Abstract
Estrogen receptors (ERs) are ligand-activated transcription factors that regulate gene expression and cell growth. Two ERs now have been identified: ERα and the more recently discovered ERβ. The physiological function of ERβ remains unclear, but evidence from vascular injury studies and from ERβ knockout mice suggests that ERβ may be involved in the regulation of cellular proliferation. Here we show a direct and specific interaction between ERβ and the cell cycle mitotic spindle assembly checkpoint protein, MAD2 (mitosis arrest-deficient 2). The ERβ-MAD2 interaction was identified by screening of a yeast two-hybrid system vascular endothelial cell library with ERβ and confirmed with glutathione S-transferase-fusion protein interaction studies. In contrast, ERα did not interact with MAD2 in either the two-hybrid system or in the protein–protein interaction experiments. Amino acids 173–208 in the hinge region of ERβ were sufficient to mediate the interaction with MAD2 in the two-hybrid system and in glutathione S-transferase-fusion protein studies. These data identify a link between ERβ and MAD2 of potential importance to regulation of the cell cycle and support a function of ERβ distinct from the established role of ERs as transcription factors.
The steroid hormone receptors are members of a large superfamily of ligand-activated transcription factors that regulate gene expression and influence the growth and function of target cells and tissues (1–4). There are two estrogen receptors (ERs) known at present: ERα (3) and ERβ (4), which share significant homology in their DNA and hormone binding domains (BDs). However, ERβ is structurally and functionally distinct from ERα, and its physiological role is not yet understood (4–7). ERs interact with a variety of intracellular proteins. In the unliganded state, ERs are found in protein complexes that include several heat shock proteins and immunophilin molecules (8). ERs activated by hormone binding can recruit a variety of proteins, including coactivator transcriptional proteins (9, 10), proteins that silence transcription (corepressors) (10, 11), other transcription factors (1, 10), and proteins that remodel chromatin (12). However, no proteins that interact specifically with ERβ have been identified to date.
In mammals, ERβ mRNA is found in many tissues, including uterus, ovary, testes, bladder, prostate, lung, and brain (4, 5), and in the heart and blood vessels of the cardiovascular system (refs. 7 and 13–16; M.E.M., unpublished observations). Some data suggest a specific role for ERβ in mediating the cardiovascular effects of estrogen (7), because estrogen protects against vascular injury in mice with no ERα (13), and ERβ, but not ERα, expression is markedly stimulated in vascular cells after vascular injury (14, 16). However, in mice harboring a disruption of the ERβ gene, estrogen is still able to protect against vascular injury (17). These ERβ knockout mice display arrested ovarian folliculogenesis and decreased ovarian efficiency, and older male animals show hyperplasia in the cells of the bladder and prostate (18). Thus, at present, the function of ERβ remains unclear. To better understand the function of ERβ, we sought to identify ERβ-specific binding proteins from vascular endothelial cells, which are known to express high levels of ERβ after vascular injury (14, 16). We characterize here a specific interaction between ERβ and the cell cycle spindle assembly checkpoint protein, MAD2 (mitosis arrest-deficient 2).
Methods
Construction and Screening of Endothelial Cell Yeast Two-Hybrid Library.
