Abstract
The Kaposi's sarcoma-associated herpesvirus LANA protein is expressed in all Kaposi's sarcoma-associated herpesvirus-infected cells, including the tumor cells of endemic and AIDS-associated Kaposi sarcoma, primary effusion lymphoma, and Castleman disease. LANA modulates cell gene expression, but the mechanisms of LANA-mediated transcriptional reprogramming are poorly understood. LANA-repressed cell genes were identified by using retroviral-transduced telomerase-immortalized microvascular endothelial cells. Transciptional repression of targeted genes was relieved by treatment with the methyltransferase inhibitor 5-aza-2′-deoxycytidine, suggesting a role for DNA methylation in repression. We found that LANA coprecipitated with DNA methyltransferases (Dnmts) and recruited endogenous DNA methyltransferase activity from the cell extract. LANA preferentially relocalized Dnmt3a from the nuclear matrix into the chromatin fraction. Further, LANA associated with repressed cellular promoters, recruited Dnmt3a to DNA, and facilitated de novo promoter methylation of a down-regulated gene, cadherin 13 (H-cadherin). The data provide an example of promoter-specific epigenetic DNA modification through viral protein recruitment of de novo Dnmt activity.
Keywords: cancer, CpG methylation, epigenetic modification, transcription repression
Kaposi's sarcoma-associated herpesvirus (KSHV) first was identified in tissues of AIDS-associated Kaposi sarcoma (1) and also is associated with the AIDS-related malignancies primary effusion lymphoma and variant multicentric Castleman disease (2). The tumor types associated with KSHV reflect the dual tropism of the virus for endothelial cells and B cells. Unlike other herpesviruses, KSHV is not ubiquitous. Serological studies find high seroprevalence rates in Africa, intermediate levels in the Mediterranean, and low levels in other parts of the world (3). All tumor cells carrying KSHV express the viral latency-associated nuclear antigen LANA and a small proportion of the cells additionally express viral genes such as vIL-6 and vGPCR that are expressed as part of the viral lytic replicative program (4–6). The model for KSHV-associated tumor development thus combines viral latency protein effects with paracrine signaling emanating from nearby cells that are undergoing viral lytic reactivation.
KSHV LANA is functionally pleiotropic being essential for the replication and persistence of KSHV genomes during latency (7–11) and also contributing to KSHV pathogenesis by stimulating S phase entry (12, 13), blocking p53-induced apoptosis (14–16), interacting with pRb (17), interacting with glycogen synthase kinase 3 to stabilize β-catenin (12, 18), and modulating cellular gene expression. Gene array analyses of LANA-expressing cells found both up-regulation and down-regulation of cell genes (13, 19, 20). Whereas many of the changes mediated by LANA appear to occur indirectly via activation of β-catenin and E2F target genes (13), direct binding of LANA to DNA leads to transcriptional repression in reporter assays (21–23). Interactions between LANA and proteins such as the corepressors mSin3, SAP30, and CIR, the methyl CpG-binding protein MeCP2, and the histone methyltransferase SUV39H1 (21, 24, 25) are consistent with a direct role for LANA in transcriptional repression, but the mechanisms of LANA-mediated repression are poorly understood.
Methylation of CpG residues in the regulatory region of cell genes is associated with transcriptional repression. CpG islands contain small stretches enriched for CpG dinucleotide pairs that are unmethylated in the promoter regions of euchromatic DNA in normal tissues but are frequently hypermethylated in cancer cells (26). Methylation of the cytosine residue in the CpG dinucleotide is carried out by the DNA methyltransferases Dnmt1, Dnmt3a, and Dnmt3b (27). Dnmt1 is responsible primarily for reestablishing the methylation pattern during DNA replication and, hence, functions in the maintenance of the established DNA methylation signature, whereas Dnmt3a and Dnmt3b can methylate the CpG cytosine residue in a background of unmethylated DNA and function as de novo methyltransferases. In cancer cells, the context of CpG islands may render certain loci particularly susceptible to de novo DNA methylation with the ultimate establishment of a particular pattern being driven by the growth advantage provided by repression of the targeted genes (28, 29). On the other hand, de novo DNA methyltransferases also may be targeted to particular loci through interactions with chromatin- associated factors. For example, the PML-RAR fusion protein generated by a chromosomal translocation in acute promyelocytic leukemia recruits Dnmts (30).
We provide evidence for LANA-mediated recruitment of Dnmt3a from the nuclear matrix and the association of LANA with DNA methyltransferase activity. The binding of Dnmts suggests that LANA coordinates de novo DNA methylation with the recruitment of DNA methyl CpG-binding proteins and histone modifiers to establish epigenetic silencing of LANA-targeted cell genes.
Results
Identification of LANA-Repressed Genes in Endothelial Cells.
