Abstract
While much study has gone into characterizing virulence factors that play a general role in disease, less work has been directed at identifying pathogen factors that act in a host-specific manner. Understanding these factors will help reveal the variety of mechanisms used by pathogens to suppress or avoid host defenses. We identified candidate Pseudomonas syringae host-specific virulence genes by searching for genes whose distribution among natural P. syringae isolates was statistically associated with hosts of isolation. We analyzed 91 strains isolated from 39 plant hosts by DNA microarray-based comparative genomic hybridization against an array containing 353 virulence-associated (VA) genes, including 53 type III secretion system effectors (T3SEs). We identified individual genes and gene profiles that were significantly associated with strains isolated from cauliflower, Chinese cabbage, soybean, rice, and tomato. We also identified specific horizontal gene acquisition events associated with host shifts by mapping the array data onto the core genome phylogeny of the species. This study provides the largest suite of candidate host-specificity factors from any pathogen, suggests that there are multiple ways in which P. syringae isolates can adapt to the same host, and provides insight into the evolutionary mechanisms underlying host adaptation.
PATHOGENS cannot cause disease indiscriminately, but are generally capable of avoiding or suppressing defenses in only a relatively small set of hosts. Both pathogen factors and host factors govern these interactions, and which of these two is dominant likely depends on the specific interaction. Nevertheless, there is a growing body of data that clearly demonstrates that pathogens have evolved compatibility factors that facilitate the disease process in a host-specific manner. (Cornelis 2002; Abramovitch and Martin 2004; Alfano and Collmer 2004; Espinosa and Alfano 2004; Nomura et al. 2005). However, there are several key issues about pathogen host-specificity factors that are still unknown. For example, the general relationship between virulence factors and host-specificity factors is unclear, although it is likely that the latter are a subset of the former required only on select hosts. It is also not known if the factors that are responsible for pathogen compatibility on different host species are fundamentally different from the factors that determine compatibility among different cultivars of the same host species. We do not even know at what level host specificity acts most strongly. For example, host-specificity factors may play important roles during initial colonization, during pathogen migration to the appropriate tissue or cell type, during the initiation of cellular interactions, during the maintenance of these interactions, or even at the point where the pathogen disperses to a new host (Levin 1996).
To address these questions, we first must have a way of identifying host-specific virulence factors. A reasonable hypothesis is that strains that are able to infect and cause disease on common host will share a common set of host-specificity factors. Bacterial genomes are remarkably flexible with respect to their specific complements of genes (Hacker and Kaper 2000; Dobrindt and Hacker 2001; Hacker and Carniel 2001; Dobrindt et al. 2002); consequently, associations between the genetic complement of pathogens and their hosts can be used to guide the search for host-specificity factors. One method that can be employed to identify these associations is microarray-based comparative genomic hybridization (CGH) (Behr et al. 1999; Malloff et al. 2001; Taboada et al. 2005). In this approach, genomic DNA from a variety of tester strains is hybridized against the arrayed genome of a reference or target strain. The relative hybridization intensity at each arrayed locus corresponds to the likelihood that the gene is present in the tester strains. Bacterial CGH studies have been extraordinarily successful in quantifying genome variability and characterizing the differences between pathogenic and nonpathogenic strains, but to date this approach has not been used for the large-scale identification of host-associated virulence factors (Behr et al. 1999; Salama et al. 2000; Hakenbeck et al. 2001; Israel et al. 2001; Malloff et al. 2001; Perrin et al. 2002; Saunders et al. 2004; Taboada et al. 2004; Tsolaki et al. 2004; Zhong et al. 2004; Lindroos et al. 2005; Porwollik et al. 2005; Taboada et al. 2005; van Leeuwen et al. 2005; Yao et al. 2005).
We used microarray-based CGH to identify host-associated factors from the model plant pathogen Pseudomonas syringae. P. syringae is an important agricultural pathogen that infects a tremendous diversity of important crop species. Although the host breadth of the species complex as a whole is extremely broad, individual P. syringae isolates typically have a very narrow range of hosts with which they are compatible (Wiebe and Campbell 1993; Cuppels and Ainsworth 1995; Morris et al. 2000). P. syringae is phylogenetically well characterized (Sawada et al. 1999; Sarkar and Guttman 2004; Hwang et al. 2005) and is well known for the large suite of virulence-associated (VA) systems that it uses during pathogenesis. These VA systems are responsible for the production of toxins (Bender et al. 1999), antimicrobials (McDermott et al. 2003), attachment polysaccharides (Yu et al. 1999), quorum-sensing molecules (Von Bodman et al. 2003), nutrient acquisition proteins (Cody and Gross 1984), and secreted effector proteins (Jin et al. 2003). Of these, the factors most commonly implicated in host specificity are the effector proteins secreted through the type III secretion system (T3SS).
The T3SS is a specialized protein secretion system used by both plant and animal pathogens to inject type III secreted effectors (T3SEs) directly into host cells. These effectors are the most promising candidate host-specificity factors since their role in suppressing the host defense response and promoting pathogen growth and transmission is well established (Abramovitch and Martin 2004; Alfano and Collmer 2004; Espinosa and Alfano 2004; Beth Mudgett 2005; Kim et al. 2005; Nomura et al. 2005). Work on bacterial avirulence proteins (which we now know to be predominantly T3SEs) and their cognate plant resistance proteins (Flor 1971) has clearly demonstrated that T3SEs are central to both the promotion and the restriction of host specificity (Jackson et al. 1999; Tsiamis et al. 2000; Guttman and Greenberg 2001; Abramovitch et al. 2003; Losada et al. 2004; Pitman et al. 2005).
P. syringae as a species has the largest known arsenal of T3SEs of any pathogen (Fouts et al. 2002; Guttman et al. 2002; Petnicki-Ocwieja et al. 2002; Zwiesler-Vollick et al. 2002; Greenberg and Vinatzer 2003; Chang et al. 2005). Functional studies have shown that while the T3SS as a whole is required for pathogenesis, many T3SEs have redundant or subtle roles (Lorang and Keen 1995; Charkowski et al. 1998; Guttman and Greenberg 2001; Badel et al. 2002, 2003; Losada et al. 2004; Lim and Kunkel 2005; Lin and Martin 2005). These findings suggest that host-specific virulence may require specific combinations or suites of T3SEs. This hypothesis is supported by preliminary studies demonstrating that P. syringae isolates from different hosts contain distinct suites of T3SEs (Guttman et al. 2002). Consequently, it is important to study T3SE repertoires in toto across the natural range of P. syringae hosts to understand their net function and role in host-specific interactions. CGH provides a powerful method for achieving this goal.
While it is generally assumed that horizontal gene transfer plays an important role in the evolution of these genes, the particular impact of this process and its importance relative to pathoadaptation (Sokurenko et al. 1999) has rarely been quantified. The few studies that have investigated T3SEs in a phylogenetic context have found support for both horizontal gene transfer and pathoadaptation (Prager et al. 2000; Lavie et al. 2004; Rohmer et al. 2004; Lindeberg et al. 2005; Guttman et al. 2006). While this debate still needs to be resolved, one issue that is clear is that many T3SEs are under intense selective pressures and that evolutionary changes in these genes can profoundly impact pathogen–host interactions (Rohmer et al. 2004; Pitman et al. 2005; Guttman et al. 2006; Ma et al. 2006).