A yeast two-hybrid system cDNA library was constructed in the pGAD10 Gal4 activation domain (AD) vector according to the manufacturer's instructions (CLONTECH), by using a total of 10 μg of poly(A) RNA from ovine pulmonary artery endothelial cells (PAEC) (19). The full-length coding region of mouse ERβ (13) was cloned into the EcoRI site of the Gal4 DNA BD vector and mERβ-GalBD was transformed with the PAEC library into Saccharomyces cerevisiae strain Y190 (CLONTECH) by using the lithium acetate method. Polypeptides interacting with ERβ were detected by their ability to reconstitute the GAL4 transcription factor and activate transcription of HIS3 and lacZ reporter genes. Colonies able to grow on HIS-deficient media were assayed for β-galactosidase activity by using a colony lift nitrocellulose filter (MSSI, Westboro, MA) assay. PGAD10 plasmids from His+,LacZ+ colonies were isolated, and cDNA inserts were sequenced in standard fashion. The N-terminal mERβ mutants (all mutants described by amino acid residues), mERβ 1–208 and 1–172, were constructed from full-length mERβ cDNA by digestion with SmaI or BamHI, respectively. A C-terminal mERβ truncation mutant was constructed by excising a BamHI mERβ fragment to create mERβ 173–485. The mERβ 173–208 mutant was created by PCR and verified by DNA sequencing. The MAD2 7–160 truncation mutant of the ovine MAD2 clone EC-1 was constructed by an EcoRI/Bgl2 digestion of the EC-1-PGAD construct and subsequent subcloning of this N-terminal fragment of MAD2 into the Gal-AD plasmid. The MAD2 45–205 truncation mutant of the ovine MAD2 clone EC-1 was constructed by an XhoI/EcoRI digestion of the EC-1-PGAD construct and subsequent subcloning of this C-terminal fragment of MAD2 into the pGAD Gal4-AD plasmid. In some instances, protein expression of mERβ truncation mutants was confirmed by immunoblotting of yeast lysates using a rabbit polyclonal anti-Gal4 DNA BD antibody (Upstate Biotechnology, Lake Placid, NY).
Glutathione S-Transferase (GST)-Fusion Protein Assays.
EC-1, mERβ1–208, and the C-terminal mERβ truncation mutant mERβ173–485 were ligated into the GST-fusion protein expression vector pGEX-4T-1 (Amersham Pharmacia Biotech). The mERβ 173–208 mutant was constructed from a BamHI digest of the mERβ cDNA fragment corresponding to amino acids 1–208. Full-length rat ERβ ligated into pGEX-2TK was created as described (9). The GST-mERβ1–208, GST-mERβ 173–485, GST-mERβ 173–208, and full-length GST-rERβ proteins were expressed in Escherichia coli (XL10-Gold/Stratagene, BL 21/Novagen), expression was confirmed and quantified by SDS/PAGE, and GST-fusion proteins were immobilized on glutathione Sepharose beads for pull-down assays, as described (20). Recombinant [S35]mERβ, [S35]hERα, and [S35]hsMAD2 were produced by in vitro transcription/translation (TNT Coupled Reticulocyte Lysate Systems Kit, Promega) from ERα, ERβ, and human MAD2 cDNA templates [pET28a(+)hsMAD2, kind gift of R. Benezra, Cornell University, New York]. Recombinant proteins were incubated with GST-fusion proteins at 4°C for 1 hr in suspension buffer (20 mM Tris, pH 7.4/137 mM NaCl/2 mM EDTA/1% Triton X-100/10% glycerol/25 mM β-glycerolphosphate/1 mM Na vanadate/10 μg/ml leupeptin/10 μg/ml aprotinine/1 mM PMSF). Beads then were washed three times, and associated proteins were resolved by SDS/PAGE and visualized by autoradiography.
Results
Yeast Two-Hybrid Screen.
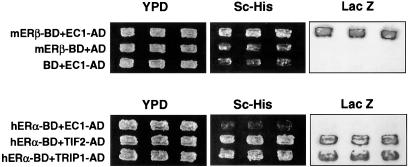
To identify proteins that associate with ERβ, a vascular endothelial cell cDNA yeast two-hybrid library was constructed from ovine pulmonary artery endothelial cells (19, 21), and 2.5 × 10 5 library clones were screened with the full-length coding sequence of mouse ERβ (13). Forty positive clones were identified during the screening, and of these 35 proved by sequencing and restriction enzyme mapping analyses to be identical to clone EC-1. EC-1 is a 2.3-kb clone that, with ERβ, consistently transactivated both histidine and β-galactosidase reporters in the two-hybrid system (Fig. 1 and Table 1). The ERβ interaction with EC-1 did not require estradiol, nor was it disrupted or enhanced by the hormone (up to 10−6 M estradiol). The interaction between ERβ and EC-1 was specific, as ERα consistently was unable to interact with EC-1 under conditions in which ERα interacted with both of the known ER coactivator proteins, TIF2 and TRIP1 (Fig. 1 Lower and Table 1). Sequence analysis of EC-1 revealed an ORF with 93% homology to nucleotides 19–618 of the human MAD2 mitotic checkpoint cDNA (22) (Genbank accession no. number U65410), encoding an ovine protein of 199 aa with 98% homology to amino acids 7–205 of human MAD2 (Fig. 2). MAD2 is a 24-kDa protein that associates with unattached kinetechores (23, 24) and coimmunoprecipitates with the anaphase-promoting complex, a ubiquitin ligase that regulates progression through the mitotic cycle by targeting destruction of the proteins that allow the onset of anaphase to proceed and mitotic exit to occur (23–26) (see Discussion).