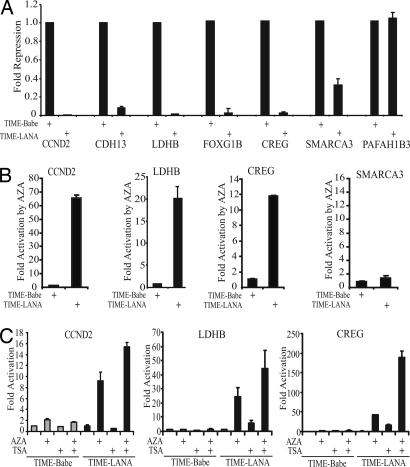
LANA has been shown to both up-regulate and down-regulate cellular genes in array analyses (13, 19, 20). To study the mechanism by which LANA expression transcriptionally represses cellular genes, we used retrovirus transduction to establish LANA-converted, telomerase-immortalized, microvascular endothelial (TIME-LANA) cell lines and vector converted controls (TIME-Babe). The TIME-LANA cells expressed LANA as shown by immunoblot and immunofluorescence assays (Fig. 6, which is published as supporting information on the PNAS web site). Array analyses of gene expression identified 80 cellular genes that were differentially down-regulated by LANA in these cell lines (Table 1, which is published as supporting information on the PNAS web site). The results were confirmed by real-time RT-PCR on a panel of the highly repressed genes [CCND2 (cyclin D2), CDH13 (H-cadherin), LDHB (Lactate dehydrogenase B), FOXG1B/FKHL1 (Forkhead box protein G1B/Forkhead-related protein1) and CREG (cellular repressor of E1A-stimulated genes)], a moderately repressed gene [SMARCA3 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 3)] and a control unaffected gene [PAFAH1B3 (platelet-activating factor acetylhydrolase, isoform IB, gamma subunit)] (Fig. 1A). In this assay, the transcriptional repression of CCND2, CDH13, LDHB, FOXG1B, and CREG in the LANA-expressing cells was dramatic, ranging from 12-fold (CDH13) to 73-fold (LDHB) in contrast to SMARCA3, which was down-regulated 2.1-fold, and PAFAH1B3, which was not affected. Such strong repression is suggestive of an epigenetic modification.
Fig. 1.
LANA represses cell gene expression. Real-time RT-PCR analyses. (A) Down-regulation of selected genes in TIME-LANA cells. PAFAH1B3 is an unaffected control gene. Reactions were performed in triplicate. (B) Reactivated expression of CCND2, LDHB, and CREG genes but not SMARCA3 in TIME-LANA cells treated with 5′-aza-2′-deoxycytidine. Fold activation is the ratio of expression in treated versus untreated cells. Reactions were performed in triplicate. (C) Gene reactivation in cells treated with 5′-aza-2′-deoxycytidine plus TSA.
CpG methylation in vertebrates is linked to changes in chromatin structure and gene silencing (31), and epigenetic alterations in DNA methylation are a common feature of carcinogenesis (32). To examine the contribution of DNA methylation to LANA-mediated transcriptional repression, cells were treated with 5-aza-2′-deoxycytidine (AZA), an inhibitor of DNA methyltransferases. This treatment did not affect expression of the tested genes in TIME-Babe cells but resulted in a strong reactivation of the highly repressed CCND2, LDHB, and CREG genes (12-fold, 20-fold, and 65-fold) in TIME-LANA cells (Fig. 1B). The SMARCA3 gene was not reactivated, suggesting that moderate repression is mediated by other mechanisms. The effect of treatment with AZA was compared with treatment with the histone deacetylase inhibitor trichostatin A (TSA) (Fig. 1C). Neither TSA nor AZA treatment affected expression in the control TIME-Babe cells. TSA alone was an ineffective activator in TIME-LANA cells compared with AZA, suggesting that DNA methyltransferase activity was particularly important for LANA-mediated repression. The combination of AZA and TSA was more effective than either agent alone. This synergy previously has been shown for other hypermethylated cell genes (33).
LANA Associates with the Repressed Promoters.
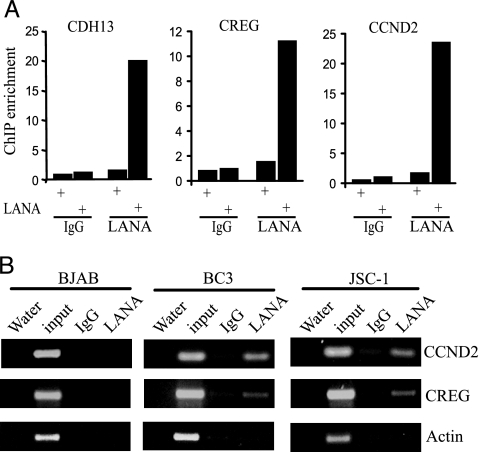
We next asked whether LANA was associated with the CDH13, CREG, and CCND2 promoters. ChIP analyses performed on LANA-transfected HEK293T cells detected LANA bound to all three endogenous promoters (Fig. 2A). Immunoprecipitates generated with control antibody or precipitates with anti-LANA antibody generated from cells receiving vector DNA were negative. LANA association with the CCND2 and CREG promoters also was examined in the KSHV-positive PEL cell lines BC3 and JSC-1 and the virus-negative BJAB B cell line (Fig. 2B). These ChIP experiments detected LANA bound to the CCND2 and CREG promoters in the PEL cell lines but not in BJAB cells. LANA was not detected on the actin promoter, indicating that there was specificity to the promoter association.
Fig. 2.
LANA associates with repressed promoters. (A) ChIP assays performed on LANA or vector-transfected HEK293T cell extracts precipitated with anti-LANA antibody or control IgG. The promoters were amplified by real-time PCR. The signal from vector-transfected cells was set at 1. (B) KSHV-positive BC3 and JSC-1 PEL cells and virus-negative BJAB B cells. Cell extracts were precipitated with anti-LANA antibody or control IgG. The input DNA was 1/40 of that used for immunoprecipitation.
LANA Interacts with DNA Methyltransferases.