We used microarray-based CGH to identify candidate P. syringae host-associated virulence factors. We assayed for the presence of 353 putative VA genes, including 53 T3SEs or putative T3SEs in 91 diverse P. syringae strains isolated from 39 unique hosts. We concentrated most of our analyses on T3SEs since they are the most promising host-specificity candidates. Our reference strain was the sequenced tomato pathogen P. syringae pv. tomato DC3000 (PtoDC3000). We tested for associations between the distribution of specific VA genes and VA gene profiles among isolates and the hosts of isolation to identify candidate host-specific virulence factors. We also mapped our findings onto the core genome phylogeny of the species to clarify the role of horizontal gene transfer in the evolution of these loci and the acquisition of new host specificities. This is the first large-scale population-level study of the association between natural genetic variation among VA genes and the host of isolation for a bacterial pathogen.
MATERIALS AND METHODS
Bacterial strains:
Ninety-one isolates of P. syringae were generously donated by members of the research community as detailed in Table 1. These isolates span the geographic, phenotypic, and evolutionary diversity of the species and have representatives from all five major monophyletic groups (phylogroups) (Sarkar and Guttman 2004; Hwang et al. 2005). The collection includes 27 pathovars isolated from 39 different hosts, contained within 16 plant host families. Given the scope of the study, it was not possible to quantitatively measure host range for the collection. Nevertheless, published studies (Morris et al. 2000; Hunter and Taylor 2006) and our own analyses (W. Ma, R. L. Morgan and D. S. Guttman, unpublished results) have found that the host of isolation quite accurately reflects the ability of particular strains to grow on specific hosts and therefore is a useful surrogate for host specificity. Strains were grown in King's B (King et al. 1954) medium at 30°.
TABLE 1.
Strains
| Pathovar | Designation | Strain name | Place of isolation | Year of isolation | Host | Sourcea | Accession no.a |
|---|---|---|---|---|---|---|---|
| aceris | ATCC10853 | Pac A10853 | Maple | J. Dangl | ATCC10853 | ||
| actinidiae | FTRS_L1 | Pan FTRS_L1 | Japan | 1984 | Kiwi | MAFF | MAFF302091 |
| aesculi | 0893_23 | Pae 0893_23 | United States | Horse chestnut | D. Cooksey | ||
| apii | 1089_5 | Pap 1089_5 | United States | Celery | D. Cooksey | ||
| aptata | 601 | Ptt 601 | 1966 | Sugarbeet | MAFF | MAFF301008 | |
| aptata | DSM50252 | Ptt DSM5022 | Wheat | J. Dangl | |||
| aptata | G733 | Ptt G733 | 1976 | Brown rice | MAFF | MAFF302831 | |
| atrofaciens | DSM50255 | Paf DSM5025 | Wheat | J. Dangl | |||
| broussonetiae | KOZ8101 | Pbr KOZ8101 | Japan | 1980 | Paper mulberry | MAFF | MAFF810036 |
| cilantro | 0788_9 | Pci 0788_9 | United States | Cilantro | D. Cooksey | ||
| coronafaciens | KN221 | Pcn KN221 | 1984 | Oats | MAFF | MAFF302787 | |
| glycinea | BR1 | Pgy BR1 | 1989 | Soybean | MAFF | MAFF210373 | |
| glycinea | KN127 | Pgy KN127 | 1982 | Soybean | MAFF | MAFF302751 | |
| glycinea | KN166 | Pgy KN166 | 1982 | Soybean | MAFF | MAFF302770 | |
| glycinea | KN28 | Pgy KN28 | 1981 | Soybean | MAFF | MAFF302676 | |
| glycinea | KN44 | Pgy KN44 | Japan | 1981 | Soybean | MAFF | MAFF301683 |
| glycinea | LN10 | Pgy LN10 | 1989 | Soybean | MAFF | MAFF210389 | |
| glycinea | MAFF301765 | Pgy M301765 | 1982 | Soybean | MAFF | MAFF301765 | |
| glycinea | MOC601 | Pgy MOC601 | 1994 | Soybean | MAFF | MAFF311113 | |
| glycinea | R4a | Pgy R4a | Soybean | J. Dangl | |||
| glycinea | UnB647 | Pgy UnB647 | Kidney bean | MAFF | MAFF210405 | ||
| japonica | MAFF301072 | Pja M301072 | Japan | 1951 | Barley | MAFF | MAFF301072 |
| lachrymans | 107 | Pla 107 | Cucumber | J. Dangl | MAFF301315 | ||
| lachrymans | 1188_1 | Pla 1188_1 | United States | Zucchini | D. Cooksey | ||
| lachrymans | ATCC7386 | Pla A7386 | Cucumber | J. Dangl | MAFF302278 | ||
| lachrymans | N7512 | Pla N7512 | Japan | 1975 | Cucumber | MAFF | MAFF301315 |
| lachrymans | YM7902 | Pla YM7902 | 1979 | Cucumber | MAFF | MAFF730057 | |
| lachrymans | YM8003 | Pla YM8003 | 1980 | Cucumber | MAFF | MAFF730069 | |
| maculicola | 4981 | Pma 4981 | Zimbabwe | Cauliflower | D. Cuppels | ||
| maculicola | AZ85297 | Pma AZ85297 | 1985 | Chinese cabbage | MAFF | MAFF302539 | |
| maculicola | ES4326 | Pma ES4326 | 1965 | Radish | J. T. Greenberg | ||
| maculicola | H7311 | Pma H7311 | Japan | 1973 | Chinese cabbage | MAFF | MAFF301174 |
| maculicola | H7608 | Pma H7608 | 1976 | Chinese cabbage | MAFF | MAFF301175 | |
| maculicola | KN203 | Pma KN203 | 1983 | Chinese cabbage | MAFF | MAFF302783 | |
| maculicola | KN84 | Pma KN84 | 1982 | Radish | MAFF | MAFF302724 | |
| maculicola | KN91 | Pma KN91 | 1982 | Radish | MAFF | MAFF302731 | |
| maculicola | M6 | Pma M6 | United Kingdom | 1965 | Cauliflower | J. Dangl | |
| maculicola | YM7930 | Pma YM7930 | 1979 | Radish | MAFF | MAFF301419 | |
| mellea | N6801 | Pme N6801 | 1968 | Tobacco | MAFF | MAFF302303 | |
| mori | MAFF301020 | Pmo M301020 | Japan | 1966 | Mulberry | MAFF | MAFF301020 |
| morsprunorum | 19322 | Pmp 19322 | European plum | J. Dangl | ATCC19322 | ||
| myricae | AZ84488 | Pmy AZ84488 | 1984 | Bayberry | MAFF | MAFF302460 | |
| myricae | MAFF302941 | Pmy M302941 | 1989 | Bayberry | MAFF | MAFF302941 | |
| oryzae | 36_1 | Por 36_1 | 1983 | Rice | MAFF | MAFF301538 | |
| oryzae | I_6 | Por I_6 | 1991 | Rice | MAFF | MAFF311107 | |
| phaseolicola | 1302A | Pph 1302A | Ethiopia | 1984 | Kidney bean | D. Arnold | |
| phaseolicola | 1449B | Pph 1449B | Ethiopia | 1985 | Hyacinth bean | D. Arnold | |
| phaseolicola | HB10Y | Pph HB10Y | Snap bean | P. Turner | ATCC21781 | ||
| phaseolicola | KN86 | Pph KN86 | Japan | 1982 | Kidney bean | MAFF | MAFF301673 |
| phaseolicola | NPS3121 | Pph NPS3121 | Kidney bean | J. T. Greenberg | |||
| phaseolicola | NS368 | Pph NS368 | 1992 | Kidney bean | MAFF | MAFF311004 | |
| phaseolicola | R6a | Pph R6a | Kidney bean | J. Dangl | |||
| phaseolicola | SG44 | Pph SG44 | United States | 1980 | Snap bean | S. Hirano | |
| phaseolicola | Y5_2 | Pph Y5_2 | Kudzu | MAFF | MAFF311162 | ||
| pisi | 895A | Ppi 895A | Pea | D. Arnold | |||
