Abstract
Many benthic marine invertebrates, like barnacles, have a planktonic larval stage whose primary purpose is dispersal. How these species colonize suitable substrata is fundamental to understanding their evolution, population biology, and wider community dynamics. Unlike larval dispersal, settlement occurs on a relatively small spatial scale and involves larval behavior in response to physical and chemical characteristics of the substratum. Biogenic chemical cues have been implicated in this process. Their identification, however, has proven challenging, no more so than for the chemical basis of barnacle gregariousness, which was first described >50 years ago. We now report that a biological cue to gregarious settlement, the settlement-inducing protein complex (SIPC), of the major fouling barnacle Balanus amphitrite is a previously undescribed glycoprotein. The SIPC shares a 30% sequence homology with the thioester-containing family of proteins that includes the α2-macroglobulins. The cDNA (5.2 kb) of the SIPC encodes a protein precursor comprising 1,547 aa with a 17-residue signal peptide region. A number of structural characteristics and the absence of a thioester bond in the SIPC suggest that this molecule is a previously undescribed protein that may have evolved by duplication from an ancestral α2-macroglobulin gene. Although the SIPC is regarded as an adult cue that is recognized by the cyprid at settlement, it is also expressed in the juvenile and in larvae, where it may function in larva–larva settlement interactions.
Keywords: arthropod, larva, pheromone, settlement cue
Charles Darwin (1) was among the first to appreciate the biological diversity of the cirripedes, whose evolution is likely to be influenced strongly by their settlement behavior. Barnacles that settle gregariously have since featured prominently in studies that have shed light on ecological processes in the marine environment (2). Some barnacle species are also major components of hard fouling communities on ship hulls and other artificial marine structures. After the recent ban on the application of organotin-containing antifouling coatings that were linked to major environmental problems (3), interest in the settlement behavior of these barnacles and other fouling organisms has intensified. Whether in a fundamental or applied context, there have been many attempts to characterize chemical cues that modulate the settlement behavior of marine invertebrate larvae. Such studies have generally met with limited success, and the reasons for this have been explained in detail by others (e.g., ref. 4).
A century after Darwin (1), Knight-Jones and Crisp (5) proposed that a factor associated with the cuticle of adult barnacles induces gregarious settlement of cypris larvae. Exhaustive studies on temperate species, particularly Semibalanus balanoides, suggested that the inductive cue, which could be extracted from tissues with seawater and partially purified by boiling, was a polymorphic system of closely related, water-soluble glycoproteins. The cue was named “arthropodin” (6, 7) because of its physicochemical similarity to insect cuticular protein (8). However, progress toward identifying arthropodin was hampered by the short settlement season of S. balanoides and thus the limited availability of larvae for settlement assays.
In only a few instances has a complete characterization of a cue to marine invertebrate larval settlement been claimed (4, 9, 10). To the best of our knowledge, the identity of only one inductive cue, histamine in the sea urchin Holopneustes purpurescens (11), is unequivocal. The fact that this cue was originally claimed to be a complex of the sugar floridoside and isethionic acid (12) (with which histamine coeluted) only serves to emphasize the difficulties associated with characterizing invertebrate settlement cues.
No structurally complex inductive cue, such as barnacle arthropodin, has been characterized. In our studies on barnacle gregariousness, we have focused on the tropical/semitropical species Balanus amphitrite, first because it is an economically important hard fouler and second for the ability to manipulate its reproduction. Importantly, these features mean that there is considerable commercial interest in controlling its settlement, and a year-round supply of settlement-stage cypris larvae can be obtained for bioassay-directed isolation of the inductive cue (13). This latter advantage over temperate species such as S. balanoides has enabled relatively rapid progress to be made toward isolating the cue.
Using a previously undescribed nitrocellulose membrane settlement assay (14), Matsumura et al. (15) isolated a glycoprotein from whole adult B. amphitrite that induced gregarious settlement of conspecific cyprids. It is important to stress that boiled extracts were not used as the starting point for the isolation, which was a major departure from the earlier work on S. balanoides (7, 16). Not surprisingly, therefore, the major bands of B. amphitrite-inductive glycoprotein detected on SDS/PAGE, of 76-, 88-, and 98-kDa molecular mass, bore no resemblance to the reported subunit composition of S. balanoides arthropodin (16). Unlike the subunits of S. balanoides, the three major SDS/PAGE bands of B. amphitrite glycoprotein were isolated and bioassayed. These bioassays revealed that each subunit was as active as the intact molecule, which was termed the settlement-inducing protein complex (SIPC) (15). Polyclonal antibodies were raised to each of the subunits; the anti-76-kDa polyclonal was used to probe other species for SIPCs by Western blotting (17). All balanomorph and lepadomorph species examined thus far, including S. balanoides (unpublished observations), contained SIPC-like immunoactivity associated with a band of comparable molecular mass. Given the positive S. balanoides result, it is doubtful that there is a relation between arthropodin and the authentic settlement cue, the SIPC, unless the former comprised degradation products of SIPC.