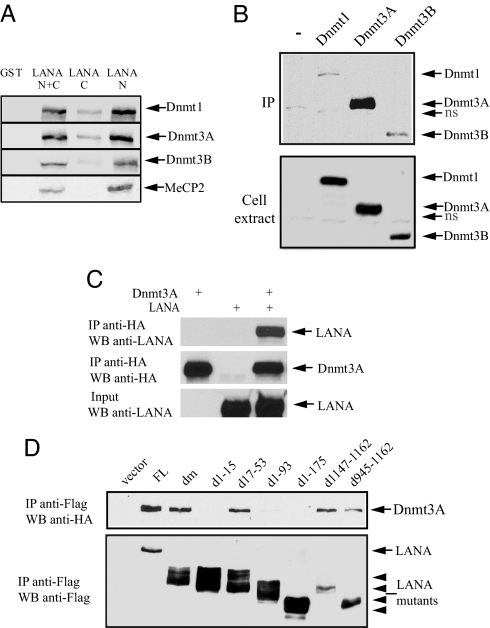
The association of LANA with repressed promoters along with the derepression seen with an inhibitor of Dnmt activity led us to examine whether there was an interaction between LANA and cellular DNA methyltransferases. GST-affinity assays were performed by using bacterially expressed GST-LANA proteins and in vitro-translated Dnmt proteins (Fig. 3A). Dnmt1, Dnmt3a, and Dnmt3b each interacted with GST-LANA N + C, which contains the unique N-terminal and C-terminal domains of LANA. No interaction was seen with the GST protein itself. The three DNMT proteins bound strongly to the LANA N terminus (LANA N), and a weak interaction also was detected to the LANA C terminus (LANA C). In vitro-translated MeCP2 served as a positive control.
Fig. 3.
LANA interacts with Dnmts. (A) LANA interacts directly with Dnmts. In vitro translated [35S]methionine-labeled Dnmt1, Dnmt3a, Dnmt3b, and MeCP2 (positive control) were incubated with equal amounts of bacterially expressed GST, GST-LANA C+ N (amino acids 1–340 and 924-1162), LANA C (amino acids 924-1162), or LANA N (amino acids 1–340). Bound proteins were eluted, separated by SDS/PAGE, and detected by autoradiography. (B) Western blots examining interaction in extracts of cotransfected HEK293T cells. (Upper) Coprecipitation of HA-Dnmts with Flag-LANA on anti-Flag M2 beads. (Lower) Detection of HA-Dnmts in the transfected cell extract. (C) (Top) Western blot showing a reciprocal coprecipitation of Flag-LANA with HA-Dnmt3a in an anti-HA immunoprecipitate of a cotransfected cell extract. (Middle and Bottom) Expression of HA-Dnmt3a and Flag-LANA were detected with anti-HA and anti-LANA antibodies. (D) LANA amino acids 1–15 are required for Dnmt3a interaction. Western blot examining coprecipitation of HA-Dnmt3a with Flag-LANA variants in extracts of cotransfected cells. (Upper) Coprecipitation of HA-Dnmt3a with Flag-LANA on anti-Flag beads. (Lower) Western blot of the same samples showing expression of the Flag-LANA proteins.
The interaction between LANA and DNMTs also was examined by using immunoprecipitations performed on Flag-LANA and HA-Dnmt cotransfected HEK293T cell extracts (Fig. 3B). Coprecipitation of each of the HA-Dnmt proteins was observed in anti-Flag-LANA precipitates, but the coprecipitation of HA-Dnmt3a was particularly strong. Binding of LANA to HA-Dnmt3a also was demonstrated in a reverse immunoprecipitation in which LANA was detected in immunoprecipitates generated against HA-Dnmt3a (Fig. 3C). The region of LANA required for interaction with Dnmt3a was mapped by immunoprecipitation (Fig. 3D). Deletion of the first 15 aa and larger deletions that encompassed LANA amino acids 1–15 (d1–93 and d1–175) destroyed the interaction. Interestingly, this deletion overlaps with the amino acids 5–22 chromatin-binding motif (14, 34, 35) and the region required for binding to MeCP2 and histones (8, 24).
LANA Preferentially Relocalizes Dnmt3a into the Chromatin Fraction.
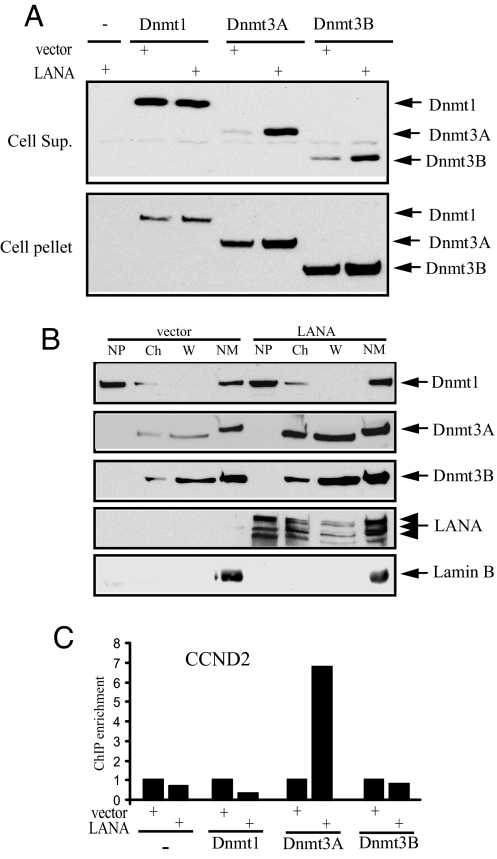
To investigate the basis for the apparent preference for interaction with Dnmt3a in transfected cells, Dnmt levels in low-salt extracts of transfected cells were compared in the presence and absence of transfected LANA. Western blotting showed little effect of LANA on the amount of the Dnmts in the cell pellet (Fig. 4A Lower). However, Dnmt3a was significantly more readily extracted into the supernatant by a low-salt buffer in the presence of LANA, and LANA also had some effect on the extraction of Dnmt3b. Dnmt1 was present in the low-salt extract at the same levels in the presence or absence of LANA (Fig. 4A Upper).