Figure 1.
Identification of an interaction between ERβ and MAD2 in the yeast two-hybrid system. (Upper) ERβ fused to the Gal4 BD (mERβ-BD) interacts with clone EC-1 (EC-1-AD) isolated from the ovine pulmonary artery endothelial cell yeast two-hybrid system library. ERβ-BD and the Gal AD vector alone fail to interact, as do the BD vector alone and EC-1-AD. The positive interaction of mERβ-BD with EC-1-AD occurs similarly in the presence and the absence of estrogen (data not shown). (Lower) ERα-BD failed to interact with EC-1-AD in the two-hybrid system in the presence of 10−8 M estradiol under conditions in which ERα-BD interacted with both of the known nuclear hormone receptor coactivator proteins TIF2 and TRIP1.
Table 1.
MAD2 interactions with ERβ and ERα in the yeast two-hybrid system
| BD | AD | LacZ assay |
|---|---|---|
| mERβ | EC-1 (MAD2 7-205) | + |
| mERβ | MAD2 45-205 | − |
| mERβ 1-208 | EC-1 | + |
| mERβ 1-172 | EC-1 | − |
| mERβ 173-208 | EC-1 | + |
| BD alone | EC-1 | − |
| EC-1 | mERβ | + |
| EC-1 | mERβ 173-485 | + |
| EC-1 | mERβ 209-485 | − |
| EC-1 | — | − |
| EC-1 | AD alone | − |
| mERβ | MAD2 7-160 | + |
| mERβ 1-208 | MAD2 7-160 | − |
| mERβ 173-208 | MAD2 7-160 | − |
| hERα | EC-1 | − |
| hERα | mERβ | + |
| hERα | TIF2 | + |
| hERα | TRIP1 | + |
Summary of histidine and β-galactosidase reporter activation for various ERβ and ERα constructs tested for their interaction in the yeast two-hybrid system with the MAD2 clone EC-1 (corresponding to amino acids 7-205 of hMAD2) and MAD2 mutants.
Figure 2.
Sequence of clone EC-1 and comparison of predicted amino acid sequence to human MAD2. Alignment of the predicted amino acid sequence of ovine EC-1 with human MAD2 reveals 98% identity between the two proteins. Amino acid identities are shown by the shading and amino acid mismatches by a gap. The EC-1 clone did not contain sequence corresponding to the first 6 aa of hMAD2 (Genbank accession no. U65410).
Yeast two-hybrid system studies were used next to define the ERβ domain that interacts with the MAD2 clone EC-1 (Table 1). Full-length ERβ-Gal4 BD interacted with clone EC-1 in the Gal4 AD in these studies. In contrast, a fragment of EC-1 corresponding to amino acids 45–205 of MAD2, when placed in the Gal4 AD, was not able to interact with full-length ERβ-Gal4 BD, supporting the idea that the N-terminal domain of EC-1 is necessary to allow the interaction with ERβ in the yeast two-hybrid system. An N-terminal fragment of ERβ of 208 aa in the Gal4 BD construct also was able to interact with EC-1 in the Gal4 AD construct. Furthermore, ERβ-Gal4 BD truncation mutants lacking amino acids 173–207 were unable to interact with EC-1 in the two-hybrid studies, whereas the cDNA encoding the peptide for amino acids 173–208 of ERβ in the Gal4 BD was able to interact with EC-1 (Table 1). The amino acids 173–208 of ERβ correspond to the hinge region of the receptor (4) and have a sequence distinct from the corresponding region in other steroid hormone receptors, including the closely related ERα. In other experiments, EC-1-Gal4 BD was able to interact with full-length ERβ or ERβ 173–485 in the Gal4 AD, but not with ERβ 209–485. When the EC-1 fragment corresponding to N-terminal amino acids 7–160 of MAD2 was expressed in the Gal4 AD, it was able to interact with full-length ERβ-Gal4 BD, but not with mutants of ERβ lacking amino acids C-terminal to amino acid 208 (Table 1). Taken together, these studies raise the possibility that amino acids C-terminal to residue 208 of ERβ and/or amino acids 160–205 of MAD2 somehow contribute to the interaction between the two proteins in this Gal4-based yeast two-hybrid system. Finally, ERα was unable to interact with EC-1 under conditions in which it interacted with ERβ in the expected heterodimer interaction between the two ERs (Table 1).