Here we report the cloning and the characterization of the full-length SIPC cDNA from B. amphitrite and provide evidence using semiquantitative RT-PCR that the SIPC mRNA is expressed widely in adults, juveniles, and larvae.
Results
Cloning and Gene Structure of SIPC.
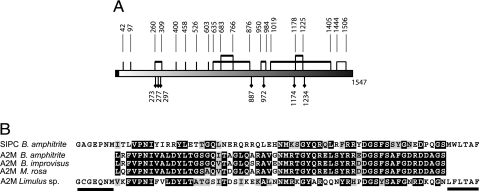
Identical N-terminal and internal peptide sequences were obtained for both the 98- and 88-kDa subunits in the SIPC sequence (Table 1). Homology-based searches revealed that the 98-, 88-, and 76-kDa peptide fragments shared significant homology with members of the α2-macroglobulin (A2M), complement factor, and insect thioester-containing protein (TEP) families (Table 1). The full-length SIPC cDNA we obtained by RT-PCR was 5,202 bp comprising a 5′ region (UTR) of 39 bp, an ORF of 4,644 bp (GenBank accession no. AY423545), and a 3′ UTR of 519 bp. The full-length ORF encoded a protein of 1,547 amino acid residues (Fig. 5, which is published as supporting information on the PNAS web site). Neglecting potential posttranslational modification, the estimated molecular mass of the SIPC protein calculated from the amino acid sequence, and excluding the 17-residue signal peptide, is 169 kDa. All of the internal and N-terminal peptide sequences we obtained previously for the 98-, 88-, and 76-kDa subunits of the purified SIPC occur in this sequence. Comparison of the predicted SIPC amino acid sequence to other sequences in the GenBank database using the BLAST algorithm (18) indicated that the SIPC of B. amphitrite most closely resembles the TEP family. The overall sequence homology was 25%, with highest homology shared with the A2M of the tick Ornothodoros moubata (31%) and the horseshoe crab Limulus sp. (29%). Fig. 1A shows the predicted structure of the SIPC. Analysis of the SIPC using the computer program SignalP V1.1 (19) indicated the presence of a signal peptide-like, hydrophobic N-terminal segment of 17 residues, which is not seen in the mature 98- and 88-kDa subunits (Fig. 5). The SIPC also contains a QTD motif (142–144), a FXVXXYVLPXFE region (228–239), and a STQDT region (1334–1338) (20). The B. amphitrite SIPC, however, does not contain the motif GCGEQ (21, 22), the anaphylatoxin-like domain (23), or a double tyrosine site near the C terminus (1530) in the ligand recognition domain, which are characteristics of different members of the TEP family. The SIPC contains 20 cysteine residues and seven potential N-glycosylation sites (Asn-Xxx-Ser/Thr and Asn-Xxx-Cys) (Fig. 1A). Enzymatic deglycosylation by N-glycosidases F and H led to a reduction in mass of 15% in the 76-, 88-, and 98-kDa SIPC subunits confirming that the SIPC contains N-glycans and indicating an approximate combined molecular mass for the three SIPC subunits of ≈200 kDa. The amino acid residues 962-PVNFLGLGEVNITV-975 of the SIPC comprise a putative SRP54-type GTP-binding site (Fig. 5). Fig. 1B shows the alignment of B. amphitrite SIPC to partial putative sequences of the thioester region of A2M from B. amphitrite and two other related balanomorph taxa.
Table 1.
Amino acid sequences of selected fragments
| Sequence | Subunit, kDa | Fragment | Similarity |
|---|---|---|---|
| vKVPEsGYLFTApKvlqa | 88 | N-terminal | A2M family |
| vKVPEsGYLFtAPKvlqaet | 98 | N-terminal | A2M family |
| k/EVsQAYsFInLRQsdnaqk | 88 and 98 | Internal | — |
| k/KPTPDDPVYsthk | 88 and 98 | Internal | — |
| k/FLKPKLPFYGEYTLsMrDGk | 88 and 98 | Internal | A2M family |
| k/TSITTFKPFFTEVSLPYsmk | 76 | Internal | A2M family |
| k/SPTGAGEPNMITLVPNIYIRRYLETTGQLNERQ | 76 | Internal | A2M family |
| k/VHDFYRPEErNIQEYELt | 76 | Internal | — |
Upper and lower case letters indicate residues identified unambiguously and weakly, respectively. The symbol k/indicates sequences that are preceded by lysine based on the strict specificity of Achromobacter protease I.
Fig. 1.