Fig. 4.
LANA relocalizes Dnmt3a and recruits it to a repressed promoter Western blotting to detect HA-Dnmts. (A) Transfected HEK293T cells were treated with low-salt lysis buffer to generate supernatant (Upper) and pellet (Lower) fractions. (B) LANA increases chromatin association of Dnmt3a. Western blots of cotransfected cells fractionated into nucleoplasm (NP), chromatin (Ch), chromatin wash (W), and nuclear matrix (NM) and probed with anti-HA antibody, anti-LANA antibody, or anti-Lamin B antibody. Lamin B is a control NM-associated protein. (C) LANA recruits Dnmt3a to the repressed CCND2 promoter. ChIP assays performed on extracts of cells cotransfected with LANA or vector plus HA-Dnmts. The chromatin was immunoprecipitated with anti-HA antibody, and the promoter was amplified by real-time PCR. The signal from vector transfected cells was set at 1.
Examination of the nuclear compartmentalization of the Dnmt proteins (Fig. 4B) found that Dnmt1 was present predominantly in the nuclear matrix (NM) and nucleoplasmic (NP) fractions, and this distribution was not affected by LANA. In contrast, LANA dramatically increased the presence of Dnmt3a in the chromatin and chromatin wash (W) fractions. LANA also slightly increased the relative amount of Dnmt3b present in the chromatin wash (W) and chromatin fractions (Ch). Localization of a control nuclear matrix protein, Lamin B, was not affected by LANA (Fig. 4B Bottom).
To further explore the linkage between LANA-mediated repression and the chromatin association of Dnmt proteins, ChIP analyses were performed on the LANA-responsive CCND2 promoter in cells cotransfected with HA-Dnmts and either LANA or vector DNA (Fig. 4C). Dnmt3a was enriched 7-fold on the endogenous CCND2 promoter in the presence of LANA. No enrichment of Dnmt3b or Dnmt1 was detected. Thus, the ChIP analysis indicated a preferential recruitment of Dnmt3a to the LANA-repressed CCND2 promoter.
LANA Recruits Enzymatically Active Dnmts and Facilitates Promoter Methylation.
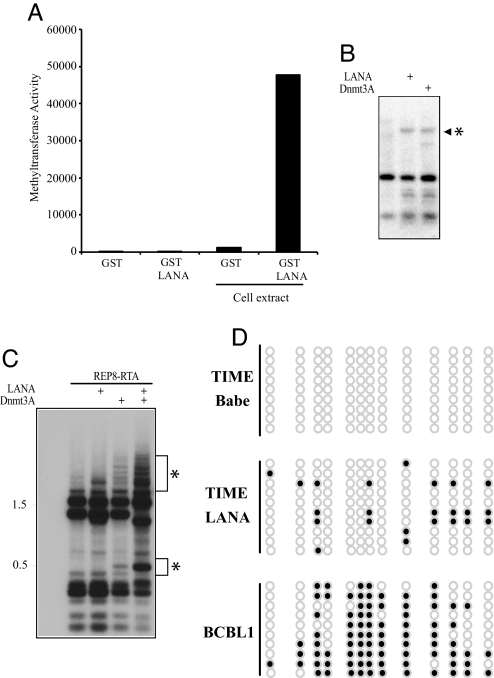
The ability of LANA to relocate de novo Dnmts into the chromatin fraction could have profound implications for gene expression in KSHV-infected cells. To test whether LANA-associated Dnmts were enzymatically active, GST-LANA was incubated in the presence or absence of cell extract, washed extensively, and the eluted protein was subjected to an in vitro methyltransferase assay (Fig. 5A). GST-LANA did not have any intrinsic DNA methyltransferase activity, but GST-LANA was able to recruit methyltransferase activity from the cell extract. The control GST protein showed minimal interaction with Dnmts.
Fig. 5.
LANA associates with DNA methyltransferase activity and induces DNA methylation. (A) Purified bacterially expressed GST or GST-LANA proteins were assayed for DNA methyltransferase activity in the presence or absence of HCT 116 extract. (B and C) Induction of DNA methylation. The methylation induced bands are indicated by an asterisk (∗). Plasmid DNAs transfected into HEK293T cells with or without LANA or Dnmt3a were isolated 7 days after transfection, and the HpaII (B) HhaI (C) digestion pattern was examined by Southern blotting. (B) Analysis of a CREG promoter plasmid probed with a radiolabeled CREG promoter DNA fragment. (C) Analysis of an episomal pREP8 plasmid carrying the KSHV RTA promoter probed with whole-plasmid DNA. (D) Methylation of the CDH13 promoter. Bisulfite sequencing analysis of the CDH13 gene amplified from TIME-Babe, TIME-LANA, and BCBL1 PEL cells. Each row represents one sequenced clone. Open circles represent unmethylated CpG sites, and filled circles represent methylated CpG sites.