| pisi | H5E1 | Ppi H5E1 | 1993 | Pea | MAFF | MAFF311141 | |
| pisi | H5E3 | Ppi H5E3 | 1993 | Pea | MAFF | MAFF311143 | |
| pisi | H6E5 | Ppi H6E5 | 1994 | Pea | MAFF | MAFF311144 | |
| pisi | H7E7 | Ppi H7E7 | 1995 | Pea | MAFF | MAFF311146 | |
| pisi | PP1 | Ppi PP1 | Japan | 1978 | Pea | MAFF | MAFF301208 |
| pisi | R6a | Ppi R6a | Pea | J. Dangl | |||
| savastanoi | 4352 | Psv 4352 | Yugoslavia | Olive | A. Colmer | ||
| sesami | HC_1 | Pse HC_1 | Sesame | MAFF | MAFF311181 | ||
| syringae | 1212R | Psy 1212R | Pea | D. Arnold | |||
| syringae | A2 | Psy A2 | Ornamental pear | C. Bender | |||
| syringae | B48 | Psy B48 | United States | Peach | T. Denny | ||
| syringae | B64 | Psy B64 | United States | Wheat | T. Denny | ||
| syringae | B728A | Psy B728A | United States | Snap bean | S. Hirano | ||
| syringae | B76 | Psy B76 | United States | Tomato | T. Denny | ||
| syringae | FF5 | Psy FF5 | United States | 1998 | Ornamental pear | C. Bender | |
| syringae | FTRS_W6601 | Psy FTRS_W6 | 1966 | Japanese apricot | MAFF | MAFF301429 | |
| syringae | FTRS_W7835 | Psy FTRS_W7 | 1978 | Japanese apricot | MAFF | MAFF301430 | |
| syringae | L177 | Psy L177 | 1983 | Lilac | MAFF | MAFF302085 | |
| syringae | LOB2_1 | Psy LOB2_1 | Japan | 1986 | Lilac | MAFF | MAFF301861 |
| syringae | NCPPB281 | Psy NCPPB28 | United Kingdom | Lilac | B. Hancock | ATCC19310 | |
| syringae | Ps9220 | Psy Ps9220 | 1992 | Spring onion | MAFF | MAFF730125 | |
| syringae | PSC1B | Psy PSC1B | United States | Corn | T. Denny | ||
| tabaci | 6606 | Pta 6606 | Japan | 1967 | Tobacco | MAFF | MAFF301612 |
| thea | K93001 | Pth K93001 | 1993 | Tea | MAFF | MAFF302851 | |
| tomato | 487 | Pto 487 | Greece | Tomato | D. Cuppels | ||
| tomato | 1318 | Pto 1318 | Switzerland | Tomato | D. Cuppels | ||
| tomato | 2170 | Pto 2170 | 1984 | Tomato | MAFF | MAFF301591 | |
| tomato | DC3000 | Pto DC3000 | United Kingdom | Tomato | J. T. Greenberg | ||
| tomato | DC84_1 | Pto DC84_1 | Canada | Tomato | D. Cuppels | ||
| tomato | DC89_4H | Pto DC89_4H | Canada | Tomato | D. Cuppels | ||
| tomato | DCT6D1 | Pto DCT6D1 | Canada | Tomato | D. Cuppels | ||
| tomato | KN10 | Pto KN10 | 1981 | Tomato | MAFF | MAFF302665 | |
| tomato | PT23 | Pto PT23 | United States | 1986 | Tomato | N. T. Keen | |
| tomato | TF1 | Pto TF1 | United States | 1997 | Tomato | S. Hirano | |
| — | Cit7 | Ps Cit7 | Navel orange | S. Lindow | |||
| — | TLP2 | Ps TLP2 | Potato | S. Lindow |
MAFF, Japanese Ministry of Agriculture, Forestry, and Fisheries. ATCC, American Type Culture Collection.
DNA isolation and PCR:
We selected 353 genes for the array on the basis of their bioinformatic assignment (Buell et al. 2003) to classes of loci known or suspected to play roles in virulence (supplemental Table 8 at http://www.genetics.org/supplemental/). These included genes responsible for the biosynthesis or assembly of alginate (a); coronatine (c); flagellar apparatus (f); general secretion pathway (g); siderophore (h); general metabolism (m); hemolysin (o); pyoverdine (p); quorum sensing (q); transcriptional regulation (r); polysaccharide (s); T3SS apparatus (t); T3SEs, putative T3SEs, and associated genes (te); unknown function (u); type IV pilus (v); and yersiniabactin (y). The genes of unknown function included on the array were upregulated in preliminary microarray studies of P. syringae during infection of Arabidopsis (G. B. Martin, unpublished results). Putative T3SEs and associated genes include those that encode some of the T3SS chaperones, genes located in the conserved effector locus (CEL) (Alfano et al. 2000), and some unannotated ORFs that have characteristics of T3SEs (Lindeberg et al. 2005). Only unique sequences of each locus were used, and in many cases only part of the coding sequence was used to eliminate cross-hybridization with paralogs or xenologs. The array was also spotted with four housekeeping genes from each of 24 P. syringae strains. These loci from recent multilocus sequence typing (MLST) studies (Sarkar and Guttman 2004; Hwang et al. 2005) were used as internal hybridization controls (discussed below). It is important to note that although some of the VA genes may not be functional in PtoDC3000, they still may have functional orthologs in other strains. Additionally, it is recognized that using only PtoDC3000 genes does result in an ascertainment bias since there are undoubtedly other VA genes not found in this strain that will be important for host-specific virulence. Nevertheless, the aim of the study is to identify genes associated with host-specific virulence, not to identify the gene(s) that strictly determine host specificity. While the latter goal is more lofty, it is also limited by our very incomplete understanding of the composition of the flexible genome of the P. syringae species complex. Nonetheless, this limitation in no way precludes us from identifying VA genes that have statistically supported associations with particular hosts and therefore putatively play a role in host-specific virulence.
Genomic DNA was isolated using the PureGene DNA isolation kit (Gentra Systems, Research Triangle Park, NC) according to the manufacturer's instructions and spectrophotometrically quantified. PtoDC3000 genes were PCR amplified using a 185-ng PtoDC3000 template, primers at a concentration of 0.36 μm, and the cycling parameters 94° for 3 min, 40 cycles at 94° for 1 min, 62° for 1 min, 72° for 2 min, and 72° for 15 min. Over 99% successfully yielded a single product as analyzed on 96-well Ready-To-Run precast agarose gels (Amersham, Buckinghamshire, UK). Clean up was performed using the QIAquick 96 PCR purification kit (QIAGEN, Chatsworth, CA). Primer sequences are presented in supplemental Table 9 at http://www.genetics.org/supplemental/.
Microarray techniques:
Samples were vacuum dried, resuspended in 10 μl of CorningPronto! spotting solution, and spotted onto γ-amino-propyl-silane-coated UltraGAPS slides (Corning, Corning, NY) using a MicroGrid Pro arrayer (BioRobotics, Ann Arbor, MI) with 32 MicroSpot2500 printing pins. PCR products were fixed to the modified glass slides by treatment with 250 mJ of UV irradiation followed by a 2-hr incubation at 85°. Each gene was spotted twice per slide.