Predicted structure of B. amphitrite SIPC and similarity to the thioester region of Limulus sp. and other barnacles. (A) Predicted structure of the SIPC showing the putative disulfide bridge pattern deduced from the conservation of specific cysteine residues between SIPC and A2M. Thick lines indicate disulfide bridges that are conserved in the Limulus and human sequences. Light lines denote the disulfide bridge showing similarity only to Limulus. The 17-aa signal peptide is denoted by the black box. The potential N-linked glycosylation sites (Asn-Xxx-Ser/Thr and Asn-Xxx-Cys) are marked by black diamonds. (B) Amino acid sequence alignment of the thioester-like region from SIPC and A2M. Black and gray shading denotes identical and functionally similar amino acids, respectively. The horizontal lines indicate the amino acid sequences to which the forward and reverse PCR primers were designed. GenBank accession numbers are as follows: Limulus sp., T18544; B. amphitrite, AY423545 and AY781062; B. improvisus, AY781061; and M. rosa, AY781063.
Expression of SIPC.
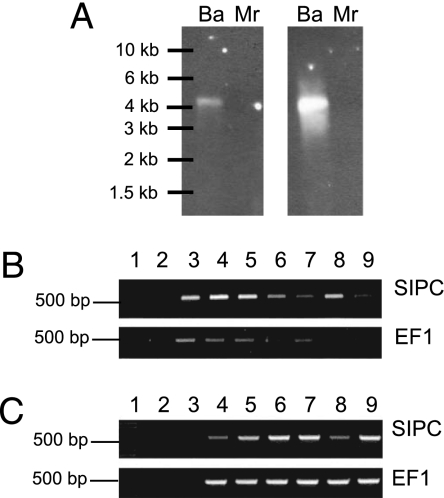
Northern blot analysis indicated a single SIPC transcript of ≈5.2 kb (Fig. 2A), in agreement with that predicted from the B. amphitrite SIPC cDNA we obtained by RT-PCR. The SIPC mRNA was present in all of the adult tissues that we examined (Fig. 2B), with the highest level of expression [normalized to elongation factor 1α (EF1) expression] in cirri and very low expression of the SIPC in the hypodermis. The SIPC transcript was also expressed in all larval developmental stages (Fig. 2C).
Fig. 2.
Expression of the SIPC transcript. (A) Northern blot. Total RNA extracted from the prosomas of B. amphitrite (Ba) and M. rosa (Mr) was hybridized with two probes corresponding to the N-terminal (Left) and the C-terminal (Right) regions of the protein. The RNA ladder is indicated on the left. (B) Expression of B. amphitrite SIPC and EF1 mRNA in different tissues. Lane 1, molecular weight marker (1 kb plus DNA ladder; Invitrogen); lane 2, epidermis; lane 3, egg mass; lane 4, ovary; lane 5, penis; lane 6, gut; lane 7, muscle; lane 8, cirri; lane 9, seminal vesicles. For the no-template control see lane 3 in C. (C) Expression of B. amphitrite SIPC and EF1 mRNA in different development stages. Lane 1, molecular weight marker (1 kb plus DNA ladder; Invitrogen); lane 2, blank; lane 3, no-template control; lane 4, nauplius stage 1; lane 5, nauplius stage 2; lane 6, cyprid (immediately after metamorphosis from the stage 6 nauplius); lane 7, cyprid 2 days after metamorphosis; lane 8, juvenile; lane 9, adult.
Phylogenetic Analysis.
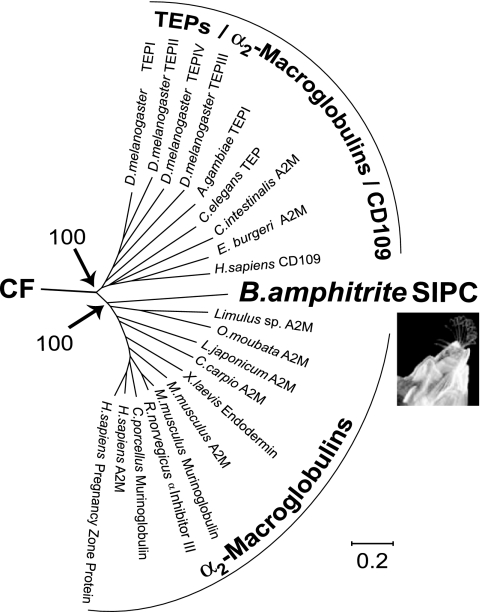
Two major clusters are defined in an unrooted phylogenetic tree (Fig. 3 and Fig. 6, which is published as supporting information on the PNAS web site), which is based on the full-length sequence alignment of members of the TEPs, the A2Ms, the complement factors, and the SIPC of B. amphitrite. One cluster comprises the deuterostome complement system (C3 and C4-C5) sequences exclusively, and the second cluster comprises both the protostome and deuterostome sequences of the A2Ms, the insect TEPs, and the SIPC of B. amphitrite.
Fig. 3.