To test for LANA-induced de novo DNA methylation, HEK293T cells were transfected with a CREG promoter plasmid together with LANA or Dnmt3a. The plasmid DNA was isolated by Hirt extraction 7 days after transfection and was subjected to methylation-sensitive restriction digestion (HpaII) and analyzed by Southern blotting by using a fragment of the CREG promoter as the probe. Consistent with de novo promoter methylation, a previously unseen HpaII-resistant CREG promoter fragment was observed upon digestion of DNA from cells cotransfected with LANA. The same DNA fragment also was generated in cells cotransfected with Dnmt3a (Fig. 5B).
The KSHV RTA (ORF50) promoter is methylated in latently infected PEL cells (36), and treatment with AZA induces RTA expression and lytic reactivation (37). De novo methylation of an RTA promoter containing episomal vector (pREP8) also was examined. A more complex banding pattern was generated in this assay, which used whole-plasmid DNA to probe the Southern blot. The plasmid methylation induced at multiple sites by transfected Dnmt3a was increased significantly in cells cotransfected with LANA, suggesting that LANA and Dnmt3a collaborate in the induction of de novo DNA methylation (Fig. 5C).
A more detailed analysis of promoter methylation was performed on the endogenous CDH13 promoter by using bisulfite sequencing (Fig. 5D). In vector-transduced TIME cells, there was no CpG methylation detected in the region of the CDH13 gene that is known to be subject to epigenetic modification in cancers (38). In TIME-LANA cells, we found increased but incomplete and scattered CpG methylation of this regulatory region. This dispersed CpG methylation pattern is reminiscent of promoter methylation generated by Dnmt3a (39). Bisulfite sequencing of the CDH13 regulatory region in KSHV-positive BCBL-1 PEL cells showed a more intense CpG methylation, which may reflect long-term selection during tumor development.
Discussion
Epigenetic inactivation of cell genes is an integral feature of the development of cancer. The mechanisms that target DNA for CpG methylation by de novo Dnmts are incompletely understood and include structural features such as the presence of repeat sequences, the influence of flanking sequences on the substrate preferences of the de novo Dnmts, and specific targeting of the Dnmts through interaction with chromatin-interacting factors. In addition to the PML–RAR interaction with Dnmts, transcription factor targeting through c-Myc, Pu.1, and p53 has been described in refs. 40–42. In the case of p53, the DNA methyltransferase activity of Dnmt3a was not required for p53-mediated repression of the p21Cip1 gene, indicating that recruitment of the Dnmt3a-associated histone deacetylases can be sufficient for establishing repression. The interaction between KSHV LANA and Dnmts described here provides evidence for viral-induced retargeting of the de novo DNA methyltransferases. LANA relocalized Dnmt3a, and to a lesser extent Dnmt3b, from the nuclear matrix into chromatin, increased plasmid CpG methylation associated with cotransfected Dnmt3a, specifically increased the association of Dnmt3a with the CCND2 promoter, and increased CpG methylation of a cellular promoter, that for cadherin 13.
The LANA repressed genes CCND2, LDHB, and CREG were dramatically derepressed by treatment with the methyltransferase inhibitor AZA, indicating that a subset of LANA-repressed genes are highly reliant on recruitment of Dnmt activity. The de novo CpG methylation induced in the CDH13 promoter in LANA-expressing TIME cells was less dense than that seen in the CDH13 promoter in PEL cells. This difference may derive from growth of LANA-expressing cells in tissue culture versus long-term tumor selection. However, it should be noted that methylation of a single nucleotide can be sufficient for promoter repression (43). Alternatively, LANA-mediated repression may represent a coordination of CpG methylation with other protein–protein interactions that produce profound transcriptional repression without the requirement for dense DNA methylation. Introduction of a small number of CpG methylations at sites that had the preferred environment for Dnmt3a (44) would produce binding sites for the methyl CpG-binding protein MeCP2, which also interacts with LANA (24). LANA, MeCP2, and Dnmts each interact with HDACs (21, 45, 46). An additional coordination between DNA methylation and repressive histone modifications would be provided by LANA- and MeCP2-mediated recruitment of the histone methyltransferase SUV39H1 (25, 47), which introduces histone H3 K9 modifications. The ability of LANA to interact with this set of proteins thus would generate a very strong localized repressive environment. This model leaves open the question of how LANA itself is targeted to promoters. LANA associated with the endogenous CCND2, CDH13, and CREG promoters in ChIP analyses. The recognized LANA interactions with chromatin-associated proteins such as MeCP2, DEK (24), core histones (8), or Brd2/RING3 (48) would not individually provide specificity, but LANA also has been found to interact with transcription factors such as Sp1 and CBF1/RBP-Jk (49, 50), and combinatorial interactions are likely to be involved in promoter-specific LANA targeting. The Dnmt3a interaction required the same region of LANA that has been implicated in binding of MeCP2 and core histones. However, the ability of Dnmt proteins generated in a cytosolic in vitro translation system to bind to bacterially expressed LANA suggests that the Dnmt proteins can contact LANA directly.
