Abstract
Mushroom bodies (MBs) are the centers for olfactory associative learning and elementary cognitive functions in the Drosophila brain. As a way to systematically elucidate genes preferentially expressed in MBs, we have analyzed genome-wide alterations in transcript profiles associated with MB ablation by hydroxyurea. We selected 100 genes based on microarray data and examined their expression patterns in the brain by in situ hybridization. Seventy genes were found to be expressed in the posterodorsal cortex, which harbors the MB cell bodies. These genes encode proteins of diverse functions, including transcription, signaling, cell adhesion, channels, and transporters. Moreover, we have examined developmental functions of 40 of the microarray-identified genes by transgenic RNA interference; 8 genes were found to cause mild-to-strong MB defects when suppressed with a MB-Gal4 driver. These results provide important information not only on the repertoire of genes that control MB development but also on the repertoire of neural factors that may have important physiological functions in MB plasticity.
Keywords: hydroxyurea, RNAi, brain development, learning and memory
Mushroom bodies (MBs) are the centers for higher-order functions in the Drosophila brain, participating in diverse behaviors such as olfactory associative learning and elementary cognition (1). In the adult brain, MBs consist of ≈2,500 neurons per brain hemisphere. The cell bodies of their intrinsic neurons [Kenyon cells (KCs)] are located in the posterodorsal cortex above a prominent dendritic structure called the calyx, which receives olfactory information from the antennal lobes (ALs) via the inner antennocerebral tract. KCs extend axons through the peduncles, which split dorsally into two lobes, α and α′, and medially into three lobes, β, β′, and γ (Fig. 1A). During development, the Drosophila MB neurons originate from four neuroblasts per brain hemisphere (2–5), with each neuroblast giving rise to an indistinguishable set of neurons and glia (6). The adult Drosophila MB comprises three types of axonal projection groups, γ, α′/β′, and α/β (7, 8), which are sequentially generated by the division of the MB neuroblasts on the basis of birth order (9). Notably, this structural subdivision is proposed to reflect functional distinctions (10–12). Moreover, consistent with their behavioral functions, many of the genes required for learning and memory are preferentially expressed in MBs (13–15).
Fig. 1.
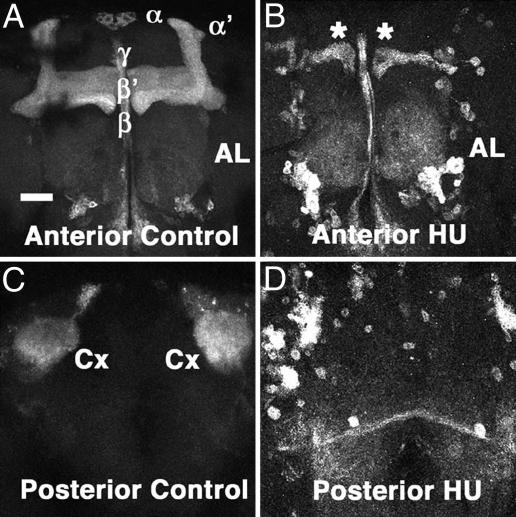
MB structures in control and HU-treated brains. (A and C) Nonablated control brain. (B and D) HU-treated brain. Reconstruction from anterior (A and B) and posterior (C and D) optical sections. MBs and ALs are visualized with UAS-mCD8::GFP driven by OK107-Gal4. The two dorsal lobes (α and α′) and the three medial lobes (β, β′, and γ) are indicated in A. Cx, MB calyx. Asterisks in B indicate residual MB lobes of embryonic origin. Note that MBs were almost completely abolished in the HU-treated brain, with only a small number of residual neurons. (Scale bar, 50 μm.)
Intriguingly, studies have revealed that the Drosophila Pax-6 homologs, eyeless (ey) and twin of eyeless, which were originally implicated as master control genes in eye development (16), are expressed in MBs and have important functions in structural differentiation of MB neurons (3, 4, 17). In particular, ey is expressed in MB neuroblasts and their progeny through development, and mutations in ey disrupt MB neuropil structures. Furthermore, molecular studies have shown that, unlike the feedback mechanism in the eye, ey and toy are independently controlled in MBs, with ey being controlled by multiple brain-specific enhancer modules that are distinct from the eye-specific enhancer (18).
