congenics. The curve at the top is a depiction of the linkage peak for Ctrq-3, including the shaded ±1.5 logarithm of odds support interval for this QTL (see ref. 14). Filled bars in the lower portion indicate C3H donor DNA, and open bars indicate recipient B6 DNA. Where breakpoints are not precisely mapped, the congenic interval giving the most conservative (i.e., largest) genetic interval is shown. Also, the congenic intervals have not been mapped beyond D11Mit20 proximally or D11Mit5 distally. The scale bar (in the upper right corner) corresponds to 10 megabases on the schematic of the chromosome. (B) The Chlamydia susceptibility phenotypes of the corresponding B6.C3H MEF lines. Indicated MEFs were pretreated in culture with IFN-γ and then infected with C. trachomatis L2 for 28 h before harvest. Each bar represents the mean of four different embryos. Bars with asterisks are significantly different from bars without asterisks (P < 0.05).
Ctrq-3 Contains Two Members of the p47 Family of GTPases.
The interval bounded by these two markers contains 18 annotated genes according to Build 36.1 of the mouse genome assembly (Fig. 2). At the distal end of the interval lie two members of the p47 family of IFN-γ-inducible GTPases. Members of this gene family are strongly induced by IFN-γ, localize to pathogen-containing vacuoles, and play critical roles in resistance to a variety of intracellular pathogens in mice (16–18). One of the p47 GTPases in our critical genetic interval is Igtp; the other is annotated as a gene model in the current genome build but has recently been designated Irgb10 (19). Although Irgb10 had been thought to be truncated, we successfully reverse-transcribed what appears to be full-length Irgb10 coding sequence from RNA isolated from the spleens of mice infected for 4 h with C. trachomatis L2. This sequence codes for a protein that is 417 aa in length with a predicted molecular weight of 47 kDa (Fig. 3). The full-length coding sequences corresponding to the two alleles of Irgb10 have been deposited in the GenBank database under accession nos. DQ508486 (B6) and DQ508487 (C3H).
Fig. 2.
Physical map of Ctrq-3. The schematic shows the position and orientation of genes in the critical genetic interval for Ctrq-3 between D11Mit164 and D11Zbh12 on chromosome 11. All information was obtained from Build 36.1 of the mouse genome at www.ncbi.nlm.nih.gov/genome/guide/mouse. Regions of alternating black and white correspond to 50-kb intervals on the chromosome. The centromeric/proximal end of the interval is to the left, and the telomeric/distal end is to the right.
Fig. 3.
Protein sequence of Irgb10. A conceptual translation of B6 Irgb10 cDNA is shown. Coding polymorphisms between B6 and C3H are in bold, with the C3H residue indicated below the corresponding B6 residue. The highly conserved G1 (GXXXXG[K/M]S), G3 (DXXG), and G4 (TXXD) domains are underlined.
Mice that are deleted for members of the p47 GTPase family are rendered dramatically more susceptible to infection with certain intracellular bacteria and protozoa (16, 17). Thus, we reasoned that polymorphisms in one or both of the p47 GTPases in our critical genetic interval might be involved in the Chlamydia susceptibility difference controlled by Ctrq-3. To test this hypothesis, we analyzed the coding sequence for both genes from B6 and C3H. Igtp contained two coding polymorphisms (DB6113EC3H and LB6321VC3H), whereas Irgb10 contained three (MB615TC3H, KB6108MC3H, and DB6161NC3H). None of these polymorphisms appears to disrupt any conserved motifs that might be critical to p47 function, such as the G domains (see Fig. 6, which is published as supporting information on the PNAS web site).
We then evaluated IFN-γ-induced mRNA expression of these two genes in MEFs isolated from B6 and C3H and from the heterozygous congenic lines whose Chlamydia susceptibility phenotypes we had tested (Fig. 4). Igtp expression did not consistently differ in the MEF lines after 15 h of IFN-γ treatment. In contrast, Irgb10 was 20-fold more induced in B6 MEFs relative to C3H MEFs after IFN-γ treatment. Additionally, an ≈2-fold difference was observed between heterozygous congenic MEFs that were relatively resistant to Chlamydia and those that were relatively susceptible. These data strongly suggest that differential expression of Irgb10 is responsible for the effect of Ctrq-3 on susceptibility to C. trachomatis in our model and that haploinsufficiency renders MEFs heterozygous for Ctrq-3 more susceptible to C. trachomatis than B6 MEFs.