The LANA-repressed gene CDH13 is down-regulated through methylation in breast cancer, non-small cell lung cancer, colorectal cancer, invasive squamous cell carcinoma, and chronic myeloid leukemia (38, 51–54). Promoter methylation and loss of expression of lactate dehydrogenase B has been described in prostate and gastric cancers (55, 56), and promoter hypermethylation of the CCND2 gene occurs in several cancers including prostate (57), acute lymphoblastic leukemia (58), and breast (59). The loss of expression of these genes in cancers through epigenetic modification strengthens the biological relevance of the LANA-mediated repression. CREG is relatively uncharacterized, and its regulation in human cancers has not been addressed. CREG binds to pRb in vitro and inhibits activation by E2F (60). E2F-responsive genes are up-regulated by LANA and, hence, repression of CREG would reinforce E2F responses in KSHV-infected cells. Moderately LANA-repressed genes such as SMARCA3 showed little response to AZA treatment, indicating that LANA also mediates repression through other, possibly indirect, mechanisms.
Virus infection has been linked to increased expression of DNA methyltransferases in cells infected with HIV-1 (61), hepatitis B virus (62), the human polyomavirus BKV, adenovirus (63), and Epstein–Barr virus (64). The interaction between KSHV LANA and Dnmts described here, and the evidence for viral protein-targeted CpG methylation, raise the possibility of a broader role for viruses in epigenetic modification in human cancers. Further, CpG methylation is known to be a factor in establishment of viral latency by contributing to transcriptional down-regulation of immunological targets and key viral lytic cycle regulators such as the KSHV RTA protein. Viral-directed CpG methylation also could place this aspect of immune evasion and latency maintenance under viral control.
Materials and Methods
Cell Culture and Plasmid DNAs.
Immortalized human dermal microvascular endothelial (TIME) cells were grown in EGM-2 medium plus MV Singlequots. TIME cells were transduced with LANA and vector pBabe recombinant retroviruses and selected with puromycin (2.5 μg/ml). Cells were treated with AZA (5 μM; Sigma, St. Louis, MO) for 5 days and the addition of trichostatin A (300 nM) was for the last 24 h. GST-LANA and Flag-LANA deletion mutants have been described (18, 21), as have HA-Dnmt vectors (65). Flag-LANA (d1–15;pMS24) and (d17–53;pMS25) were generated from pMF24.
GST-Affinity and Immunoprecipitation Assays.
GST and GST-LANA proteins were prepared, and assays were performed as described in ref. 24 by using equal amounts of 35S-labeled in vitro translated proteins (Promega, Madison, WI). For immunoprecipitations, HEK293T cells in 10-cm dishes were transfected with 10 μg of total DNA by using calcium phosphate precipitation. Forty-eight hours after transfection, cells were resuspended in 1 ml of lysis buffer (50 mM Tris, pH 7.9/100 mM NaCl/0.5 mM EDTA/2% glycerol/0.2% Nonidet P-40/0.5 mM PMSF/2 μg/ml aprotinin and leupeptin), sonicated for 10 s, and cleared by centrifugation. Extracts were diluted with wash buffer (lysis buffer minus glycerol and Nonidet P-40), precleared by using protein G beads, and immunoprecipitated with anti-Flag M2 agarose (Sigma) or anti-HA antibody (Roche, Indianapolis, IN) and protein A/protein G beads overnight at 4°C. Beads were washed six times, and bound proteins were detected by Western blotting with anti-HA (Sigma) or anti-LANA (NovaCastra, Newcastle, U.K.) antibodies.
RT-PCR and Bisulfite Sequencing.
Total RNA was isolated with the RNeasy mini kit (Qiagen, Valencia, CA) and DNase I treatment with RNase-free DNase. RNA was reverse transcribed by using AMV reverse transcriptase (Promega). Bisulfite treatment, methylation-specific PCR, and real-time PCR were performed as described in refs. 66 and 67. CDH13 DNA was amplified by using the primers 5′-TTGGAAAAGTGGAATTAGTTGG and 5′-CCTCTTCCCTACCTAAAACA. PCR products were ligated into an AT-vector (Invitrogen, Carlsbad, CA) and sequenced by Macrogen (Seoul, Korea).
Cell Fractionation.
For low-salt extraction, cells were resuspended in buffer [50 mM Tris, pH 7.6/50 mM NaCl/0.5 mM EDTA/1% glycerol/0.2% Nonidet P-40/0.5 mM PMSF/2 μg/ml aprotinin and leupeptin). After 10 min on ice, the extract was centrifuged for 10 min at 15,800 × g. The supernatant was collected, and the cell pellet was solubilized in 8 M urea and 1% SDS and clarified by centrifugation at 15,800 × g for 5 min. Nuclear matrix fractionation was performed as described in ref. 68.
DNA Methyltransferase Assays.
Assays were performed as described in ref. 69 by using extracts from HCT116 cells. The extract was added to the GST-fusion proteins bound to glutathione Sepharose beads and rotated at 4°C for 4 h. The beads were washed six times, and bound proteins were eluted with glutathione. Activity was measured by using 3 μCi (1 Ci = 37 GBq) S-adenosyl-l-[methyl-3H] methionine (Amersham, Piscataway, NJ) and 0.5 μg of poly(dI-dC)-poly(dI-dC) (Amersham). Control reactions were minus DNA. Assays in transfected HEK293T cells were performed as described in ref. 39. The cells were harvested 7 days after transfection, and the DNA was purified by Hirt extraction and digested by HhaI or HpaII before Southern blotting.