Genomic DNA was sheared by sonication (Branson sonifier) to generate fragment sizes between 0.5 and 1 kb. Approximately 1 μg of gDNA per microarray experiment was Klenow-labeled (Promega, Madison, WI) with either Cy3 or Cy5 using random primers (Promega). Reactions were purified using QIAGEN Qiaquick spin columns. Labeling efficiency was on average 1 pmol/μl and 0.7 pmol/μl of incorporated Cy3 or Cy5 fluorescent nucleotide, respectively. Equal amounts of Cy3- and Cy5-labeled genomic DNA representing two different strains were mixed, vacuum dried, dissolved in 65 μl of universal hybridization solution, and denatured at 95° for 5 min. Hybridization was performed for 13–16 hr at 42° using 0.25 pmol/μl of labeled sheared genomic DNA using the Promega Pronto! kit. Scanning of slides was performed using a two-channel confocal microarray scanner (ScanArray5000; GSI Lumionics, Boston) and ScanArray software (v3.1; Packard BioChip Technologies, Boston). All microarray experiments were performed at the Center for Gene Expression Profiling at the Boyce Thompson Institute for Plant Research at Cornell University. All hybridization experiments were replicated two or three times.
Data processing:
Numerical signal intensity data were adjusted to account for hybridization inconsistencies across the surfaces of individual slides by implementing the “spatial lowess” algorithm (Cui et al. 2002), which is based on linear scaling of average background values per channel followed by surface correction. Individual spots within Cy3 and Cy5 channels were also corrected for differences in amount of DNA spotted among the genes by linear scaling relative to the signal obtained from the hybridization of the reference strain PtoDC3000. The median hybridization signal intensity was obtained from two independent Cy3-labled hybridizations of PtoDC3000 genomic DNA (the standard deviation between these replicates was negligible). This value was used to determine a spot-specific scale factor, which transformed each signal to a similar intensity. Only one Cy5-labled PtoDC3000 hybridization was performed, so the Cy5 scale factor was calculated from this single value. The signal intensities for each spot were transformed by this scale factor for all subsequent hybridizations.
Scoring and validation:
We developed a new analytical method for identifying homologs of PtoDC3000 VA genes among strains using housekeeping genes as positive hybridization controls. These housekeeping genes were the same loci used in the MLST phylogenetic analysis of the P. syringae core genome (Sarkar and Guttman 2004; Hwang et al. 2005) and are therefore known to be present in all P. syringae isolates. Each chip contained four housekeeping genes from each of 24 strains that span the diversity of the P. syringae species complex. An objective cutoff was determined by identifying the signal intensity of these housekeeping genes from all of the strains not within the phylogroup of the query (probe) strain. The exclusion of strains from within the same phylogroup as the query strain removed a potential source of phylogenetic and sample size bias from the analysis.
The objective cutoff was determined on the basis of an analysis of the fully sequenced genome of the bean pathogen P. syringae pv. syringae B728A (PsyB728A) (Feil et al. 2005). First we bioinformatically determined which PtoDC3000 genes had homologs in PsyB728A by BLASTN analysis using an expected value of <e−10 and a normalized bitscore of >75 (to remove gene fragments). We used this analysis to bin the PsyB728A VA genes into those that are present or divergent. We then analyzed the data from the microarray experiment in which PsyB728A was used as the probe strain and identified a hybridization signal intensity that minimized the type I and type II errors on the basis of bioinformatic analysis. This signal intensity corresponded to the 1.25th percentile value for the housekeeping genes and resulted in a reasonable error rate of 12.9% false presents (type I) and 4.8% false absents (type II). We used the hybridization signal intensity corresponding to the 1.25th percentile for the housekeeping genes as the cutoff for each hybridization, with the specific cutoff determined independently for each hybridization experiment. We raised the housekeeping gene cutoff to the 1.4th percentile for the phylogroup 1 strains on the basis of an analysis of the PtoDC3000 hybridization results and to compensate for the evolutionary closeness of the phylogroup 1 strains to the reference strain PtoDC3000.
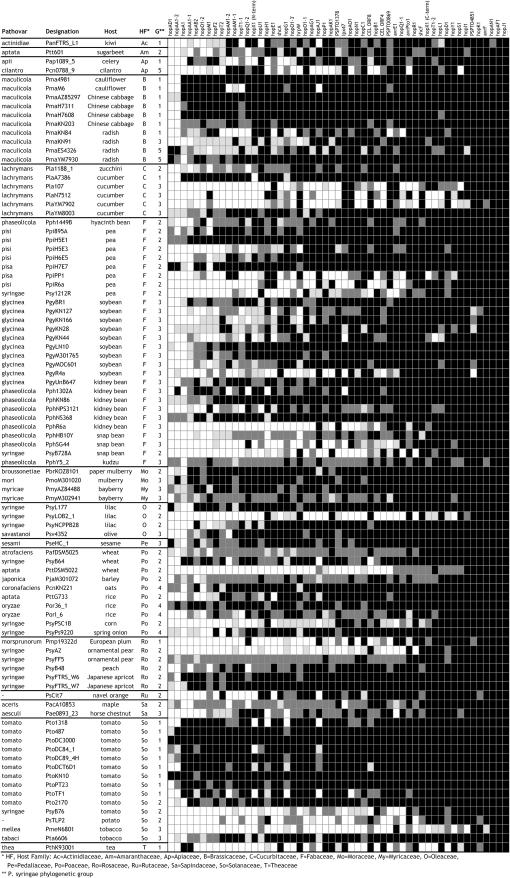
Following the standard convention used in CGH studies, genes with hybridization signal intensities below the cutoff were designated as “divergent” (Chan et al. 2003). In the case of two replicate microarray experiments with two replicate spots per gene, we called genes “present” if all four spots had signal intensities above the cutoff threshold described above. Genes were called “divergent” if none of the four spots had signal intensities above the cutoff. Present genes are solid in Figures 1–3, while divergent genes are open. We also coded genes with light and dark shading to indicate genes that were called divergent or present, respectively, with lower confidence. The genes in the former category had one or two of the four spots above the cut-off threshold, while those in the latter group had three of the four spots above the cutoff.