Fig. 4.
Expression of Irgb10 and Igtp after IFN-γ treatment. MEFs from indicated mouse strains were treated for 15 h with 10 units/ml IFN-γ and then harvested to test for mRNA expression of either Igtp (filled bars) or Irgb10 (striped bars). Data are expressed relative to B6. Each bar represents the mean of three separate experiments. Asterisks indicate that expression of the gene differs from the 9L line at P < 0.05.
Complementation of Irgb10 Expression Reduces Susceptibility to C. trachomatis in Vitro.
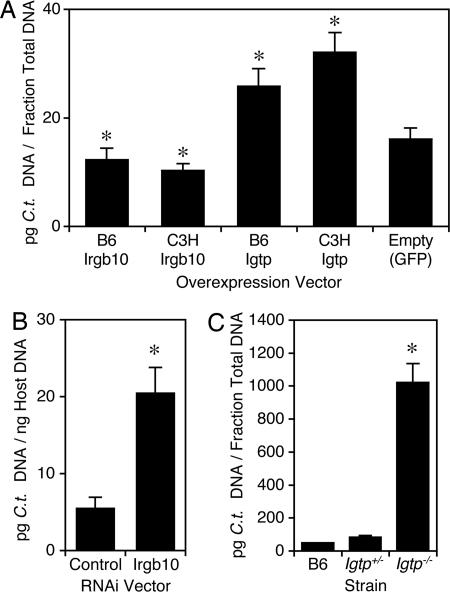
To test our hypothesis that diminished expression of Irgb10 is responsible for the effect of Ctrq-3 on susceptibility to C. trachomatis in MEFs, we attempted to render susceptible MEFs more resistant by expressing Irgb10 ectopically. We used retroviral transduction to overexpress Irgb10 or Igtp in MEFs from the 2B congenic line in the presence of IFN-γ. Overexpression of either the B6 or C3H allele of Irgb10 rendered cells more resistant to C. trachomatis in the presence of IFN-γ (Fig. 5A). Surprisingly, overexpression of either allele of Igtp actually seemed to render cells more susceptible (Fig. 5A).
Fig. 5.
Susceptibility of MEFs that are manipulated for Irgb10 and Igtp expression. (A) Effect of overexpressing alleles of Irgb10 or Igtp on susceptibility to C. trachomatis in MEFs. MEFs from the 2B congenic line were transduced with indicated vector and infected for 28 h with C. trachomatis L2 after pretreatment with IFN-γ. Bars with asterisks are significantly different from control (P < 0.05). (B) Effect of RNAi-mediated inhibition of Irgb10 expression on susceptibility in MEFs. B6 MEFs were transduced with indicated vector, pretreated with IFN-γ, and infected for 28 h with C. trachomatis L2; means are significantly different at P < 0.05. (C) Susceptibility of MEFs deleted for Igtp. Indicated MEFs were pretreated with IFN-γ and infected for 28 h with C. trachomatis L2; means for Igtp+/− and Igtp−/− are significantly different at P < 0.05. In each panel, bars represent the mean of at least three data points.
Irgb10 and Igtp Are Both Required for IFN-γ-Induced Cellular Resistance to C. trachomatis.
In complementary experiments, we also tested the effect of inhibition of Irgb10 or Igtp expression on susceptibility to C. trachomatis after IFN-γ treatment in MEFs. RNAi-based inhibition of Irgb10 expression rendered B6 MEFs more susceptible to C. trachomatis (Fig. 5B). Similarly, MEFs isolated from Igtp−/− embryos were more susceptible to C. trachomatis than those isolated from B6 or Igtp+/− embryos (Fig. 5C). Because removal of either Irgb10 or Igtp from the IFN-γ response pathway renders MEFs more susceptible to C. trachomatis, we suggest that both genes are important to the host response to C. trachomatis infection.