GST–Protein-Protein Interaction Studies.
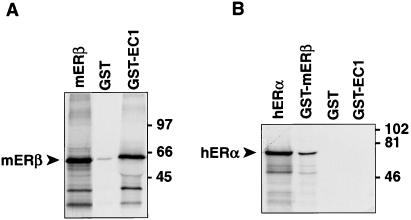
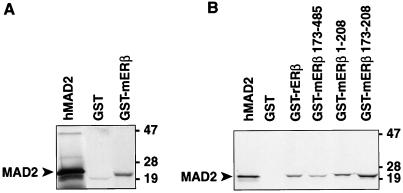
The interaction between ERβ and MAD2 also was examined by GST-fusion protein–protein interaction (GST pull-down) studies (Figs. 3 and 4). A GST fusion protein expressing the MAD2 amino acids 7–205 encoded by clone EC-1 was able to bind ERβ (Fig. 3A). In contrast, GST-MAD2 protein was not able to bind ERα under conditions in which the recognized heterodimer interaction (27, 28) between ERα and ERβ was demonstrable (Fig. 3B). These experiments confirm the interaction between MAD2 and ERβ but not ERα seen in the two-hybrid system studies and the specificity of the ERβ-MAD2 interaction. GST-ERβ also was able to interact with MAD2 protein in GST fusion protein–protein interaction studies (Fig. 4A). To explore what domain of ERβ is sufficient to support the interaction between ERβ and MAD2, a series of experiments also was performed by using full-length ERβ and the various truncation mutants of ERβ studied above in the yeast two-hybrid system (Fig. 4B). GST fusion proteins expressing full-length ERβ, ERβ amino acids 173–485, and ERβ amino acids 1–208 all bound MAD2 in these studies (Fig. 4B). Furthermore, the small peptide corresponding to amino acids 173–208 of ERβ also bound MAD2, as it did in the two-hybrid system, confirming that this domain is sufficient for the ERβ-MAD2 interaction (Fig. 4B).
Figure 3.
Protein–protein interaction studies with ERβ, ERα, and MAD2. (A) Protein–protein interaction studies with [S35]mERβ and GST-EC-1. [S35]mERβ was incubated with GST beads alone or GST-EC-1 beads followed by SDS/PAGE and autoradiography. The first lane shows 10% of [S35]mERβ input. (B) Protein–protein interaction studies with [S35]hERα and ERβ or EC-1. [S35]hERα was incubated with GST-mERβ (positive control), GST beads alone, or GST-EC-1, followed by SDS/PAGE and autoradiography. The first lane shows 7% of [S35]hERα input.
Figure 4.
Protein–protein interaction studies with full-length or truncated ERβ and MAD2. (A) Protein–protein interaction studies with [S35]hMAD2 and mERβ. [S35]MAD2 was incubated with GST beads alone or GST-mERβ beads followed by SDS/PAGE and autoradiography. The first shows 15% of [S35]MAD2 input. (B) Protein–protein interaction studies with [S35]hMAD2 and full-length ERβ or several ERβ truncation mutants. [S35]MAD2 was incubated with GST beads alone, GST-full-length ERβ (GST-rERβ), GST-mERβ173–485, GST-mERβ1–208 beads, or GST-mERβ 173–208, followed by SDS/PAGE and autoradiography. The first lane shows 7% of [S35]MAD2 input.