In this article, we present a genome-wide survey of the Drosophila transcripts preferentially expressed in MBs. By comparing the transcriptional profiles of MB-ablated and nonablated brains, we first identified 100 genes that exhibited down-regulation as a result of MB ablation. The expression patterns of the identified genes were examined by in situ hybridization to the adult brain, to yield 70 genes that exhibited preferential expression in the posterodorsal cortex, which harbors the MB cell bodies. Furthermore, we have examined developmental functions of 40 of the identified genes with transgenic RNAi and found 8 genes whose suppression causes mild-to-strong MB defects in the adult brain. These results provide systematic information on the transcriptional profile of MBs and deepen our insights on the genetic mechanisms controlling the development and plasticity of this seminal structure in the fly brain.
Results
Chemical Ablation of MBs with Hydroxyurea (HU).
To identify genes preferentially expressed in MBs, we analyzed genome-wide alteration of the gene-expression profiles between MB-ablated and nonablated brains using the Drosophila GeneChip (DrosGenome 1). To ablate MB neurons, we used a pharmacological technique with HU, an inhibitor of ribonucleotide reductase that blocks DNA synthesis and kills dividing cells (19). The specificity of ablation depends on the unique proliferation pattern of the MB neuroblasts (20–22). For the first 8–12 h after larval hatching, only five neuroblasts are active in each brain hemisphere. Four of them are located in the dorsal protocerebrum and correspond to the MB neuroblasts. The other single neuroblast located anterolaterally is a progenitor of a subset of the AL neurons (23). HU fed to newly hatched larvae is preferentially incorporated into these five neuroblasts and, thus, ablates the four MB neuroblasts, effectively deleting most MB neurons, with the exception of a small number of neurons born during the course of embryonic development (Fig. 1). Otherwise, the HU-mediated ablation was reproducible at the HU concentration used (25 mg/ml), because all of the brains examined (n = 20) lacked most MB structures. Although the lateral neuroblast was also ablated with HU as described (23), we noticed little reduction or abnormality of the ALs, consistent with the previous observation that HU ablation is less effective for the ALs (19).
Identification of MB Genes by Comparative Microarray Analysis.
To have an overview on the number of the expressed genes in the Drosophila brain, we first determined the transcriptional profile of the wild-type (Canton S) brain using the DrosGenome1 GeneChip, which represents virtually the entire Drosophila genome, with >13,500 transcripts; 9,637 transcripts were identified above a threshold (signal intensity >0.5, after normalization by the MAS 5.0 software). This corresponds to ≈70% of the total Drosophila genes and agrees with the previous genetic screen by enhancer detection, in which >80% of lines showed reporter gene expression in the adult brain (24).
We then compared gene-expression profiles between the control and MB-ablated brains; 1,465 transcripts were identified above a fold-change criterion (Control/HU >1.5). Statistical filtering of the data (P < 0.05 by Welch t test for Control/HU) then identified 103 transcript probes on the microarray, which correspond to 100 genes (excluding two existence-uncertain genes and one duplication) (see Table 2, which is published as supporting information on the PNAS web site). It should be noted that the set of transcripts that can be identified by the microarray data varies with different Control/HU criteria, with the majority belonging to the fold-change range 1.0–2.0 (P < 0.05) (Fig. 5, which is published as supporting information on the PNAS web site). Importantly, by the filtering criteria used in this work (Control/HU >1.5, P < 0.05), we recovered ey (Control/HU = 1.84, P = 0.0006) and ultraspiracle (usp) (Control/HU = 1.55, P = 0.0185), the latter of which had been identified in a genetic mosaic screen as a regulator of MB remodeling during metamorphosis (25). We also recovered transformer 2 (tra2) (Control/HU = 1.57, P = 0.0264), which is expressed in female MBs to control female-specific behaviors (26), and trio (Control/HU = 2.66, P = 0.018), which encodes a Rho guanyl-nucleotide exchange factor required for axodendritic extension of MB neurons (27). We consequently decided to focus on the 100 genes identified by these criteria for further analyses.
Expression Patterns of MB Genes in the Brain.