Discussion
Chlamydia is an obligate intracellular pathogen that requires a specialized vacuolar compartment termed an inclusion for growth and replication (4, 5). Although it is well established that Chlamydia itself plays a critical role in the development of this intracellular niche (6), less is known about how the host cell attempts to subvert Chlamydia replication. Some of the better-characterized intracellular defense mechanisms are linked to the release of proinflammatory cytokines that accompanies Chlamydia infection. A key inducer of cellular resistance in this regard is IFN-γ, a cytokine that is released primarily by T cells at the site of Chlamydia infection and that induces intracellular cascades that inhibit Chlamydia replication (15, 20).
The intracellular IFN-γ-induced effectors that limit Chlamydia growth are not completely understood. In cultured human cells, depletion of intracellular tryptophan stores secondary to activation of the indoleamine-2,3-dioxygenase pathway is responsible for inhibiting Chlamydia replication (10). Indeed, exogenous tryptophan rescues Chlamydia growth in human cells treated with IFN-γ. In mice, production of reactive nitrogen species (especially NO) via the inducible NO synthase (iNOS/Nos2) pathway is known to play a critical role in restricting the growth of many intracellular pathogens (21), but the relevance of NO production to Chlamydia infection and clearance is still controversial. For example, although Nos2 is strongly induced by IFN-γ treatment and/or Chlamydia infection in most mouse cells in vitro, NO production does not consistently correlate with inhibition of Chlamydia growth (10). Additionally, Nos2 knockout mice clear genital tract infection with C. trachomatis as efficiently as WT mice (9), suggesting that IFN-γ-inducible molecules other than Nos2 serve to restrict Chlamydia growth in mice.
Recently, RNAi analysis was used to identify the IFN-γ-inducible p47 GTPase Iigp1 as a critical component of the inhibitory effect of IFN-γ on the growth of C. trachomatis L2 in murine epithelial cells in vitro (9). We used a forward genetic approach in relatively resistant (B6) and susceptible (C3H) mouse strains to screen for host factors operative during the acute phase of systemic C. trachomatis infection in vivo. One of the QTL we identified, Ctrq-3, also affects growth of C. trachomatis in vitro, and, remarkably, this QTL maps to a small genomic interval containing two additional members of the same family of IFN-γ-inducible p47 GTPases, Irgb10 and Igtp.
Although our data suggest a role for both Irgb10 and Igtp in the intracellular host response to C. trachomatis infection, we favor the hypothesis that naturally occurring differences in Irgb10 expression underlie the effect of the Ctrq-3 QTL. This hypothesis is based on the correlation between Irgb10 expression and Chlamydia susceptibility, both in congenic MEFs that carry DNA from the susceptible parent and in MEFs manipulated for Irgb10 expression. Also, the fact that both the B6 and C3H alleles of Irgb10 confer increased resistance to C. trachomatis when overexpressed in MEFs supports our hypothesis that the difference between the two alleles is related to expression only. Ultimately, however, it will be important to establish which polymorphisms are responsible for the strain difference in susceptibility through use of targeted “knockin” approaches.
The Irgb10 expression difference that we observe between B6 and C3H maps in the vicinity of Irgb10 itself, suggesting that IFN-γ-responsive promoter elements might be polymorphic between these two strains. Indeed, sequencing of the genomic region upstream of Irgb10 reveals an AB6-to-GC3H transition that mutates one of two IFN-stimulated response elements (GCTTTCAGTTTC, where the site of the polymorphism is underlined) that lie within 100 bp of the putative transcription initiation site (19). The effect of this polymorphism on expression is unclear because this promoter element will accept either an A or a G at this central nucleotide position and still be bound by the Irf1 and Irf2 transcription factors (22). Still, analysis of a promoter:reporter fusion system may demonstrate that the B6 promoter is more efficiently stimulated by IFN-γ than the C3H promoter.
There do not appear to be significant interstrain differences in Igtp expression between B6 and C3H, nor are there any biologically meaningful polymorphisms in the Igtp coding sequence. Therefore, it seems that B6 and C3H do not differ for Igtp function. Nonetheless, Igtp knockout MEFs are highly susceptible to C. trachomatis infection, indicating that Igtp does in fact play a role in the intracellular host response to Chlamydia. This conclusion is concordant with previous studies showing an increase in susceptibility to the protozoa Toxoplasma gondii and Leishmania major in the Igtp knockout (16). However, our results demonstrate a role for Igtp in susceptibility to a bacterial pathogen and suggest that infections with T. gondii, L. major, and C. trachomatis have features in common that elicit a response from the same protein. Additionally, it is noteworthy that Nelson et al. (9) failed to find an effect of RNAi-mediated inhibition of Igtp expression on susceptibility to C. trachomatis in murine epithelial cells in vitro. This contradiction is possibly related to a cell-type specificity of Igtp function or insufficient knockdown of Igtp expression in their study.