Unrooted phylogenetic tree showing the relationship of B. amphitrite SIPC to the TEP. CF indicates the branch leading to the complement factor proteins. The full tree comprising A2M, SIPC, complement factor, and other TEPs is in Fig. 6. Numbers by major nodes represent bootstrap values for 100 replicates. The scale bar indicates the length of each branch.
Settlement-Inducing Activity of SIPC and Human A2M (hA2M).
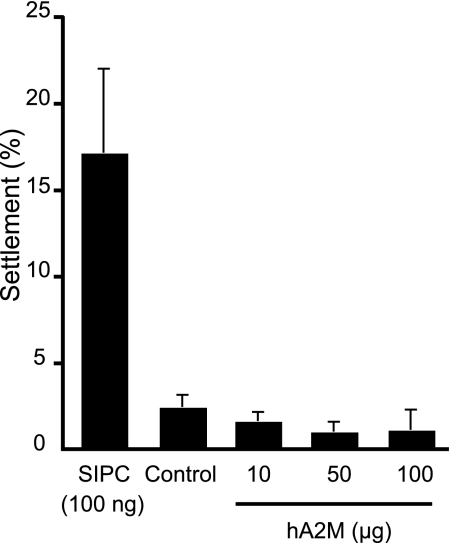
Western blotting using the SIPC-N antibody raised against a peptide of the cloned SIPC confirmed the presence of the SIPC in the purified SIPC protein fraction (data not shown). The amount of protein in the final fraction was 40 mg, which represents ≈43.5 μg per barnacle. A one-way ANOVA detected highly significant differences in the data (F = 0.28, d.f. = 4, P < 0.001). Post hoc comparisons of all pairs of group means revealed that settlement on the SIPC treatment was significantly higher (P < 0.05) than the hA2M treatments and the buffer control (Fig. 4). No other comparisons were significantly different (P > 0.05); i.e., hA2M was not active in the settlement assay.
Fig. 4.
Settlement choice assay of B. amphitrite cyprids with B. amphitrite SIPC and hA2M. B. amphitrite cyprid settlement was significantly higher (P < 0.05) on the B. amphitrite SIPC (applied at 100 ng/0.8 cm2) compared with the control or hA2M (applied at 10, 50, or 100 μg/0.8 cm2), which were not significantly different from each other (P > 0.05).
Discussion
Specific surface-bound and waterborne cues have been implicated in the gregarious settlement behavior of a number of benthic marine invertebrates (4, 9, 10, 13). The key molecule responsible for gregarious settlement in B. amphitrite is the SIPC (13, 15). This study has shown that the B. amphitrite SIPC is a previously undescribed protein encoded by a single transcript of ≈5.2 kb. Because all of the peptide sequences of the three SIPC subunits observed on SDS/PAGE occurred in the translated SIPC cDNA sequence that we obtained (Table 1 and Fig. 5), we are confident that the correct cDNA was isolated. Furthermore, by using an antibody raised against a predicted peptide of the cloned SIPC cDNA we were able to confirm that the extracted native SIPC protein induces settlement of B. amphitrite cyprids.
The concentration of adsorbed SIPC that induced settlement (100 ng per 0.8 cm2) is 100-fold lower than that reported previously for crude barnacle extract (14). If we assume that the surface area of a barnacle equates roughly to a cylinder without its base, it is possible to calculate the effective concentration. For an average-sized adult of 1 cm basal diameter and 1 cm height, and using our estimate of 43.5 μg of SIPC per barnacle, the surface concentration (assuming 100% is expressed at the surface) would be 11 μg/cm2, approximately two orders of magnitude higher than the effective concentration assayed here. Thus, the isolated SIPC is likely to be active in laboratory assays at ecologically realistic concentrations, lending further support to our contention that the SIPC is the authentic settlement cue.
The predicted molecular mass of the three SIPC subunits, including sugar chains, is ≈200 kDa, which agrees with previous estimates obtained by Western blotting (24). Gel filtration, however, indicates a molecular mass for the native protein of 200–400 kDa, so SIPC may be a dimer (15). Phylogenetic analysis of the translated SIPC cDNA revealed it to be a previously undescribed glycoprotein belonging to the A2M subgroup of TEPs; its phylogenetic placement within the A2M subgroup is supported by the absence of the anaphylatoxin-like domain specific to complement proteins (23). In what may be a remarkable parallel, a 70-kDa contact protein bearing some sequence similarity to A2M has been implicated in mate recognition in the copepod Tigriopus japonicus (25). However, our isolation of putative A2M sequences from B. amphitrite and other barnacles suggests that the SIPC is not barnacle A2M (Fig. 1B). We have also shown that hA2M is inactive in the barnacle settlement assay. Nevertheless, the SIPC shares motifs and shows high homology with regions that are structurally and functionally important to the TEP family (Fig. 5). These include the QTD motif (142–144), the FXVXXYVLPXFE region (228–239), and the STQDT region (1334–1338) (20).