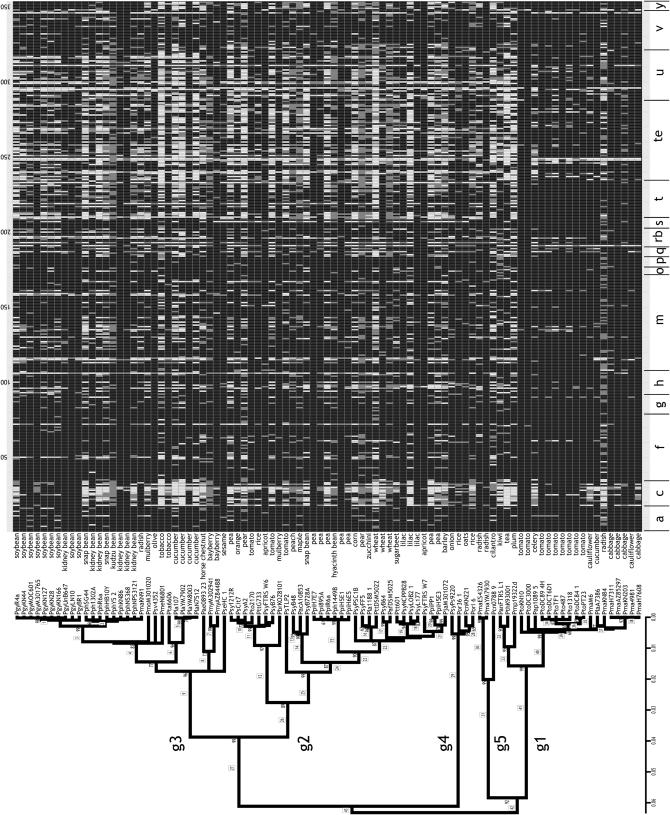
Figure 1.—
Comparative genomic microarray data for 91 P. syringae strains. Strains are arrayed by their evolutionary relationships as determined by MLST (Sarkar and Guttman 2004; Hwang et al. 2005) (neighbor-joining phylogenetic analysis and bootstrap scores from 1000 replicates are presented above each node). The host of each strain is presented after the strain designation. See Table 5 for strain details. The five major P. syringae phylogroups are designated g1–g5. PtoDC3000, which was used as the reference strain for the microarray design, falls into group 1. Numbers in boxes are clade numbers (see text). The solid and open grid represents the microarray data for genomic DNA hybridized against 353 VA genes. Solid, present; open, divergent; dark and light shading, present and divergent with lower confidence, respectively. See materials and methods for details. The VA genes are arranged by functional groups (Table 1). Supplemental Table 10 at http://www.genetics.org/supplemental/ presents the data in a more accessible form.
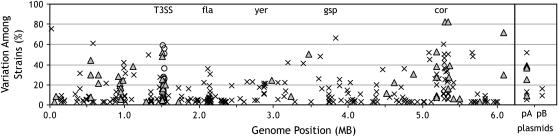
Figure 3.—
Genetic variation and genome organization of VA genes ordered along the PtoDC3000 chromosome. The percentage of variation for each VA gene is plotted according to the genome location of its PtoDC3000 ortholog. The x-axis shows the genome location in megabases; the y-axis shows the variation among strains (0%, no variation and present in all strains). Triangles, effectors; circles, hrp/hrc genes; ×, all other VA genes. T3SS, fla, yer, gsp, and cor approximate locations of the genes encoding T3SS apparatus, flagella apparatus, yersiniabactin biosynthesis, general secretory pathway, and coronatine biosynthesis. pA and pB represent the two endogenous plasmids of PtoDC3000.
Figure 2.—
Presence/divergence data for T3SEs. Strains are arranged on the basis of their host of isolation and the family of that host. T3SEs are arranged by frequency among the strains. See Figure 1 for information on the shading.
A conservative cutoff is essential for CGH studies in that it ensures that all of the genes called present are in fact orthologs of the target sequences. The extremely high frequency of chimeric and truncated T3SEs (Stavrinides et al. 2006) makes a conservative assignment of orthologs even more critical. We empirically determined that the hybridization cutoff as dictated by the signal intensity of the housekeeping genes generally resulted in calling genes present only if they were at ∼80% or more identical to the target sequence. This approach is consistent with that taken in other CGH studies (Evertsz et al. 2001; Malloff et al. 2001; Taboada et al. 2005) and in fact is less conservative than the 88–92% range proposed by Falkow and colleagues (Chan et al. 2003).
Data analysis:
Hierarchical clustering was performed using Cluster (Eisen et al. 1998) by average linkage clustering. Statistical tests for overrepresentation of profiles or genes in isolates of a given host species or family were implemented using bootstrapped permutation tests performed with dedicated PERL scripts (available from D.S.G. upon request).
The bootstrap permutation test for population structure used Watterson's θ, a measure of polymorphism based on the number of segregating sites as a measure of the average genetic distance among strains, and was calculated on the basis of the core genome's MLST housekeeping data set. To determine the statistical significance of these comparisons, we generated a null distribution of θ-values by random bootstrap resampling strains irrespective of the VA genes that they carried. Each bootstrapped data set was composed of the same number of strains as was empirically observed for each effector. One thousand bootstrapped data sets were generated for each effector, and this was compared to the observed θ-value for each individual effector. We performed this analysis for the entire strain set, as well as separately for the three largest P. syringae phylogroups. The latter analysis was performed to remove biases introduced by shared evolutionary history and to control for the fact that comparisons between phylogroups could easily mask any signal that may be present within any one group.
A likelihood-ratio test was used to determine if specific VA genes were statistically associated with specific hosts of isolation or host families. Each VA gene was tested with each host or host family independently. This test compared two models. The first model assumed that there was a strict positive association between individual VA genes and individual hosts or host families, with the probabilities for present and divergent calls being 0.999 and 0.001, respectively. A probability of 0.0 could not be used for the divergent calls since they would have zeroed out the likelihood estimates. The second model assumed that there was no association between the individual VA genes and individual hosts or host families. The probabilities for present and divergent calls were determined empirically from the observed frequency of the gene of interest among all of the strains not isolated from the host of interest. The likelihood-ratio statistic was compared to the chi-square distribution with 1 d.f.
RESULTS AND DISCUSSION
Functional associations:
We grouped each VA gene into a functional group on the basis of its annotation (Buell et al. 2003) (Figure 1, Table 2, supplemental Table 8 at http://www.genetics.org/supplemental/) and assessed which VA functional groups are generally conserved among P. syringae isolates. We used the frequency of individual VA genes among the 91 isolates as an indicator of variation. Following standard population genetics convention used for identifying polymorphic loci, we designated VA genes as “variable” if they were present in <95% of the strains (≤86 strains) and as “conserved” if they were present in >95% of the strains. Conserved genes are likely to encode functions necessary for the survival of all strains regardless of host, while variable genes may include those that encode host- or niche-specific functions.
TABLE 2.
VA gene functional categories
| All strainsa | Phylogroupa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Code | Functional category | N | 1 | 2 | 3 | 4 | 5 | |
| a | Alginate biosynthesis | 17 | 97.4 | 98.9 | 99.1 | 97.4 | 100.0 | 96.1 |
| c | Coronatine biosynthesis | 18 | 75.6 | 76.3 | 74.7 | 74.7 | 88.9 | 96.3 |
| f | Flagellar apparatus | 43 | 95.9 | 98.7 | 95.9 | 96.7 | 98.8 | 93.8 |
| g | General secretion pathway | 13 | 96.8 | 99.0 | 97.2 | 97.6 | 100.0 | 97.4 |
| h | Siderophore | 16 | 87.6 | 96.3 | 84.8 | 86.9 | 93.8 | 81.3 |
| m | Metabolism | 64 | 92.1 | 96.9 | 93.1 | 90.0 | 98.0 | 90.1 |
| o | Hemolysin | 5 | 95.4 | 98.2 | 96.4 | 95.6 | 95.0 | 93.3 |
| p | Pyoverdine | 7 | 98.6 | 100.0 | 100.0 | 99.6 | 100.0 | 95.2 |
| q | Quorum sensing | 7 | 86.5 | 89.6 | 86.6 | 85.3 | 96.4 | 90.5 |
| r | Transcriptional regulation | 10 | 86.9 | 81.8 | 81.2 | 78.8 | 95.0 | 93.3 |
| s | Polysaccharide synthesis | 7 | 90.7 | 98.7 | 87.9 | 90.6 | 89.3 | 95.2 |
| t | T3SS apparatus | 25 | 83.3 | 97.6 | 81.6 | 77.8 | 92.0 | 73.3 |
| te | T3SE putative T3SE and associated genes | 53 | 76.6 | 85.8 | 73.4 | 74.5 | 89.2 | 75.5 |
| u | Unknown function | 36 | 77.0 | 87.2 | 76.7 | 73.3 | 91.9 | 74.5 |
| v | Type IV pilus | 26 | 93.0 | 94.8 | 94.2 | 92.4 | 98.1 | 97.4 |
| y | Yersiniabactin biosynthesis | 6 | 85.4 | 94.7 | 82.3 | 86.5 | 91.7 | 61.1 |
Average percentage of strains carrying genes in each functional category.