We found that Igtp overexpression rendered embryonic fibroblasts more susceptible to infection with C. trachomatis. This observation is inconsistent both with the susceptibility phenotype of the Igtp knockout MEFs and with the notion that p47 GTPase expression serves an antimicrobial function. We suggest that Igtp overexpression in this context results in competition for effector molecules that might be essential for the function of other critical antichlamydial p47 GTPases, such as Irgb10 or Iigp1. The only binding partner identified to date for any of the p47 GTPases is the microtubule-associated protein Hook3 (23); thus, it is not certain whether these proteins share common binding partners or to what extent overexpression of one p47 GTPase might influence the activities of others.
Neither Igtp nor Irgb10 has been previously implicated in the host response to Chlamydia infection, and it is not yet clear how these proteins might function to restrict Chlamydia growth. Results from studies involving other p47 GTPases suggest that this class of proteins may influence trafficking events around the parasitophorous vacuole in which many intracellular pathogens reside (24, 25). Indeed, our own preliminary studies involving epitope-tagged alleles of Irgb10 suggest that this protein (but not Igtp) localizes to the inclusion membrane itself (data not shown). Additionally, others have found that C. trachomatis L2 inclusions do not efficiently acquire sphingolipids in IFN-γ-treated cells, suggesting that one or more of the p47 GTPases may act to restrict proper trafficking to the inclusion (9). Our MEF model, in which expression of Irgb10 and Igtp can be freely manipulated, presents a unique opportunity to investigate the involvement of these two p47 GTPases in processes related to inclusion development. More generally, we can begin to explore the molecular and functional interactions among Irgb10, Igtp, and other IFN-γ-inducible molecules (including Iigp1). In this regard, for example, we have found that exogenous expression of Irgb10 can, even in the absence of IFN-γ treatment, modestly reduce intracellular replication of Chlamydia in MEFs (data not shown). Other combinatorial experiments will continue to yield insight into the function of this remarkable class of proteins.
Genetic loci underlying complex, multigenic traits are difficult to localize and identify. In fact, only a handful of such QTL have been cloned (26). We have used the straightforward approach of fine structure mapping in congenics, combined with sequencing, expression, and functional complementation analyses, to identify a very strong candidate for a gene that underlies the effect of a QTL mapped in a relatively small population of cross animals. Additionally, we were able to identify a close paralogue of our cloned QTL that is also involved in processes relevant to our phenotype. These results suggest the utility of forward mouse genetics in identifying novel loci that have not previously been linked to the phenotype being studied.
Materials and Methods
Animals.
All mice were maintained and bred under specific pathogen-free conditions. WT B6 mice for experiments and breeding were obtained from The Jackson Laboratory (Bar Harbor, ME). The initial B6.C3H congenic used for derivation of the subcongenic lines was described previously (14). The Igtp knockout was described previously (27).
Marker Selection and Genotyping.
Simple sequence length polymorphism markers for fine mapping were either selected from the Broad Institute (Cambridge, MA) or designed around polymorphic repeat expansions identified in genomic sequences obtained from the National Center for Biotechnology Information (Bethesda, MD). Markers were amplified by standard PCR methods and electrophoresed on 4% agarose gels. Physical positions of all markers and genes were obtained from the genomic assembly at the National Center for Biotechnology Information. Primer sequences for markers described here are as follows: D11Zbh3 forward, 5′-CAACAAACACAAGGCAGACAAC-3′; D11Zbh3 reverse, 5′-AACTTTAAAGGCAACAGCGAAC-3′; D11Zbh9 forward, 5′-AACAGATCAAGTCTTGGTTGAGTG-3′; D11Zbh9 reverse, 5′-TGGCTAAGGTCTTTTTCTTCTGTC-3′; D11Zbh12 forward, 5′-GGTGAATCATGGTACAACACTTG-3′; D11Zbh12 reverse, 5′-GAGCCACGACTCAGAAATCTAAC-3′.