Perhaps the most interesting structural finding, however, is that the B. amphitrite SIPC does not contain the unique β-cysteinyl γ-glutamyl thioester signature sequence (GCGEQ) that defines most, but not all, described proteins of the TEP family (21, 22, 26, 27). In the SIPC, the nonpolar amino acids alanine and proline replace the cysteine and glutamine residues, respectively. TEPs play specific roles in innate immunity (20, 28), and in A2M proteins the internal thioester bond is essential for their native conformation and mechanism of action (29). Further analysis of the SIPC is therefore needed to determine how the absence of the GCGEQ motif affects this protein. SIPC also lacks the catalytic asparagine (A2M) and histidine (C3 and C4) residues that are involved in the molecular reaction mechanisms of the TEPs.
All classes of A2Ms contain a highly variable bait region (28). Our analyses suggest that the SIPC may contain a bait region between residues 764 and 842, where the flanking amino acid sequences and the position of the cysteine residue (766) are similar. The position of disulfide bridges and glycosylation sites has been determined for horseshoe crab A2M (30). Our alignment of the SIPC to horseshoe crab A2M indicates that 14 of the cysteine residues appear to be conserved (Fig. 1A). Because these proteins appear to share a common ancestor, it is not unreasonable to suggest that they have a similar structure. The putative GTP-binding site present in the SIPC could control its activity during settlement. GTP-binding proteins coordinate interactions during signaling and increase the fidelity of signal recognition (31), and G protein-linked receptors have been implicated in the transduction of settlement cues of some, but not all, benthic marine invertebrates, including barnacles (13).
Although the B. amphitrite SIPC is close to the A2M superfamily, a number of structural characteristics and the absence of the thioester bond suggest that the SIPC is a previously undescribed protein, which might have evolved by duplication from an ancestral barnacle A2M gene. Because we have not yet explored the potential antiprotease activity of the SIPC, we cannot yet fully determine its functional properties. Nevertheless, we found that the SIPC transcript is expressed in all adult tissues examined but comparatively weakly in the epidermis (= hypodermis), gut, muscle, and seminal vesicles. Previous immunological studies in our laboratory revealed that the SIPC was expressed in extracts of both the shell and soft body (prosoma plus ovary and egg mass) (24). Because the shell fraction comprised only epidermis, cuticle, and possible muscle contamination, this result supported the earlier suggestion that the gregarious settlement cue is a cuticular glycoprotein (6, 32). The immunological detection of the SIPC in shell and soft tissues together with the present expression results suggest, however, that the SIPC is more widely expressed in body tissues, in accord with results of biological assays (32). It is reasonable to expect, however, that the cyprid is only able to make contact with the SIPC during surface exploration on the outer surface of the shell, perhaps associated with the cuticular covering (33). All developmental stages, including the cyprid, expressed the SIPC. The latter result was not surprising because extracts of cyprids induce settlement of conspecifics (32). Evidence has also been presented that the cyprid temporary adhesive deposited as footprints on nitrocellulose membrane, and which has a secondary function as a settlement pheromone (13), is related immunologically to SIPC (24, 34). Interestingly, the protein (24) and mRNA expression (this study) results are not in complete accord because SIPC was not evident in immunoblots of nauplii that had just hatched. The SIPC mRNA is evidently expressed in stage I nauplii, however, so this apparent contradiction may simply reflect the greater sensitivity of our methods.
Proteins do not feature prominently in the literature on cues to marine invertebrate larval settlement (4, 9, 10). However, in common with barnacles, surface-associated proteins have been implicated in the gregarious settlement of the tubeworm Phragmatopoma lapidosa californica (35) and the oyster Crassostrea virginica (36), although it is far from clear that these are the authentic cues (4, 9, 10). Interestingly, a water-soluble peptide cue is also associated with induction of settlement of both these species. Whether there is a relation between the surface-associated and waterborne cues is unknown, but it has been suggested that the waterborne cue of B. amphitrite, which may also be a peptide (13), is derived from the SIPC (37). The molecular identification of the SIPC of B. amphitrite reported here thus represents a major advance in our understanding of barnacle gregariousness. Information on the carbohydrate component of this glycoprotein is sparse but will be essential to realize given that sugar chains are considered important to the activity of the SIPC (14).
Barnacles respond at settlement to allospecific barnacles (6, 13, 16, 17, 32), and, moreover, the magnitude of the settlement induction corresponds to their systematic affinity (38). Continued study of the SIPC of B. amphitrite and the sequences of other barnacle species will advance understanding of the evolution of these organisms and their gregarious behavior. A more complete understanding of barnacle settlement may suggest new ways to control barnacle fouling.
Materials and Methods
Animals.
B. amphitrite and Megabalanus rosa were collected at Lake Hamana and at Uwajima (Japan), respectively. Balanus improvisus were collected from Tjarnö, Sweden. B. amphitrite larvae were cultured in the laboratory from adult broodstock (39) in seawater collected at Tynemouth in North East England.