The functional groups with the greatest number of conserved VA genes include genes associated with pyoverdine and alginate biosynthesis, the general secretion pathway, and the flagellar apparatus (Figure 1). The conservation of these genes across diverse isolates of P. syringae indicates that these functions are indispensable regardless of host and therefore may be candidate pathogen-associated molecular patterns (PAMPs) (Nurnberger and Brunner 2002). This is supported by the fact that bacterial flagellin (PSPTO1949), the prototypical PAMP (Felix et al. 1999), is conserved across all isolates.
Interesting VA genes include those that are individually highly variable, but that are associated with a functional group consisting of genes that are largely conserved across strains. These loci belong to essential systems, but may be adaptive in specific hosts. Examples include gspD, a general secretory pathway outer membrane protein that has homology to a gatekeeper for secreted exotoxins in enterotoxigenic Escherichia coli (Tauschek et al. 2002); hlyB, potentially involved in hemolysin iron uptake (Expert 1999); and the flagellar hook genes fliD, fliK, and flgE-2, which have homology to proteins that act as adhesins in Pseudomonas aeruginosa (Yonekura et al. 2002).
The functional groups that contain the greatest number of highly variable VA genes include the coronatine biosynthetic genes and T3SEs (Figures 1 and 2), which is consistent with previous studies (Guttman et al. 2002; Hwang et al. 2005). Additionally, many genes of unknown function are also highly variable, indicating potentially important roles in host adaptation. Interesting exceptions are those highly conserved genes in functional groups that are largely composed of variable VA genes. These genes may be essential components of otherwise flexible systems. Examples include coronatine biosynthetic genes cfa5, corR, cfl, cmaC, cfa2, cmaT, and cfa3 and 10 T3SEs, putative T3SEs, or associated genes including shcF, hopS1, hopI1, PSPTO4851, hopD1, avrF, hopK1, hopAN1, hopAF1, and hopJ1. The high conservation of these genes in diverse strains may reflect selective constraint more typical of core genes and suggests that these loci may play central roles in the general disease process.
Genes encoding or associated with the type IV pilus, general metabolism, polysaccharide synthesis, siderophore, transcriptional regulation, quorum sensing, and T3SS apparatus are intermediate in their level of variation. The interpretation of these intermediate-variable genes is more complicated, given the inherent difficulty of differentiating between alleles that are highly divergent and alleles that are absent. This limitation is of course not limited to CGH studies, but is an inherent limitation of all hybridization-based approaches when there is not 100% probe-target identity (Taboada et al. 2005) (see materials and methods). Some intriguing highly variable genes found within the functional groups that are primarily composed of intermediate-variable genes include general metabolism genes such as glutathione S-transferase (gst), polysaccharide metabolism genes such as the endogluconase wssD, tonB-dependent siderophore receptors, and regulatory genes such as σ-factor rpoS, the global activator sensor gacS, and the T3SS regulator hrpS. This latter result was initially surprising since previous studies indicate that hrpS is ubiquitous among P. syringae strains (Sawada et al. 1999); nevertheless, we confirmed that the hrpS alleles are highly polymorphic by comparing our calls to the nucleotide variation observed in hrpS alleles. All of the alleles that we called divergent were <80% identical to the PtoDC3000 allele. These results are consistent with those of other CGH studies; for example, Tiedje and colleagues called gyrB divergent in their CGH study of Shewanella oneidensis due to its relatively high level of polymorphism (Murray et al. 2001). In general, these results on intermediate-variable loci are useful in that they provide information about which loci are polymorphic and suggest further investigation into their possible roles in adaptation.
Phylogenetic associations:
We first attempted to identify those VA genes that evolved primarily through vertical descent vs. horizontal gene transfer. These two modes of evolution are not mutually exclusive, since genes that are acquired via horizontal exchange may then be transmitted vertically within a lineage indefinitely. Using the most simplistic approach, we note that four of the T3SEs (7.5%) are present in all strains, 10 T3SEs (18.9%) are present in 95% of strains, and 36 T3SE (67.9%) are present in 75% of the strains. Unfortunately, this approach cannot distinguish horizontal gene transfer from gene loss and therefore is of limited use. A better approach is to test for positive statistical associations between the distribution of individual genes and the core genome phylogeny developed by MLST of housekeeping genes (Sarkar and Guttman 2004; Hwang et al. 2005) (Figure 1 and supplemental Figure 1 at http://www.genetics.org/supplemental/). VA genes that did not display such an association were considered horizontally transferred. We then attempted to identify whether any horizontal transfer events were associated with apparent host shifts.
The most obvious association between the P. syringae core genome phylogeny and the VA gene profiles is in P. syringae phylogroup 1, which contains the reference strain PtoDC3000. Phylogroup 1 is the most highly conserved of the three largest P. syringae phylogenetic groups and, consequently, phylogroup 1 isolates are likely to carry a disproportionate proportion of homologs closely related to the PtoDC3000 target strain. However, despite this conservation, substantial variation is still obvious in the coronatine biosynthetic genes, T3SEs, and genes of unknown function in phylogroup 1 isolates. No distinct phylogenetic association is seen in the distribution of variable VA genes among the four other P. syringae phylogroups, indicating a prominent role for horizontal processes in the evolution of these genes.
We then identified potential VA gene horizontal acquisition and loss events by character mapping the distribution of each VA gene onto the core genome phylogeny. Each clade on the core genome tree was identified (boxed numbers along branches of the core genome phylogeny in Figure 1 and supplemental Figure 1 at http://www.genetics.org/supplemental/), and the percentage of strains within each clade that contained each VA gene was determined. Supplemental Tables 4 and 5 at http://www.genetics.org/supplemental/ present the percentage of occurrence of each VA and T3SE gene in each clade along with the total number of strains in each clade. Of particular interest are the clades that contain strains with similar hosts of isolation. For example, clade 40, which includes most of the strains in P. syringae phylogroup 1, is dominated by strains isolated from either tomato or Brassica hosts (radish, cabbage, cauliflower). The strains in this clade all carry hopO1-2, hopM1, hopF2, hopU1, hrpW, and other T3SEs. hopF2 and hopU1 are closely linked on the PtoDC3000 genome. It is interesting to speculate that the acquisition of some of these T3SEs was responsible for the expansion into this new niche—namely the ability to infect tomato and Brassica hosts. This is supported by the fact that there are no isolates from tomato or Brassica hosts outside of clade 40 within phylogroup 1.
Other illustrative examples include clade 29, which defines phylogroup 4. This small clade is strictly composed of strains that are pathogenic on monocot hosts. These strains share numerous T3SEs, including hopF2, hopX1, hopU1, and hopH 1. Clearly, the ancestor of this group acquired a large number of effectors that have been maintained by its descendants. It will be interesting to determine which of these T3SEs was instrumental in the movement to monocot hosts. Likewise, clade 3 defines a tight cluster of predominantly bean pathogens in phylogroup 3. The strains in this clade all contain T3SEs hopAJ1, hopQ1-1, and hopR1. These are likely candidate factors for the shift to bean hosts.