C. trachomatis Infection.
C. trachomatis serovar L2 434/Bu was propagated in McCoy cells and purified as described (28). Primary embryonic fibroblasts were grown and infected with C. trachomatis L2 as described (14). Briefly, cells were isolated from embryonic day 12.5–14.5 embryos, pretreated for 15 h with 10 units/ml recombinant mouse IFN-γ (Invitrogen, Frederick, MD), and then spin-infected for 1 h at 37°C with C. trachomatis L2 at a multiplicity of infection of 2:1. To measure chlamydial load, total DNA was isolated from trypsinized cell pellets and subjected to quantitative real-time PCR to assess the amount of Chlamydia DNA present in the culture, as described (14). In some instances, the amount of Chlamydia DNA was normalized against host GAPDH DNA.
Sequence and Expression Analyses.
Primers for amplification and sequencing of Igtp and Irgb10 were selected based on sequences derived from the National Center for Biotechnology Information. Igtp coding sequence was amplified through standard PCR from B6 and C3H genomic DNA. Irgb10 coding sequence was amplified by reverse transcription and PCR by using the SuperScript RT-PCR kit (Invitrogen) on splenic RNA from B6 and C3H mice infected systemically with C. trachomatis L2 (14). Sequencing was performed by using standard methods. To analyze RNA expression of Igtp and Irgb10, total RNA was isolated from MEFs treated for 15 h with 10 units/ml recombinant mouse IFN-γ by using the RNeasy kit from Qiagen (Valencia, CA). RNA was then subjected to real-time RT-PCR on an ABI 7000 Sequence Detection System using the QuantiTect SYBR Green RT-PCR kit from Qiagen and the following primer sequences: Igtp forward, 5′-AAGTTGCCACAAAATATCTGGAAGAC-3′; Igtp reverse, 5′-GGCTGATGAGGCGCTTGA-3′; Irgb10 forward, 5′-ACCTCTTTCTCAAAGGAGCTGTGT-3′; Irgb10 reverse, 5′-GCATCGGGTTTAGAAGAAGACTGA-3′. Expression was normalized against Rpl19 expression by using the following primer sequences: forward, 5′-ATCCGCAAGCCTGTGACTGT-3′; reverse, 5′-TCGGGCCAGGGTGTTTTT-3′.
Manipulation of p47 Expression.
The template for Irgb10 hairpin RNAi was generated by ligating the annealed primers 5′-GATCCCCTGAAGTTCCAGGAGTATGATTCAAGAGATCATACTCCTGGAACTTCATTTTTGGAAA-3′ and 5′- AGCTTTTCCAAAAATGAAGTTCCAGGAGTATGATCTCTTGAATCATACTCCTGGAACTTCAGGG-3′ into the BglII and HindIII sites of a modified pSUPER vector pSUPER_Xho (generous gift of Radek Skoda, University Hospital, Basel, Switzerland). The H1 expression cassette for Irgb10 short hairpin RNA was excised from pSUPER_Xho as an XhoI fragment and subcloned into the XhoI site of a mouse stem cell virus-based vector containing a GFP expression cassette under the control of the EF1-α promoter. For the overexpression studies, WT Irgb10 or Igtp coding sequence was subcloned into mouse stem cell virus upstream of an IRES-GFP cassette. Retroviral production and transduction of MEFs were carried out by using standard protocols. At 48 h after transduction, MEFs were treated with IFN-γ followed by protein expression analysis and infection with C. trachomatis, as described above.
Statistical Analyses.
All comparisons were evaluated for statistical significance through use of unpaired, two-tailed t tests.
Supplementary Material
Acknowledgments
We thank Kerry McAuliffe and David Grotsky for assistance with our mouse colony and James Nicholas Lafave and Jeff Dougherty for technical support. This work was supported by National Institutes of Health Grants AI062827 (to W.F.D.), AI039558 (to M.N.S.), and AI055900 (to M.N.S.); National Institute of Allergy and Infectious Diseases Grant AI57831 (to G.A.T.); and a Department of Veterans Affairs Merit Review Grant (to G.A.T.). J.C. was supported by a research fellowship from the Deutsche Forschungsgemeinschaft.