Purification and Amino Acid Sequencing of the SIPC.
SIPC from B. amphitrite was purified according to Matsumura et al. (15). Internal peptide sequences of three putative SIPC subunits (76, 88, and 98 kDa) were determined after digestion by Achromobacter protease I (API/LYS-C) in the presence of 0.1% SDS. The proteolytic fragments were separated by HPLC by using tandem columns (TSK gel DEAE5PW and Mightysil RP-18) in a linear gradient of 0.075% trifluoroacetic acid/80% CH3CN (0–60%) in 0.085% trifluoroacetic acid. All amino acid sequences were determined by automated Edman degradation.
RNA Extraction.
The prosomas of adult barnacles were ground under liquid nitrogen, and the total RNA was extracted by using TRIzol (Invitrogen, Paisley, United Kingdom). The concentration was determined spectrophotometrically, and mRNA was purified from the total RNA pool by using Oligotex (Qiagen, Valencia, CA). The mRNA was obtained from B. amphitrite larvae by using an RNeasy mini kit (Qiagen).
Amplification and Cloning of B. amphitrite SIPC cDNA.
A cDNA template for PCR was synthesized by reverse transcription of B. amphitrite mRNA using Thermoscript (Invitrogen) and an oligo(dT)17 primer. A partial SIPC cDNA was then amplified by nested PCR using degenerate oligonucleotide primers designed from peptide fragments obtained from the 76-kDa subunit (Table 1). This partial cDNA sequence was then used to obtain the 5′ and 3′ UTRs by RACE methods. The full-length SIPC ORF was then amplified by PCR using a proofreading polymerase (Pfu; Stratagene, Amsterdam, The Netherlands) and primers located in the 5′ and 3′ UTRs. The full-length SIPC ORF was cloned, and DNA sequences were determined on both strands by automated sequencing.
Settlement Assay of the SIPC and hA2M.
To confirm that the SIPC is involved in barnacle settlement, we partially purified SIPC from adult B. amphitrite according to the method described by Matsumura et al. (15), but omitting the affinity chromatography step, which we have since found to be unnecessary. The fractions were then examined by Western blotting to detect SIPC by using a different antibody (SIPC-N) raised against a predicted epitope near the N terminus of the protein, this time determined from the putative SIPC amino acid sequence (amino acid positions 424–437) (Fig. 5) (40). We then performed a larval choice settlement assay to confirm that we had isolated biologically active SIPC, following a method slightly modified from that of Matsumura et al. (14) with hA2M (Sigma–Aldrich, St. Louis, MO) as a protein control. Briefly, the purified SIPC at a concentration of 100 ng per 0.8 cm2; hA2M at concentrations of 10, 50, and 100 μg per 0.8 cm2; and a negative control (TBS buffer only) were adsorbed onto a nitrocellulose membrane by aspirating with a 24-well dot blot vacuum manifold. Each treatment and the control were applied randomly to one of four wells of the manifold. After adsorption, the membranes were fixed to the inside base of new polypropylene containers by using double-sided carbon adhesive tape (Agar Scientific, Stansted, United Kingdom); each of three replicate containers contained one membrane. A total of 300 B. amphitrite cyprids in 300 ml of artificial sea water (Tropic Marin, Biener GmbH, Wartenberg-Angersbach, Germany) was then added to each container. By using a binocular microscope, the number of settled larvae on the protein and control spots was determined after 48 h of incubation at 28°C in the dark. The percentage of cyprids that had settled on each treatment and the controls, expressed as the mean of the three replicate membranes, was then calculated. For statistical analyses (41), percentage values were arcsine-transformed. Levene's test did not detect unequal variances (F = 0.713, P = 0.601), and a plot of sample means vs. variances showed no relationship (r2 = 0.086, P = 0.633). The data were thus analyzed by one-way ANOVA, and unplanned pairwise comparisons of means were done by using Tukey's test.
Amplification and Cloning of A2M from B. amphitrite and Related Barnacles.
To confirm that the SIPC gene is distinct from that of A2M we amplified a partial fragment of A2M from B. amphitrite, B. improvisus, and M. rosa by RT-PCR using a proofreading polymerase (Pfu; Stratagene) and degenerate amplification primers designed from the highly conserved thioester region of the A2M protein (Table 2). PCR products were cloned and sequenced on both strands.
Table 2.
Primer sequences
| Primer gene name | Gene position | Oligonucleotide sequence |
|---|---|---|
| A2M | 5′ | 5′-CCNSANACNTGGHTNTGG-3′ |
| 5′ nested | 5′-GGITGYGGIGARCARAAYATG-3′ | |
| 3′ | 5′-AYNACNGTRTCYTGNGT-3′ | |
| 3′ nested | 5′-ACRSANGCNGTNAGCCAIGT-3′ | |
| EF1 | 5′ | 5′-CTGGGTGTGAAGCAGATGAT-3′ |
| 3′ | 5′-GCAGAGATTCGTGGTGCAT-3′ | |
| SIPC | 5′ | 5′-CTACACCAAGCGATTTACGGT-3′ |
| 3′ | 5′-TGACGATTTCATTCTTCGCA-3′ |
Semiquantitative PCR.