Supplementary Material
Acknowledgments
We thank Vivien Goh for technical assistance and Kornel Schuebel, Kam-Wing Jair, and Steve Baylin for reagents and helpful advice. This work was supported by National Institutes of Health Specialized Programs of Research Excellence Grants 1P50 CA96888 and P01 CA113239 (to S.D.H).
Abbreviations
- AZA
5-aza-2′-deoxycytidine
- KSHV
Kaposi's sarcoma-associated herpesvirus
- TIME-LANA
LANA-converted, telomerase-immortalized, microvascular endothelial
- TSA
trichostatin A.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Moore PS, Chang Y. Annu Rev Microbiol. 2003;57:609–639. doi: 10.1146/annurev.micro.57.030502.090824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henke-Gendo C, Schulz TF. Curr Opin Infect Dis. 2004;17:53–57. doi: 10.1097/00001432-200402000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Boshoff C, Weiss RA. Philos Trans R Soc Lond B. 2001;356:517–534. doi: 10.1098/rstb.2000.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou CJ, Poole LJ, Kim PS, Ciufo DM, Cannon JS, ap Rhys CM, Alcendor DJ, Zong JC, Ambinder RF, Hayward GS. J Virol. 2002;76:3421–3439. doi: 10.1128/JVI.76.7.3421-3439.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore PS, Chang Y. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz TF. J Pathol. 2006;208:187–198. doi: 10.1002/path.1904. [DOI] [PubMed] [Google Scholar]
- 7.Ballestas ME, Chatis PA, Kaye KM. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 8.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 9.Grundhoff A, Ganem D. J Virol. 2003;77:2779–2783. doi: 10.1128/JVI.77.4.2779-2783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J, Garber AC, Renne R. J Virol. 2002;76:11677–11687. doi: 10.1128/JVI.76.22.11677-11687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verma SC, Choudhuri T, Kaul R, Robertson ES. J Virol. 2006;80:2243–2256. doi: 10.1128/JVI.80.5.2243-2256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimuro M, Wu FY, ap Rhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. Nat Med. 2003;9:300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- 13.An FQ, Compitello N, Horwitz E, Sramkoski M, Knudsen ES, Renne R. J Biol Chem. 2005;280:3862–3874. doi: 10.1074/jbc.M407435200. [DOI] [PubMed] [Google Scholar]
- 14.Wong LY, Matchett GA, Wilson AC. J Virol. 2004;78:10074–10085. doi: 10.1128/JVI.78.18.10074-10085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friborg JJ, Kong W, Hottiger MO, Nabel GJ. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 16.Si H, Robertson ES. J Virol. 2006;80:697–709. doi: 10.1128/JVI.80.2.697-709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radkov SA, Kellam P, Boshoff C. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 18.Fujimuro M, Hayward SD. J Virol. 2003;77:8019–8030. doi: 10.1128/JVI.77.14.8019-8030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renne R, Barry C, Dittmer D, Compitello N, Brown PO, Ganem D. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe T, Sugaya M, Atkins AM, Aquilino EA, Yang A, Borris DL, Brady J, Blauvelt A. J Virol. 2003;77:6188–6196. doi: 10.1128/JVI.77.11.6188-6196.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krithivas A, Young DB, Liao G, Greene D, Hayward SD. J Virol. 2000;74:9637–9645. doi: 10.1128/jvi.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwam DR, Luciano RL, Mahajan SS, Wong L, Wilson AC. J Virol. 2000;74:8532–8540. doi: 10.1128/jvi.74.18.8532-8540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garber AC, Hu J, Renne R. J Biol Chem. 2002;277:27401–27411. doi: 10.1074/jbc.M203489200. [DOI] [PubMed] [Google Scholar]
- 24.Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. J Virol. 2002;76:11596–11604. doi: 10.1128/JVI.76.22.11596-11604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakakibara S, Ueda K, Nishimura K, Do E, Ohsaki E, Okuno T, Yamanishi K. J Virol. 2004;78:7299–7310. doi: 10.1128/JVI.78.14.7299-7310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baylin SB. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 27.Bestor TH. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 28.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. Genomics. 2006;87:572–579. doi: 10.1016/j.ygeno.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 30.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, et al. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 31.Jaenisch R, Bird A. Nat Genet. 2003;33(Suppl 3):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 32.Herman JG, Baylin SB. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 33.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 34.Piolot T, Tramier M, Coppey M, Nicolas JC, Marechal V. J Virol. 2001;75:3948–3959. doi: 10.1128/JVI.75.8.3948-3959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbera AJ, Ballestas ME, Kaye KM. J Virol. 2004;78:294–301. doi: 10.1128/JVI.78.1.294-301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Ueda K, Sakakibara S, Okuno T, Parravicini C, Corbellino M, Yamanishi K. Proc Natl Acad Sci USA. 2001;98:4119–4124. doi: 10.1073/pnas.051004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyooka S, Toyooka KO, Harada K, Miyajima K, Makarla P, Sathyanarayana UG, Yin J, Sato F, Shivapurkar N, Meltzer SJ, Gazdar AF. Cancer Res. 2002;62:3382–3386. [PubMed] [Google Scholar]