Discussion
The studies described here identify a specific interaction between ERβ and the mitotic spindle assembly checkpoint protein, MAD2. Both yeast two-hybrid system and GST pull-down experiments demonstrate the interaction of amino acids 173–208 of ERβ with MAD2 and show that the interaction between ERβ and MAD2 is not altered by the absence or presence of estradiol. ERα does not interact with MAD2 in these studies, and thus MAD2 is the first protein identified that interacts with ERβ but not ERα.
The MAD genes were identified first in yeast (22, 29) and the vertebrate homologue of MAD2 was shown to associate with kinetechores of chromosomes in early prometaphase cells (23, 24, 30). Association of MAD2 with the anaphase-promoting complex (APC) arrests cellular progression through mitosis by binding to and inhibiting Cdc20/Fzy, an essential activator of the APC, which allows the onset of anaphase to proceed and mitotic exit to occur (25, 26, 30). Based on experiments with anti-MAD2 antibodies, MAD2 also regulates the onset of anaphase in mammalian cells without obvious spindle defects (30), suggesting that MAD2 and the signaling pathway it regulates are activated during normal cell cycles. It is therefore possible that ERβ is a component of the spindle checkpoint assembly. It is also possible that the ERβ-MAD2 interaction described here is important in protecting or promoting the fidelity of replication for cells that are proliferating rapidly, such as the blood vessel cells that are involved in the reparative process after vascular injury, in which ERβ is markedly induced (14, 16). Alternatively, the ERβ-MAD2 interaction may be important for some purpose distinct from MAD2 regulation of the spindle checkpoint. The MAD2 protein also interacts with the cytoplasmic domain of the insulin receptor (31) and with the MHC ubiquitin-like protein FAT10 (32), but the significance of these interactions is unknown at present. MAD2 also encodes the α-subunit of a prenyltransferase and is required in yeast for the membrane association of several monomeric GTP-binding proteins (29, 33). It is therefore possible that this enzymatic activity of MAD2 is recruited by ERβ for an as-yet-unidentified function.
In summary, we have identified a specific interaction between ERβ and the spindle assembly checkpoint protein MAD2. This interaction may be related to the induction of ERβ mRNA that occurs in rapidly proliferating cells, such as vascular cells after vascular injury (14, 16), and supports the hypothesis that ERβ has some function distinct from those it shares with ERα. The ERβ-MAD2 interaction identified here also supports that ERβ has a role in cellular proliferation that is distinct from its function as a transcription factor.
Acknowledgments
This paper is for Kathy Weingarten. We thank Richard Karas and Howard Surks for helpful discussions and Marc Vidal for reagents and advice regarding the two-hybrid screens. Y.K. is a Research Fellow of the Japan Society for the Promotion of Science and also is supported by the Uehara Memorial Foundation. This work was supported in part by National Institutes of Health Grants HL53546 (P.W.S.), CA57374 (M.B.), and HL56069 and HL59953 (M.E.M.). MEM is an Established Investigator of the American Heart Association.