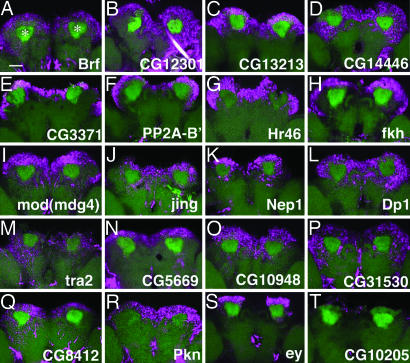
To confirm the microarray results and reveal expression patterns in the brain, we have performed in situ hybridization of the 100 genes and ranked them in four groups based on their signal intensities (Table 3, which is published as supporting information on the PNAS web site); 70 genes were found in rank A–C (25 in A, 26 in B, and 19 in C), which exhibited strong-to-weak expression in MBs (Fig. 2). Intriguingly, most of the expression patterns are broader than that of ey, even though they are restricted to the posterodorsal brain cortex, indicating that they are expressed in MBs and nearby neurons. Some of the genes are also expressed in parts of the anterior brain (Fig. 6, which is published as supporting information on the PNAS web site) and/or optic lobes (OLs) (data not shown), although, for most genes, signals in these regions are weaker than that in the posterodorsal cortex. Notably, we failed to find genes that are expressed in other brain structures but not in MBs. Moreover, consistent with the selection criteria for the microarrays (signal level in the control brain >0.5), we confirmed brain expression by RT-PCR, even for genes with background levels (rank D) (data not shown), although we do not know exactly which brain structures express them.
Fig. 2.
In situ hybridization of MB genes in the adult brain. (A–T) Expression patterns of MB genes (magenta) in the adult brain as revealed by fluorescent in situ hybridization. Reconstruction of confocal images of the posterior part of the brain. MBs are visualized with UAS-mCD8::GFP driven by OK107-Gal4 (green). Note that the specific GFP signals were compromised during hybridization, and other neuropil structures are also visible by nonspecific green fluorescence. Asterisks in A indicate the MB calyces. (Scale bar, 50 μm.) CG10205 is included as a negative control (belonging to rank-D in Table 3).
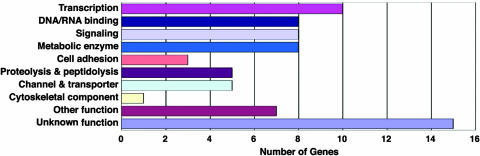
Annotations in FlyBase (28) revealed that proteins encoded by the 70 MB genes (rank A–C) belong to diverse functional categories. Among them, proteins controlling gene expression represent the largest group (Fig. 3): 10 transcription factors and eight DNA/RNA binding proteins are found. In addition, eight signaling, three cell-adhesion, and five proteolysis–peptidolysis proteins as well as five channels/transporters were also found. On the other hand, 15 genes encode proteins whose functions are unknown.
Fig. 3.
Functional classification of the MB genes. Molecular functions of the 70 MB genes detected by in situ hybridization. The classification is based on gene ontology data in FlyBase in conjunction with a homolog search using the BLAST data in FLIGHT and functional protein domains in InterPro.
Structural MB Defects Caused by Developmental RNAi.
To systematically reveal functional aspects of the identified genes, we attempted to down-regulate gene functions with RNAi using a large-scale collection of Drosophila upstream activating sequence (UAS)-RNAi stocks, which targets >6,000 genes of the Drosophila genome (K.T., K.S. and R.U., unpublished data). RNAi lines were available for 40 of the 100 genes identified with microarrays, involving 7 genes whose expression was undetectable by in situ hybridization (rank D). To suppress gene function, we induced UAS-RNAi expression by the OK107-Gal4 line (29), which has a P-element insertion at the ey locus and drives robust Gal4 expression in MBs throughout development (18). We examined the structural integrity of MBs, ALs, and OLs using mCD8::GFP, a membrane-bound reporter, and categorized the observed structural defects according to their severity (Table 4, which is published as supporting information on the PNAS web site).
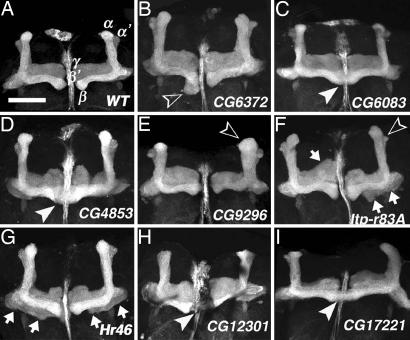
Whereas most of the RNAi lines resulted in weak or no MB abnormalities (Table 4), 8 of the 40 genes exhibited mild-to-strong structural defects with penetrances (≥24%) markedly above those of the control crossings (Table 1; also see Table 4 for specific defects). These lines caused diverse morphological defects, including malformation of MB lobes, fusion of medial lobes, and aberrant peduncles and calyces (Table 4 and Fig. 4). In particular, Hr46 (Hormone receptor-like in 46) RNAi caused MB defects in 30% of brains (Fig. 4G). Hr46 encodes an ecdysone-inducible orphan nuclear receptor required for prepupal–pupal transition and differentiation of adult cuticle structures (30). CG6083 RNAi caused MB defects in 35% of brains, many of which exhibited strong β-lobe fusion (Fig. 4C). Notably, CG6083 brain expression was detected by RT-PCR but not by in situ hybridization (data not shown). RNAi of three genes encoding cell-signaling molecules also caused a variety of MB defects: CG4853 (guanyl-nucleotide exchange factor for Ras-like GTPases) (Fig. 4D), CG9296 (cGMP-specific phosphodiesterase) (Fig. 4E), and Itp-r83A (Inositol 1,4,5,-Tris-phosphate receptor) (Fig. 4F). Intriguingly, CG9296 RNAi also caused OL defects (75% of brains) and compound eye degeneration (66% of eyes). Except that Itp-r83A is expressed in honey bee MBs (31, 32), little is known about the functional significance of these signaling molecules in MBs.