Semiquantitative RT-PCR was used to determine expression of the SIPC in different tissues and developmental stages of B. amphitrite. We used the gene coding for EF1 in B. amphitrite as a positive control. DNase 1-treated mRNAs prepared from whole adults, separate adult tissues (epidermis, egg mass, ovary, penis, gut, muscle, cirri, and seminal vesicles), juveniles, nauplius stages 1 and 2 and day 0 (day of metamorphosis), and day 2 cyprids were reverse-transcribed by using SuperScript III RNase H reverse transcriptase (Invitrogen) and incubated finally with RNase H for 20 min at 37°C (Invitrogen). Fragments of EF1 (472 bp) and the SIPC (524 bp) were then amplified by PCR using 10 pmol of each gene-specific primer (Table 2) in a standard reaction containing recombinant TaqDNA polymerase (Invitrogen), 1.5 mM MgCl2, 200 μM dNTPs, 20 mM Tris·HCl, and 50 mM KCl (pH 8.4). The PCRs involved either 25 (EF1) or 30 (SIPC) amplification cycles of denaturation at 94°C for 20 s, primer annealing at 55°C for 30 s, and strand extension at 72°C for 30 s.
Northern Blot Analysis.
We performed a Northern blot to determine the size of the B. amphitrite SIPC transcript using 25 μg of total RNA extracted from B. amphitrite and M. rosa (control) per lane. Two different probes were used corresponding to the 5′ or 3′ ends of the B. amphitrite SIPC. DNA probes were biotin-labeled by nick translation using the BioNick labeling system (Invitrogen).
Computer Search and Sequence Analysis.
DNA sequence analysis was performed by using DNAsis. The protein analyses were carried out by using web servers, which included the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) for general protein comparison, ExPAsy (http://ca.expasy.org), Infobiogen (www.infobiogen.fr) for prediction of signal peptide cleavage sites, and European Molecular Biology Laboratory–European Bioinformatics Institute (www.ebi.ac.uk) for general protein sequence/pattern searches. The predicted amino acid sequence of the B. amphitrite SIPC was then aligned to sequences of the TEP family by using Clustal-W (42). Phylogenetic trees were constructed by neighbor-joining on GZ-Gamma corrected amino acid distances (43) using version 2 of the MEGA computer program (44).
Supplementary Material
Acknowledgments
We are grateful to Dr. K. Berntsson (Göteborg University, Göteborg, Sweden) for providing B. improvisus. This work was supported by Natural Environment Research Council Grants GST/02/1436 (to A.S.C.) and NER/A/S/2001/00532 (to A.S.C. and R.R.K.) and a Ray Lankester Investigatorship from the Marine Biological Association of the United Kingdom (to K.M.) for work in R.R.K.'s molecular laboratory. R.R.K. is a Royal Society University Research Fellow in plankton molecular ecology.
Abbreviations
- A2M
α2-macroglobulin
- hA2M
human A2M
- EF1
elongation factor 1α
- SIPC
settlement-inducing protein complex
- TEP
thioester-containing protein.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Darwin C. A Monograph of the Sub-class Cirripedia, with Figures of all the Species. The Balanidae (or Sessile Cirripedes); the Verrucidae. London: Ray Society; 1854. [Google Scholar]
- 2.Leslie HM, Breck EN, Chan F, Lubchenco J, Menge BA. Proc Natl Acad Sci USA. 2005;102:10534–10539. doi: 10.1073/pnas.0503874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthiessen P, Gibbs PE. Environ Toxicol Chem. 1998;17:37–43. [Google Scholar]
- 4.Steinberg PD, de Nys R, Kjelleberg S. In: Marine Chemical Ecology. McClintock JB, Baker JB, editors. Boca Raton, FL: CRC; 2001. pp. 355–387. [Google Scholar]
- 5.Knight-Jones EW, Crisp DJ. Nature. 1953;171:1109–1110. doi: 10.1038/1711109a0. [DOI] [PubMed] [Google Scholar]
- 6.Crisp DJ, Meadows PS. Proc R Soc London Ser B; 1962. pp. 500–520. [Google Scholar]
- 7.Larman VN, Gabbott PA. Comp Biochem Physiol B. 1982;72:329–338. [Google Scholar]
- 8.Fraenkel G, Rudall KM. Proc R Soc London Ser B; 1947. pp. 111–144. [DOI] [PubMed] [Google Scholar]