Abbreviations
- ER
estrogen receptor
- GST
glutathione S-transferase
- MAD2
mitosis arrest-deficient 2
- BD
binding domain
- AD
activation domain
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050580997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050580997
References
- 1.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown M. Hematol Oncol Clin North Am. 1994;8:101–112. [PubMed] [Google Scholar]
- 3.Walter P, Green S, Krust A, Bornert J M, Jeltsch J M, Staub A, Jensen E, Scrace G, Waterfield M, Chambon P. Proc Natl Acad Sci USA. 1985;82:7889–7893. doi: 10.1073/pnas.82.23.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuiper G G J M, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J Å. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuiper G G J M, Carlsson B, Grandien K, Enmark E, Häggblad J, Nilsson S, Gustafsson J Å. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 6.Paech K, Webb P, Kuiper G G J M, Nilsson S, Gustafsson J Å, Kushner P J, Scanlan T S. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 7.Mendelsohn M E, Karas R H. N Engl J Med. 1999;340:1801–1811. doi: 10.1056/NEJM199906103402306. [DOI] [PubMed] [Google Scholar]
- 8.Pratt W B, Toft D O. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 9.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 10.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M-J, O'Malley B W. Recent Prog Horm Res. 1997;52:141–165. [PubMed] [Google Scholar]
- 11.Montano M M, Ekena K, Delage-Mourroux R, Chang W, Martini P, Katzenellenbogen B S. Proc Natl Acad Sci USA. 1999;96:6947–6952. doi: 10.1073/pnas.96.12.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichinose H, Garnier J M, Chambon P, Losson R. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- 13.Iafrati M D, Karas R H, Aronovitz M, Kim S, Sullivan T R, Jr, Lubhan D B, O'Donnell T F, Jr, Korach K S, Mendelsohn M E. Nat Med. 1997;3:545–548. doi: 10.1038/nm0597-545. [DOI] [PubMed] [Google Scholar]
- 14.Lindner V, Kim S K, Karas R H, Kuiper G G J M, Gustafsson J-Å, Mendelsohn M E. Circ Res. 1998;83:224–229. doi: 10.1161/01.res.83.2.224. [DOI] [PubMed] [Google Scholar]
- 15.Register T C, Adams M R. J Steroid Biochem Mol Biol. 1998;64:187–191. doi: 10.1016/s0960-0760(97)00155-6. [DOI] [PubMed] [Google Scholar]
- 16.Makela S, Savolainen H, Aavik E, Myllarniemi M, Strauss L, Taskinen E, Gustafsson J, Hayry P. Proc Natl Acad Sci USA. 1999;96:7077–7082. doi: 10.1073/pnas.96.12.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karas R H, Hodgin J B, Kwoun M, Krege J H, Aronovitz M A, Mackey W, Gustafsson J A, Korach K, Smithies O, Mendelsohn M E. Proc Natl Acad Sci USA. 1999;96:15133–15136. doi: 10.1073/pnas.96.26.15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krege J H, Hodgin J B, Couse J F, Enmark E, Warner M, Mahler J F, Sar M, Korach K S, Gustafsson J, Smithies O. Proc Natl Acad Sci USA. 1998;95:15677–15682. doi: 10.1073/pnas.95.26.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pace M C, Chambliss K L, German Z, Yuhanna I S, Mendelsohn M E, Shaul P W. Am J Physiol. 1999;277:L106–L112. doi: 10.1152/ajplung.1999.277.1.L106. [DOI] [PubMed] [Google Scholar]
- 20.Wang G R, Zhu Y, Halushka P V, Lincoln T M, Mendelsohn M E. Proc Natl Acad Sci USA. 1998;95:4888–4893. doi: 10.1073/pnas.95.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Yuhanna I S, Galcheva-Gargova Z I, Karas R H, Mendelsohn M E, Shaul P W. J Clin Invest. 1999;103:401–406. doi: 10.1172/JCI5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Murray A W. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 23.Chen R, Waters J C, Salmon E D, Murray A W. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Benezra R. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang G W, Yu H T, Kirschner M W. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowley S M, Hoare S, Mosselman S, Parker M G. J Biol Chem. 1997;272:19858–19862. doi: 10.1074/jbc.272.32.19858. [DOI] [PubMed] [Google Scholar]
- 28.Pettersson K, Grandien K, Kuiper G G J M, Gustafsson J Å. Mol Endocrinol. 1997;11:1486–1496. doi: 10.1210/mend.11.10.9989. [DOI] [PubMed] [Google Scholar]
- 29.Li R, Havel C, Watson J A, Murray A W. Nature (London) 1993;366:82–84. doi: 10.1038/366082a0. [DOI] [PubMed] [Google Scholar]
- 30.Gorbsky G J, Chen R, Murray A W. J Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Neill T J, Zhu Y, Gustafson T A. J Biol Chem. 1997;272:10035–10040. doi: 10.1074/jbc.272.15.10035. [DOI] [PubMed] [Google Scholar]
- 32.Liu M, Pan J, Zhang C, Fan W, Collinge M, Bender J R, Weissman S M. Proc Natl Acad Sci USA. 1999;96:4313–4318. doi: 10.1073/pnas.96.8.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Y l, Rossi G, Ferro-Novick S. Nature (London) 1993;366:84–86. doi: 10.1038/366084a0. [DOI] [PubMed] [Google Scholar]