Table 1.
Summary of the structural defects caused by the eight RNAi lines
| Gene | Control/HU (P)* | Expression rank† | RNAi line | Structural defects with OK107‡ |
Structural defect with CS§ |
Gene ontology/human homolog | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MBs, % (n) | Als, % (n) | OLs, % (n) | No. of brains | MBs, % (n) | ALs, % (n) | OLs, % (n) | No. of brains | |||||
| CG9296 | 1.771 (0.008) | A | 9296R-3 | 25 (5) | 0 (0) | 75 (15) | 20 | 7 (1) | 0 (0) | 0 (0) | 15 | Signaling: cGMP-specifc phosphodiesterase/PDE6D |
| CG6083 | 1.686 (0.045) | D | 6083R-1 | 35 (7) | 0 (0) | 0 (0) | 20 | 10 (2) | 0 (0) | 0 (0) | 20 | Metabolic enzyme: aldehyde reductase/AKR1B1 |
| CG4853 | 1.664 (0.020) | C | 4853R-1 | 24 (4) | 6 (1) | 0 (0) | 17 | 5 (1) | 0 (0) | 0 (0) | 20 | Signaling: Ras gualyl-nucleotide exchange factor/RASGEF1C |
| CG6372 | 2.026 (0.018) | C | 6372R-2 | 43 (6) | 0 (0) | 0 (0) | 14 | 13 (2) | 0 (0) | 0 (0) | 15 | Proteolysis and peptidolysis: leucyl aminopeptidase/LAP3 |
| Itp-r83A | 1.699 (0.028) | B | 1063R-2 | 24 (5) | 5 (1) | 0 (0) | 21 | 13 (2) | 0 (0) | 0 (0) | 16 | Signaling: inositol-1,4,5-triphosphate receptor/ITPR1 |
| Hr46 | 3.489 (0.022) | A | 11823R-4 | 30 (19) | 0 (0) | 0 (0) | 64 | 0 (0) | 0 (0) | 0 (0) | 20 | Transcription: ligand-dependent nuclear receptor/RORB |
| CG17221 | 1.786 (0.010) | B | 17221R-1 | 30 (6) | 0 (0) | 0 (0) | 20 | 10 (2) | 0 (0) | 0 (0) | 21 | Metabolic enzyme: zinc-containing alchohol dehydrogenase/RTN4IP1 |
| CG12301 | 2.095 (0.020) | B | 12301R-2 | 30 (6) | 0 (0) | 0 (0) | 20 | 11 (2) | 0 (0) | 0 (0) | 18 | Other function: Utp14 protein domain/UTP14A |
*Control/HU signal ratio in microarray experiments (see Table 2).
†Expression rank by in situ hybridization (see Table 3).
‡Anatomical brain defects in UAS-RNAi flies crossed with OK107. See Table 4 for specific description of the MB phenotypes. RNAi for CG4853 and Itp-r83A caused weak AL defects in single brains. Only CG9296 RNAi caused OL degeneration.
§Anatomical brain defects in UAS-RNAi flies crossed with Canton S. MB defects were rare in these control flies, and only weak β -lobe fusion was observed. Similarly, only weak β -lobe fusion was observed in 8.3% (5 of 60) of the progeny of OK107 crossed with Canton S. No abnormality was detected for ALs and OLs. Flies were raised at 28°C to stimulate the Gal4 activity. Note that β -lobe fusion is less frequent at 25°C (1 of 60 for OK107/Canton S).
Fig. 4.