We also studied the population structure of each variable VA gene relative to the core genome phylogeny. Specifically, we asked whether the set of strains that carry any particular variable VA genes are more closely related to each other than are a set of randomly selected strains of the same size. When this test is upheld we infer that there is structure in the underlying population of strains that carry that particular VA gene. This indicates that a significant number of closely related strains carry the same VA gene; in other words, the gene has been transmitted vertically within this lineage. Alternatively, lack of structure would indicate that the specific VA gene has been randomly distributed among strains by horizontal gene transfer.
The test for population structure relied upon a bootstrap permutation test in which Watterson's θ, a measure of polymorphism based on the number of segregating sites, was used to determine the average genetic distance among strains on the basis of the core genome MLST data set. Each VA gene was analyzed independently. In general, we find that there is very little population structure among the strains carrying most T3SEs, supporting an important role for horizontal gene transfer (Table 3). Unfortunately, much of the signal in the analysis of the total data set and of the phylogroup 1 strains is masked by the very close similarity of the strains in phylogroup 1. Nevertheless, there are a relatively small number of cases where vertical evolution can be inferred. For example, the T3SE hopAA1-2 is quite rare and shows no population structure in any of the three major clades, unless all of the strains are analyzed simultaneously. The significant θ appears to be due to the disproportionate number of strains from phylogroups 4 and 5 that carry this T3SE. Likewise, hopAA1-1 is a highly variable T3SE that is overrepresented within clade 12 of phylogroup 2. The simplest conclusion from this analysis is that horizontal gene transfer has dominated the evolution of nearly all of the variable T3SEs.
TABLE 3.
Population structure of T3SE
| All strains
|
Group 1 strains
|
Group 2 strains
|
Group 3 strains
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T3SE | N | (%) | θa | Pb | N | (%) | θ | P | N | (%) | θ | P | N | (%) | θ | P |
| hopAD1 | 17 | (18.7) | 0.0577 | 0.107 | 3 | (13.6) | 0.0176 | 0.429 | 7 | (21.9) | 0.0270 | 0.555 | 5 | (16.1) | 0.0198 | 0.384 |
| hopAA1-2 | 18 | (19.8) | 0.0590 | 0.015 | 4 | (18.2) | 0.0193 | 0.231 | 7 | (21.9) | 0.0324 | 0.089 | 5 | (16.1) | 0.0195 | 0.384 |
| hopA1 | 27 | (29.7) | 0.0533 | 0.224 | 7 | (31.8) | 0.0124 | 0.738 | 8 | (25.0) | 0.0302 | 0.156 | 10 | (32.3) | 0.0196 | 0.418 |
| hopAA1-1 | 47 | (51.6) | 0.0457 | 0.860 | 9 | (40.9) | 0.0060 | 0.961 | 18 | (56.3) | 0.0299 | 0.015 | 19 | (61.3) | 0.0212 | 0.144 |
| hopO1-2 | 47 | (51.6) | 0.0514 | 0.073 | 11 | (50.0) | 0.0161 | 0.213 | 12 | (37.5) | 0.0294 | 0.135 | 20 | (64.5) | 0.0220 | 0.052 |
| hopAB2 | 49 | (53.8) | 0.0495 | 0.379 | 14 | (63.6) | 0.0154 | 0.250 | 14 | (43.8) | 0.0274 | 0.275 | 18 | (58.1) | 0.0204 | 0.226 |
| hopM1 | 51 | (56.0) | 0.0490 | 0.414 | 12 | (54.5) | 0.0138 | 0.651 | 20 | (62.5) | 0.0272 | 0.201 | 17 | (54.8) | 0.0210 | 0.137 |
| hopF2 | 55 | (60.4) | 0.0495 | 0.287 | 12 | (54.5) | 0.0159 | 0.200 | 19 | (59.4) | 0.0269 | 0.250 | 21 | (67.7) | 0.0207 | 0.133 |
| hopAM1-2 | 59 | (64.8) | 0.0504 | 0.047 | 14 | (63.6) | 0.0142 | 0.525 | 23 | (71.9) | 0.0294 | 0.002 | 17 | (54.8) | 0.0193 | 0.385 |
| hopQ1-2 | 59 | (64.8) | 0.0507 | 0.025 | 16 | (72.7) | 0.0136 | 0.676 | 20 | (62.5) | 0.0279 | 0.137 | 18 | (58.1) | 0.0221 | 0.063 |
| hopT2 | 61 | (67.0) | 0.0500 | 0.096 | 17 | (77.3) | 0.0159 | 0.114 | 21 | (65.6) | 0.0298 | 0.004 | 19 | (61.3) | 0.0219 | 0.073 |
| hopAM1-1 | 61 | (67.0) | 0.0517 | 0.001 | 15 | (68.2) | 0.0164 | 0.097 | 21 | (65.6) | 0.0302 | <0.001 | 20 | (64.5) | 0.0220 | 0.052 |
| hopX1 | 61 | (67.0) | 0.0507 | 0.027 | 12 | (54.5) | 0.0157 | 0.222 | 24 | (75.0) | 0.0290 | 0.003 | 21 | (67.7) | 0.0215 | 0.059 |
| hopT1-1 | 62 | (68.1) | 0.0503 | 0.043 | 15 | (68.2) | 0.0151 | 0.341 | 21 | (65.6) | 0.0262 | 0.385 | 22 | (71.0) | 0.0218 | 0.051 |
| hopE1 | 68 | (74.7) | 0.0502 | 0.031 | 15 | (68.2) | 0.0151 | 0.341 | 24 | (75.0) | 0.0264 | 0.257 | 24 | (77.4) | 0.0215 | 0.045 |
| shcA | 68 | (74.7) | 0.0492 | 0.106 | 17 | (77.3) | 0.0148 | 0.385 | 26 | (81.3) | 0.0293 | <0.001 | 21 | (67.7) | 0.0207 | 0.133 |
| Total | 91 | 0.0510 | 22 | 0.0167 | 32 | 0.0284 | 31 | 0.0219 | ||||||||
Only highly variable (present in <75% of strains) T3SES are shown. See supplemental Tables 6 and 7 at http://www.genetics.org/supplemental/ for full data.
Watterson's θ measure of nucleotide polymorphism.
Probability that strains carrying the specific T3SE are no more closely related to each other than to random strains.
Host associations and individual VA genes:
VA genes that are statistically overrepresented in isolates from specific hosts are the strongest candidate host-specific virulence factors. We used a likelihood analysis to test for an association between the presence of specific VA genes and the host of isolation against the null hypothesis that there is no association. Table 4 and supplemental Table 3 at http://www.genetics.org/supplemental/show the P-value results of a likelihood-ratio test of these hypotheses. The most obvious pattern is among the Chinese cabbage isolates, which show seven strongly significant associations, although these patterns more likely reflect the very close relationship among these strains and their high level of similarity to the reference strain PtoDC3000 than anything biologically meaningful. More interesting examples include hopAA1-1, which is found in only 46% of all strains, but is significantly associated with the three isolates obtained from rice. Two of these three isolates are clustered closely together in phylogroup 4, while the third is very divergent and found in phylogroup 2. hopAA1-1 is also marginally significant among tomato isolates (carried by 90% of all tomato isolates) and a number of other hosts including cauliflower, Japanese apricot, and bayberry. The three rice isolates also show marginally significant associations with hopAM1-2, hopQ1-2, and hopX1 (N-term). The nine soybean isolates are strongly associated with three loci. One of these is shcA, the chaperone for hopA1 (van Dijk et al. 2002), while two others are predicted to be lytic transglycosylases (hopAJ1 and PSPTO1378). hopAJ1, which is commonly considered a T3SS helper protein, is also associated with strains isolated from kidney beans. The 11 tomato isolates are significantly associated with a version of hopO1-3 that is truncated in PtoDC3000, hopP1, hopAK1, and an allele of hopAG interrupted by an insertion element in PtoDC3000. Truncated and chimeric T3SEs are extremely common and have been shown to confer new functions and secretion apparatus specificities and even to influence the host range of the bacteria carrying them (Stavrinides et al. 2006).