- 9.Pawlik JR. Oceanogr Mar Biol Annu Rev. 1992;30:273–335. [Google Scholar]
- 10.Hadfield MG, Paul VJ. In: Marine Chemical Ecology. McClintock JB, Baker JB, editors. Boca Raton, FL: CRC; 2001. pp. 431–461. [Google Scholar]
- 11.Swanson RL, Williamson JE, de Nys R, Kumar N, Bucknall MP, Steinberg PD. Biol Bull. 2004;206:161–172. doi: 10.2307/1543640. [DOI] [PubMed] [Google Scholar]
- 12.Williamson JE, de Nys R, Kumar N, Carson DG, Steinberg PD. Biol Bull. 2000;198:332–345. doi: 10.2307/1542689. [DOI] [PubMed] [Google Scholar]
- 13.Clare AS, Matsumura K. Biofouling. 2000;15:57–71. doi: 10.1080/08927010009386298. [DOI] [PubMed] [Google Scholar]
- 14.Matsumura K, Mori S, Nagano M, Fusetani N. J Exp Zool. 1998;280:213–219. [PubMed] [Google Scholar]
- 15.Matsumura K, Nagano M, Fusetani N. J Exp Zool. 1998;281:12–20. [PubMed] [Google Scholar]
- 16.Gabbott PA, Larman VN. In: Barnacle Biology. Southward AJ, editor. Rotterdam, The Netherlands: Balkema; 1987. pp. 377–388. [Google Scholar]
- 17.Kato-Yoshinaga Y, Nagano M, Mori S, Clare AS, Fusetani N, Matsumura K. Comp Biochem Physiol A. 2000;125:511–516. doi: 10.1016/s1095-6433(00)00179-3. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Solomon KR, Sharma P, Chan M, Morrison PT, Finberg RW. Gene. 2004;327:171–183. doi: 10.1016/j.gene.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 21.Dodds AW. Immunobiology. 2002;205:340–354. doi: 10.1078/0171-2985-00137. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong PB, Quigley JP. Dev Comp Immunol. 1999;23:375–390. doi: 10.1016/s0145-305x(99)00018-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Thangamani S, Ho B, Ding JL. EMBO J. 2005;24:382–394. doi: 10.1038/sj.emboj.7600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumura K, Nagano M, Kato-Yoshinaga Y, Yamazaki M, Clare AS, Fusetani N. Proc R Soc London Ser B; 1998. pp. 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ting JH, Snell TW. Mar Biol. 2003;143:1–8. [Google Scholar]
- 26.Christophides GK, Zbobnov E, Barillus-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Domopoulos G, et al. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 27.Blandin S, Levashina EA. Mol Immunol. 2004;40:903–908. doi: 10.1016/j.molimm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Dodds AW, Laws SKA. Immunol Rev. 1998;166:15–26. doi: 10.1111/j.1600-065x.1998.tb01249.x. [DOI] [PubMed] [Google Scholar]
- 29.Isaac L, Isenman DE. J Biol Chem. 1992;267:10062–10069. [PubMed] [Google Scholar]
- 30.Husted LB, Sorensen EB, Armstrong PB, Quigley JP, Kristensen L, Sottrup-Jensen L. J Biol Chem. 2002;277:43698–43706. doi: 10.1074/jbc.M208236200. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 32.Knight-Jones EW. J Exp Biol. 1953;30:584–598. [Google Scholar]
- 33.Bourget E. Naturaliste Can. 1977;104:281–323. [Google Scholar]
- 34.Dreanno D, Kirby RR, Clare AS. Biol Lett. 2006;2:423–425. doi: 10.1098/rsbl.2006.0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen RA, Morse DE. J Chem Ecol. 1990;16:911–930. doi: 10.1007/BF01016500. [DOI] [PubMed] [Google Scholar]
- 36.Crisp DJ. J Anim Ecol. 1967;36:329–335. [Google Scholar]
- 37.Harrison P. PhD thesis. Sydney, Australia: Univ of New South Wales; 1998. [Google Scholar]
- 38.Knight-Jones EW. Nature. 1955;175:266. doi: 10.1038/175941a0. [DOI] [PubMed] [Google Scholar]
- 39.Clare AS. Biofouling. 1996;10:141–159. doi: 10.1080/08927019609386276. [DOI] [PubMed] [Google Scholar]
- 40.Dreanno C, Kirby RR, Clare AS. Proc R Soc London Ser B; 2006. [DOI] [Google Scholar]
- 41.Quinn GP, Keough MJ. Experimental Design and Data Analysis for Biologists. Cambridge, UK: Cambridge Univ Press; 2002. p. 537. [Google Scholar]
- 42.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu X, Zhang J. Mol Biol Evol. 1997;14:1106–1113. doi: 10.1093/oxfordjournals.molbev.a025720. [DOI] [PubMed] [Google Scholar]
- 44.Kumar S, Tamura K, Jakobsen IB, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.