Abbreviations
- B6
C57BL/6J
- C3H
C3H/HeJ
- QTL
quantitative trait locus
- MEF
mouse embryonic fibroblast
Footnotes
References
- 1.Schachter J. In: Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Stephens RS, editor. Washington, DC: Am Soc Microbiol; 1999. pp. 139–169. [Google Scholar]
- 2.World Health Organization. Global Prevalence and Incidence of Selected Curable Sexually Transmitted Infections: Overview and Estimates. Geneva: WHO; 2001. pp. 1–43. [Google Scholar]
- 3.Resnikoff S, Pascolini D, Etya'ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. Bull WHO. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 4.Hackstadt T. In: Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Stephens RS, editor. Washington, DC: Am Soc Microbiol; 1999. pp. 101–138. [Google Scholar]
- 5.Fields KA, Hackstadt T. Annu Rev Cell Dev Biol. 2002;18:221–245. doi: 10.1146/annurev.cellbio.18.012502.105845. [DOI] [PubMed] [Google Scholar]
- 6.Scidmore MA, Rockey DD, Fischer ER, Heinzen RA, Hackstadt T. Infect Immun. 1996;64:5366–5372. doi: 10.1128/iai.64.12.5366-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 8.Ripa KT, Mardh PA. J Clin Microbiol. 1977;6:328–331. doi: 10.1128/jcm.6.4.328-331.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, Crane DD, Steele-Mortimer O, Kari L, McClarty G, et al. Proc Natl Acad Sci USA. 2005;102:10658–10663. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roshick C, Wood H, Caldwell HD, McClarty G. Infect Immun. 2006;74:225–238. doi: 10.1128/IAI.74.1.225-238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Maza LM, Pal S, Khamesipour A, Peterson EM. Infect Immun. 1994;62:4675–4681. doi: 10.1128/iai.62.5.2094-2097.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darville T, Andrews CW, Jr, Laffoon KK, Shymasani W, Kishen LR, Rank RG. Infect Immun. 1997;65:3065–3073. doi: 10.1128/iai.65.8.3065-3073.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu H, Yang J, Bai H, Fan Y, Wang S, Han X, Chen L, Yang X. Immunology. 2004;111:453–461. doi: 10.1111/j.0019-2805.2004.01835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein-Hanley I, Balsara ZR, Ulmer W, Coers J, Starnbach MN, Dietrich WF. Genes Immun. 2006;7:122–129. doi: 10.1038/sj.gene.6364285. [DOI] [PubMed] [Google Scholar]
- 15.Rottenberg ME, Gigliotti-Rothfuchs A, Wigzell H. Curr Opin Immunol. 2002;14:444–451. doi: 10.1016/s0952-7915(02)00361-8. [DOI] [PubMed] [Google Scholar]
- 16.Taylor GA, Feng CG, Sher A. Nat Rev Immunol. 2004;4:100–109. doi: 10.1038/nri1270. [DOI] [PubMed] [Google Scholar]
- 17.MacMicking JD. Trends Immunol. 2004;25:601–609. doi: 10.1016/j.it.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.MacMicking JD. Curr Opin Microbiol. 2005;8:74–82. doi: 10.1016/j.mib.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn DM, Glowalla E, Leptin M, Howard JC. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loomis WP, Starnbach MN. Curr Opin Microbiol. 2002;5:87–91. doi: 10.1016/s1369-5274(02)00291-6. [DOI] [PubMed] [Google Scholar]
- 21.MacMicking J, Xie QW, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka N, Kawakami T, Taniguchi T. Mol Cell Biol. 1993;13:4531–4538. doi: 10.1128/mcb.13.8.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser F, Kaufmann SH, Zerrahn J. J Cell Sci. 2004;117:1747–1756. doi: 10.1242/jcs.01039. [DOI] [PubMed] [Google Scholar]
- 24.MacMicking JD, Taylor GA, McKinney JD. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 25.Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, Howard JC. PLoS Pathog. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flint J, Valdar W, Shifman S, Mott R. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 27.Taylor GA, Collazo CM, Yap GS, Nguyen K, Gregorio TA, Taylor LS, Eagleson B, Secrest L, Southon EA, Reid SW, et al. Proc Natl Acad Sci USA. 2000;97:751–755. doi: 10.1073/pnas.97.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard L, Orenstein NS, King NW. Appl Microbiol. 1974;27:102–106. doi: 10.1128/am.27.1.102-106.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.