Structural MB abnormalities caused by developmental RNAi. (A) Wild type. (B–I) RNAi flies. Expression of UAS-RNAi was induced with OK107, which drives Gal4 in most MB neurons throughout development. MB lobes are visualized with UAS-mCD8::GFP. White arrowheads indicate fused medial lobes (C, D, H, and I). Open arrowheads indicate abnormal β-lobe (B), swollen α-lobe (E), and small α′-lobe (F). Arrows in F and G indicate aberrant γ-lobes. Shown are reconstructions of confocal sections. (Scale bar, 50 μm.)
Using a quantitative PCR technique, we measured mRNA levels of the eight genes in the total brain RNA preparations from the OK107-driven RNAi flies but failed to detect down-regulation. This may, in part, be accounted for by the limited expression domain of this driver, which is only a subset of those of the eight genes. On the other hand, neither OK107 nor UAS-RNAi flies exhibited marked MB defects when each was crossed separately with Canton S (Table 1), indicating that the induced structural defects are, indeed, caused by Gal4-mediated RNAi.
Discussion
In this study, we have performed a genome-wide screen of Drosophila genes that are preferentially expressed in MBs. Although the lateral AL neuroblasts are coablated, specific removal of MB neurons was achieved by the chemical ablation protocol. Moreover, we have confirmed by in situ hybridization that 70 of the 100 identified genes are, indeed, expressed in the posterodorsal cortex, which harbors the MB cell bodies. The validity of our microarray data was also supported by the finding of several regulatory genes, such as ey, usp, tra2, and trio, that are known to be expressed in MBs. Because our experimental design depends on reliable signal comparisons between the nonablated control and MB-ablated samples, both the Control/HU and the Welch t test values are critical for gene identification. Notably, despite the fact that a large number of MB genes are successfully identified by the microarray screening, some of the known MB genes, such as dnc (Control/HU = 1.05, P = 0.742), rut (Control/HU = 1.64, P = 0.162), and toy (Control/HU = 1.37, P = 0.048) were excluded by the selection criteria used in this work (Control/HU >1.5 and P < 0.05). Many of these genes are expressed not only in MBs but also in other brain structures, such as OLs, which constitute a significant part of the brain.
By transgenic RNAi screening, we have identified eight genes whose suppression causes mild-to-strong MB defects in the adult brain. The fact that comparable MB defects were rarely observed in the control flies (UAS-RNAi/Canton S and OK107 flies/Canton S) indicates that the observed MB defects are, indeed, caused by Gal4-mediated suppression of the target-gene functions. Although we failed to detect mRNA suppression by OK107-driven RNAi, five of the lines (CG9296R-3, CG1063(Itp-r83A)R-2, CG11823(Hr46)R-4, CG17221R-1, and CG12301R-2) are lethal when driven by Actin-Gal4, a strong ubiquitous driver, demonstrating effective gene suppression by these RNAi lines in developing flies (K.T. and R.U., unpublished data). On the other hand, it should be noted that our RNAi screening might not be sensitive enough to identify all of the functional genes. Only weak-to-moderate MB abnormalities were induced by RNAi for usp (20% MB defects) and trio (14% MB defects) (see Table 4), despite the fact that both genes are known to control MB development (25, 27). For these genes, the transgenic RNAi technique might be insufficient to suppress the developmental gene functions in MBs. Alternatively, RNAi phenotypes might also be compromised by the genetic background of the fly stocks, which is known to affect anatomical phenotypes of MB mutants, both qualitatively and quantitatively (33). Complementary genetic approaches, such as screenings with systematic deficiency sets and genome-wide P-element collections (28), will be required in conjunction with microarray analysis to further identify the MB genes of potential developmental importance.
Screenings of genes expressed in MBs have been attempted (24, 34). By examining 6,000 enhancer detector lines, Han et al. (24) isolated 106 MB-Gal4 lines, representing 48 genetic loci with 12 lines inserted at the rut locus. More recently, Dubnau et al. (34) conducted a systematic screen for memory genes, using DNA microarrays to identify 42 candidate genes regulated during long-term memory formation. In parallel, by screening 6,681 P-element insertion lines, they also have isolated 60 memory mutants. Surprisingly, only two genes, mod (mdg4) (from microarray screening) and klg (from P-element screening), overlap with the 100 genes identified in this study. On the other hand, genetic screens for abnormal MB structures have been conducted with chemical mutagenesis (35), enhancer-trapping (36, 37), gain-of-function (38), and mosaic techniques (25, 39). Again, only a few genes overlap with our screen. Whereas more genes can be identified by our differential microarray screen with less stringent fold-change criteria (see Fig. 5), the limited overlap of identified genes with other screens emphasizes the importance of diverse approaches to uncover the genetic programs that control the complex development and functions of MBs.