- 39.Lin IG, Han L, Taghva A, O'Brien LE, Hsieh CL. Mol Cell Biol. 2002;22:704–723. doi: 10.1128/MCB.22.3.704-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, et al. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Hara E, Tenen DG, Hozumi N, Oikawa T. Oncogene. 2006;25:2477–2488. doi: 10.1038/sj.onc.1209272. [DOI] [PubMed] [Google Scholar]
- 42.Wang YA, Kamarova Y, Shen KC, Jiang Z, Hahn MJ, Wang Y, Brooks SC. Cancer Biol Ther. 2005;4:1138–1143. doi: 10.4161/cbt.4.10.2073. [DOI] [PubMed] [Google Scholar]
- 43.Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, et al. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oka M, Rodic N, Graddy J, Chang LJ, Terada N. J Biol Chem. 2006;281:9901–9908. doi: 10.1074/jbc.M511100200. [DOI] [PubMed] [Google Scholar]
- 45.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 46.Burgers WA, Fuks F, Kouzarides T. Trends Genet. 2002;18:275–277. doi: 10.1016/S0168-9525(02)02667-7. [DOI] [PubMed] [Google Scholar]
- 47.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 48.Viejo-Borbolla A, Ottinger M, Bruning E, Burger A, Konig R, Kati E, Sheldon JA, Schulz TF. J Virol. 2005;79:13618–13629. doi: 10.1128/JVI.79.21.13618-13629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verma SC, Borah S, Robertson ES. J Virol. 2004;78:10348–10359. doi: 10.1128/JVI.78.19.10348-10359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lan K, Kuppers DA, Robertson ES. J Virol. 2005;79:3468–3478. doi: 10.1128/JVI.79.6.3468-3478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roman-Gomez J, Castillejo JA, Jimenez A, Cervantes F, Boque C, Hermosin L, Leon A, Granena A, Colomer D, Heiniger A, Torres A. J Clin Oncol. 2003;21:1472–1479. doi: 10.1200/JCO.2003.08.166. [DOI] [PubMed] [Google Scholar]
- 52.Toyooka S, Tokumo M, Shigematsu H, Matsuo K, Asano H, Tomii K, Ichihara S, Suzuki M, Aoe M, Date H, et al. Cancer Res. 2006;66:1371–1375. doi: 10.1158/0008-5472.CAN-05-2625. [DOI] [PubMed] [Google Scholar]
- 53.Hanabata T, Tsukuda K, Toyooka S, Yano M, Aoe M, Nagahiro I, Sano Y, Date H, Shimizu N. Oncol Rep. 2004;12:177–180. [PubMed] [Google Scholar]
- 54.Ulivi P, Zoli W, Calistri D, Fabbri F, Tesei A, Rosetti M, Mengozzi M, Amadori D. J Cell Physiol. 2006;206:611–615. doi: 10.1002/jcp.20503. [DOI] [PubMed] [Google Scholar]
- 55.Leiblich A, Cross SS, Catto JW, Phillips JT, Leung HY, Hamdy FC, Rehman I. Oncogene. 2006;25:2953–2960. doi: 10.1038/sj.onc.1209262. [DOI] [PubMed] [Google Scholar]
- 56.Maekawa M, Taniguchi T, Ishikawa J, Sugimura H, Sugano K, Kanno T. Clin Chem. 2003;49:1518–1520. doi: 10.1373/49.9.1518. [DOI] [PubMed] [Google Scholar]
- 57.Rosenbaum E, Hoque MO, Cohen Y, Zahurak M, Eisenberger MA, Epstein JI, Partin AW, Sidransky D. Clin Cancer Res. 2005;11:8321–8325. doi: 10.1158/1078-0432.CCR-05-1183. [DOI] [PubMed] [Google Scholar]
- 58.Scholz C, Nimmrich I, Burger M, Becker E, Dorken B, Ludwig WD, Maier S. Ann Hematol. 2005;84:236–244. doi: 10.1007/s00277-004-0969-1. [DOI] [PubMed] [Google Scholar]
- 59.Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B, Buluwela L, Weitzman SA, Marks J, Sukumar S. Cancer Res. 2001;61:2782–2787. [PubMed] [Google Scholar]
- 60.Veal E, Eisenstein M, Tseng ZH, Gill G. Mol Cell Biol. 1998;18:5032–5041. doi: 10.1128/mcb.18.9.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mikovits JA, Young HA, Vertino P, Issa JP, Pitha PM, Turcoski-Corrales S, Taub DD, Petrow CL, Baylin SB, Ruscetti FW. Mol Cell Biol. 1998;18:5166–5177. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JO, Kwun HJ, Jung JK, Choi KH, Min do S, Jang KL. Oncogene. 2005;24:6617–6625. doi: 10.1038/sj.onc.1208827. [DOI] [PubMed] [Google Scholar]
- 63.McCabe MT, Low JA, Imperiale MJ, Day ML. Oncogene. 2006;25:2727–2735. doi: 10.1038/sj.onc.1209266. [DOI] [PubMed] [Google Scholar]
- 64.Tsai CN, Tsai CL, Tse KP, Chang HY, Chang YS. Proc Natl Acad Sci USA. 2002;99:10084–10089. doi: 10.1073/pnas.152059399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bachman KE, Rountree MR, Baylin SB. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 66.Poole LJ, Yu Y, Kim PS, Zheng QZ, Pevsner J, Hayward GS. J Virol. 2002;76:3395–3420. doi: 10.1128/JVI.76.7.3395-3420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ben-Yehoyada M, Ben-Dor I, Shaul Y. J Biol Chem. 2003;278:34475–34482. doi: 10.1074/jbc.M301051200. [DOI] [PubMed] [Google Scholar]
- 69.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.