To date, an increasing number of studies have monitored transcript catalogs of specific cell types or anatomical subregions in the brain (reviewed in refs. 40–42). Gene-expression patterns of neural stem cells have been analyzed with DNA microarrays (43, 44). Other studies have applied microarrays to identify genes that are expressed in defined brain structures, such as the hippocampus (45) and the amygdaloid complex (46), although the functional significance of the identified genes is mostly unknown. Given the conservation of the cellular components and neural networks involved in the olfactory systems of vertebrates and fruit flies (3, 13, 47), genome-wide information on the repertoire of the Drosophila MB genes will help us to understand complex brain functions in general, although elucidation of specific neural processes and functions regulated by each of the identified genes awaits further genetic and behavioral analyses.
Materials and Methods
Ablation of MBs.
Canton S was used as the wild-type stock. Flies were raised at 25°C in a 12-h light/12-h dark cycle. Ablation of MBs with HU was done as described (19). Briefly, newly hatched larvae were fed for 4 h with heat-inactivated yeast paste containing HU at 25 mg/ml. This concentration was experimentally determined and found to give reproducible MB ablation (100%, n = 20) without affecting other parts of the brain. Larvae were then rinsed and raised on a standard corn meal food at 25°C in a 12-h light/12-h dark cycle.
RNA Preparation.
Two-day-old female brains were carefully dissected in sterile PBS on ice and immediately frozen with dry ice/ethanol. Dissection was done between 2 and 5 p.m. to minimize circadian fluctuation. Total RNA was then isolated with TRIzol (Invitrogen, Carlsbad, CA) as described (48). The integrity of the extracted RNA was determined by capillary electrophoresis on an RNA6000 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Target Preparation and Microarray Analysis.
For each experiment, 100 ng of total brain RNA was amplified by using a GeneChip Two-Cycle cDNA Synthesis kit (Invitrogen), which involves two cycles of cDNA synthesis and intermediate in vitro transcription by T7 RNA polymerase. After the second round of cDNA synthesis, the resultant double-stranded cDNA was purified and used as a template in the second in vitro transcription reaction, in which biotin-labeled complementary RNA was generated by using the BioArray High Yield RNA Transcript Labeling kit (Enzo, Farmingdale, NY). After purification on RNeasy mini columns (Qiagen, Valencia, CA), 20 μg of the labeled complementary RNA was fragmented and hybridized to GeneChip microarrays (DrosGenome1; Affymetrix, Santa Clara, CA) for 16 h at 45°C. After washes on a Fluidics Station 400 (according to the EukGE-WS2v4 procedure), hybridized signals were scanned on Agilent Technologies GeneArray Scanner G2500A using the MAS5 software (Affymetrix). Data were normalized and analyzed by using GeneSpring 7.2 software (Agilent Technologies). For both control (nonablated) and HU-treated samples, the whole experiment was repeated three times with different batches of total RNA extracted from the dissected brains of independent fly cultures. The microarray data files are deposited in the Gene Expression Omnibus Database (accession no. GSE3379).
In Situ Hybridization.
Fluorescent in situ hybridization was done as described (49), with modifications. Briefly, digoxygenin (DIG)-labeled RNA probes were prepared with either cDNA clones obtained from the Drosophila Genomic Resource Center (Bloomington, IN) or with PCR-amplified genomic templates tagged by T3 promoter. Probes were examined by agarose-gel electrophoresis for integrity and by dot-blot hybridization for detection sensitivity. Dissected brains were fixed with 4% paraformaldehyde for 20 min and hybridized with a DIG-RNA probe at 65°C overnight. Proteinase treatment was omitted to minimize background. Hybridization signals were detected with an anti-DIG antibody and amplified with the TSA Biotin System (PerkinElmer, Wellesley, MA).
RNAi.
For constructing transgenic RNAi flies, cDNA fragments were amplified by PCR and inserted as a pair of inverted repeats into pUAST-R57 (50), which has an intronic fragment of the Drosophila ret oncogene to enhance RNAi efficacy. Expression of UAS-RNAi in MBs was driven by OK107-Gal4 (18, 29) with a membrane-bound reporter, UAS-mCD8::GFP. Flies were raised at 28°C after larval hatching to stimulate Gal4 activity. The structural integrity of MBs in F1 females was examined by confocal microscopy using a Zeiss (Thornwood, NJ) laser scanning microscope. Two independent UAS-RNAi lines were examined for each gene.
mRNA Quantification.
To measure transcript levels in the brain, total RNA was extracted from dissected female brains, and target mRNA levels were quantified by RT-PCR-mediated quantification. PCR cycle times were predetermined for each gene by separate control experiments to ensure linear amplification. Fluorescence intensity of specific PCR products was directly and digitally quantified in agarose gel with Molecular Imager (Bio-Rad, Hercules, CA) at three different PCR cycles in the linear amplification range. Signal intensity was standardized by using GAPDH as a control.
Supplementary Material
Acknowledgments
We thank Drs. Mitsuhiko Kurusu, Shuji Shigenobu, and Satoru Kobayashi for helpful suggestions for RNA preparation and microarray analyses; Ms. Noriko Kasai and Ayumi Hara for their help in fly brain dissection; Dr. Richard Weisburd for reading the manuscript; the Indiana Drosophila Genomics Resource Center for EST clones; and the Bloomington Drosophila Stock Center and the Kyoto Drosophila Genetic Resource Center for fly stocks. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K.F.-T. and M.K.) and by a Research Project of the Tsukuba Advanced Research Alliance (to K.F.-T.). We are also grateful for financial support from the Kantons of Basel, the Swiss National Science Foundation, and the Balzan Prize Foundation (to W.J.G. and L.M.).
Abbreviations
- AL
antennal lobe
- HU
hydroxyurea
- MB
mushroom body
- OL
optic lobe.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE3379).
References
- 1.Heisenberg M. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 2.Tettamanti M, Armstrong J, Endo K, Yang M, Furukubo-Tokunaga K, Kaiser K, Reichert H. Dev Genes Evol. 1997;207:242–252. doi: 10.1007/s004270050112. [DOI] [PubMed] [Google Scholar]
- 3.Kurusu M, Nagao T, Walldorf U, Flister S, Gehring WJ, Furukubo-Tokunaga K. Proc Natl Acad Sci USA. 2000;97:2140–2144. doi: 10.1073/pnas.040564497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noveen A, Daniel A, Hartenstein V. Development (Cambridge, UK) 2000;127:3475–3488. doi: 10.1242/dev.127.16.3475. [DOI] [PubMed] [Google Scholar]
- 5.Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K. Development (Cambridge, UK) 2002;129:409–419. doi: 10.1242/dev.129.2.409. [DOI] [PubMed] [Google Scholar]
- 6.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. Development (Cambridge, UK) 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 7.Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis R. Learn Mem. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- 8.Strausfeld NJ, Sinakevitch I, Vilinsky I. Microsc Res Tech. 2003;62:151–169. doi: 10.1002/jemt.10368. [DOI] [PubMed] [Google Scholar]
- 9.Lee T, Lee A, Luo L. Development (Cambridge, UK) 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 10.Zars T, Fischer M, Schulz R, Heisenberg M. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 11.Pascual A, Preat T. Science. 2001;294:1115–1117. doi: 10.1126/science.1064200. [DOI] [PubMed] [Google Scholar]
- 12.Isabel G, Pascual A, Preat T. Science. 2004;304:1024–1027. doi: 10.1126/science.1094932. [DOI] [PubMed] [Google Scholar]
- 13.Davis RL. Neuron. 2004;44:31–48. doi: 10.1016/j.neuron.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Davis RL. Ann Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 15.Margulies C, Tully T, Dubnau J. Curr Biol. 2005;15:R700–R713. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callaerts P, Halder G, Gehring WJ. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- 17.Callaerts P, Leng S, Clements J, Benassayag C, Cribbs D, Kang YY, Walldorf U, Fischbach KF, Strauss R. J Neurobiol. 2001;46:73–88. doi: 10.1002/1097-4695(20010205)46:2<73::aid-neu10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 18.Adachi Y, Hauck B, Clements J, Kawauchi H, Kurusu M, Totani Y, Kang YY, Eggert T, Waldport U, Furukubo-Tokunaga K, Callaerts P. Mech Dev. 2003;120:1113–1126. doi: 10.1016/j.mod.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 19.de Belle JS, Heisenberg M. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- 20.Truman J, Bate M. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, Hotta Y. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- 22.Prokop A, Technau GM. Dev Biol. 1994;162:321–337. doi: 10.1006/dbio.1994.1034. [DOI] [PubMed] [Google Scholar]
- 23.Stocker RF, Heimbeck G, Gendre N, de Belle JS. J Neurobiol. 1997;32:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Han P-L, Meller V, Davis RL. J Neurobiol. 1996;31:88–102. doi: 10.1002/(SICI)1097-4695(199609)31:1<88::AID-NEU8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 25.Lee T, Marticke S, Sung C, Robinow S, Luo L. Neuron. 2000;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- 26.O'Dell KM, Armstrong JD, Yang MY, Kaiser K. Neuron. 1995;15:55–61. doi: 10.1016/0896-6273(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 27.Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, Hama C. Neuron. 2000;26:119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- 28.Drysdale RA, Crosby MA, The FlyBase Consortium Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. http://flybase.org/ [DOI] [PMC free article] [PubMed]
- 29.Connolly JB, Roberts IJH, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- 30.Lam G, Hall BL, Bender M, Thummel CS. Dev Biol. 1999;212:204–216. doi: 10.1006/dbio.1999.9343. [DOI] [PubMed] [Google Scholar]
- 31.Kamikouchi A, Takeuchi H, Sawata M, Ohashi K, Natori S, Kubo T. Biochem Biophys Res Commun. 1998;242:181–186. doi: 10.1006/bbrc.1997.7870. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi H, Fujiyuki T, Shirai K, Matsuo Y, Kamikouchi A, Fujinawa Y, Kato A, Tsujimoto A, Kubo T. FEBS Lett. 2002;513:230–234. doi: 10.1016/s0014-5793(02)02319-0. [DOI] [PubMed] [Google Scholar]
- 33.de Belle JS, Heisenberg M. Proc Natl Acad Sci USA. 1996;93:9875–9880. doi: 10.1073/pnas.93.18.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubnau J, Chiang A-S, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 35.Heisenberg M, Böhl K. Z Naturforshung C. 1979;34:143–147. [Google Scholar]
- 36.Moreau-Fauvarque C, Taillebourg E, Boissoneau E, Mesnard J, Dura JM. Mech Dev. 1998;78:47–61. doi: 10.1016/s0925-4773(98)00147-6. [DOI] [PubMed] [Google Scholar]
- 37.Pascual A, Chaminade M, Preat T. Dev Biol. 2005;280:177–186. doi: 10.1016/j.ydbio.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 38.Nicolai M, Lasbleiz C, Dura J-M. J Neurobiol. 2003;57:291–302. doi: 10.1002/neu.10277. [DOI] [PubMed] [Google Scholar]
- 39.Reuter JE, Nardine TM, Penton A, Billuart P, Scott EK, Usui T, Uemura T, Luo L. Development (Cambridge, UK) 2003;130:1203–1213. doi: 10.1242/dev.00319. [DOI] [PubMed] [Google Scholar]
- 40.Kastel PL, Davis KL, Haroutunian V. Int Rev Neurobiol. 2005;63:41–82. doi: 10.1016/S0074-7742(05)63003-6. [DOI] [PubMed] [Google Scholar]
- 41.Mischel PS, Cloughesy TF, Nelson SF. Nat Rev Neurosci. 2004;5:782–792. doi: 10.1038/nrn1518. [DOI] [PubMed] [Google Scholar]
- 42.Henry GL, Zito K, Dubnau J. Curr Opin Neurobiol. 2003;13:570–576. doi: 10.1016/s0959-4388(03)00107-7. [DOI] [PubMed] [Google Scholar]
- 43.Geschwind DH, Ou J, Easterday MC, Dougherty JD, Jackson RL, Chen Z, Antoine H, Terskikh A, Weissman IL, Nelson SF, Kornblum HI. Neuron. 2001;29:325–339. doi: 10.1016/s0896-6273(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 44.Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 45.Zhao X, Lein ES, He A, Smith SC, Aston C, Gage FH. J Comp Neurol. 2001;441:187–196. doi: 10.1002/cne.1406. [DOI] [PubMed] [Google Scholar]
- 46.Zirlinger M, Kreiman G, Anderson DJ. Proc Natl Acad Sci USA. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strausfeld NJ, Hildebrand JG. Curr Opin Neurobiol. 1999;9:634–639. doi: 10.1016/S0959-4388(99)00019-7. [DOI] [PubMed] [Google Scholar]
- 48.Michaut L, Flister S, Neeb M, White KP, Certa U, Gehring WJ. Proc Natl Acad Sci USA. 2003;100:4024–4029. doi: 10.1073/pnas.0630561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- 50.Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, Ueda R, Lemaitre BJ. J Biol Chem. 2004;279:12848–12853. doi: 10.1074/jbc.M313324200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.