TABLE 4.
T3SE by host of isolation likelihood analysis
| Brassicaceaea
|
Cauliflowera
|
Chinese cabbagea
|
Fabaceae
|
Soybean
|
Kidney bean
|
Tomatoa
|
Cucurbitaceae
|
Cucumber
|
Poaceaea
|
Ricea
|
Moraceae
|
Japanese apricota
|
Bayberrya
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T3SE | N | 10 | 2 | 4 | 28 | 9 | 6 | 11 | 6 | 5 | 10 | 3 | 2 | 2 | 2 |
| hopA1 | 27 | ** | *** | ||||||||||||
| hopAA1-1 | 46 | * | ** | * | ** | * | * | * | |||||||
| hopo1-2 | 47 | * | ** | * | * | * | * | ||||||||
| hopAB2 | 48 | ** | |||||||||||||
| hopM1 | 50 | ** | |||||||||||||
| hopF2 | 54 | ** | |||||||||||||
| hopAM1-2 | 57 | ** | * | ||||||||||||
| hopQ1-2 | 58 | * | * | ||||||||||||
| hopX1 (N-term) | 58 | * | * | ||||||||||||
| hopT2 | 59 | * | |||||||||||||
| hopAM1-1 | 60 | * | |||||||||||||
| hopT1-1 | 61 | * | |||||||||||||
| hopE1 | 66 | ** | * | ||||||||||||
| shcA | 66 | ** | |||||||||||||
| hopAJ1 | 67 | ** | ** | ||||||||||||
| hopH1 | 69 | ** | |||||||||||||
| hopO1-3′ (N-term) | 69 | *** | |||||||||||||
| hrpW | 71 | * | |||||||||||||
| hopAG1 | 72 | ** | |||||||||||||
| hopP1 | 72 | ** | |||||||||||||
| hopAK1 | 74 | ** | |||||||||||||
| Ipx47 | 74 | ** | |||||||||||||
| PSPTO1378 | 75 | ** | ** | ||||||||||||
| hopB1 | 76 | ** | ** | ||||||||||||
| PSPTO0869 | 77 | * | |||||||||||||
| hopQ1-1 | 78 | * | * | ||||||||||||
| CEL ORF4 | 79 | * | * | ||||||||||||
| hopAO1 | 79 | * | * | ||||||||||||
| hopN1 | 79 | * | |||||||||||||
| avrE1 | 80 | * | |||||||||||||
| hopS1 | 87 | * |
P-values from likelihood-ratio test of association model vs. nonassociation model (see Materials and Methods for details). *, 0.1–0.05; **, 0.05–0.01; ***, 0.01–0.001. Only those hosts with two or more isolates were analyzed. The hosts and host families that are not presented in this table did not exhibit any significant associations. Only T3SEs with significant associations are shown. N, number of strains that carry each T3SE and are isolated from each host.
Hosts are grouped on the basis of their taxonomic family.
Host associations and VA profiles:
We attempted to determine whether specific VA gene profiles, defined as the suite of VA genes carried by any individual isolate, are associated with specific hosts of isolation. Two possible outcomes define the extremes of the expectations for this test: (1) all isolates of a given host have the same VA profile, implying that a specific complement of VA genes is required to infect a particular host and (2) all isolates of a given host have different VA profiles, implying that there are either multiple ways of infecting a given host or that there may be subprofiles that are similar among these isolates that are masked by differences in other VA genes. These differences may be due to unrecognized differences in host specificity or niche among strains.
The T3SE profiles clustered by host species and host family are shown in Figure 2. We quantified this analysis by determining the average pairwise level of profile identity shared among strains isolated from common hosts or hosts from the same families. Specifically, we asked whether the average level of profile identity was greater among strains isolated from common hosts than randomly selected strains. Profile identity was calculated by simply determining the proportion of genes that were in the same state (present or divergent) for each pairwise comparison of profiles. Statistical evaluation was performed against bootstrapped null distributions of 1000 pseudoreplicates created from randomly chosen strains. Only variable genes and hosts with three or more isolates were used in this analysis.
VA gene profiles are generally more conserved than T3SE profiles due to the presence of highly conserved functional categories (Table 5). Groups of strains isolated from Chinese cabbage, tomato, Brassicaceae, soybean, and rice have ≥75% profile identity. The profiles from Chinese cabbage and tomato isolates are statistically more similar than chance for both VA genes and T3SEs. The significant profile identity of tomato isolates is not surprising, given that most of the strains are very tightly clustered in phylogroup 1 along with the arrayed reference strain (which is also a tomato pathogen). The four cauliflower isolates are nearly identical strains that also cluster with the tomato isolates in phylogroup 1. Cuppels and Ainsworth (1995) demonstrated that all maculicola strains (Brassica pathogens) could infect tomato and that many tomato isolates could infect cauliflower, indicating that pathogens of these hosts must utilize similar suites of VA genes. Interestingly, when the larger collection of Brassica isolates is examined, significantly similar VA gene profiles are also apparent. These strains were isolated from a wide variety of Brassica hosts including radish, Chinese cabbage, and cauliflower, but are quite evolutionary divergent, being distributed in phylogroups 1, 3, and 5. This analysis suggests that there may be common selective pressures that act on all Brassica pathogens. In general, this analysis supports both predictions concerning profile identity among isolates from a common host, in that some hosts yield isolates that have very similar profiles, while many others do not.
TABLE 5.
T3SE and VA gene profile identity for hosts and host families
| T3SE profiles
|
VA gene profiles
|
||||||
|---|---|---|---|---|---|---|---|
| Host | Na | Identity | SD | P | Identity | SD | P |
| Chinese cabbage | 4 | 1.000 | 1.080 | 0.000 | 0.964 | 1.041 | 0.000 |
| Tomato | 11 | 0.808 | 0.847 | 0.041 | 0.849 | 0.877 | 0.010 |
| Brassicaceae | 10 | 0.793 | 0.818 | 0.067 | 0.814 | 0.833 | 0.047 |
| Rice | 3 | 0.783 | 0.904 | 0.264 | 0.826 | 0.954 | 0.210 |
| Soybean | 9 | 0.749 | 0.770 | 0.156 | 0.761 | 0.779 | 0.201 |
| Solanaceae | 14 | 0.686 | 0.747 | 0.386 | 0.742 | 0.782 | 0.186 |
| Kidney bean | 6 | 0.681 | 0.734 | 0.408 | 0.707 | 0.751 | 0.376 |
| Fabaceae | 28 | 0.675 | 0.698 | 0.460 | 0.676 | 0.693 | 0.657 |
| Poaceae | 11 | 0.658 | 0.691 | 0.537 | 0.679 | 0.713 | 0.511 |
| Radish | 4 | 0.651 | 0.705 | 0.485 | 0.666 | 0.724 | 0.512 |
| Pea | 8 | 0.635 | 0.671 | 0.622 | 0.639 | 0.674 | 0.724 |
| Beanb | 9 | 0.628 | 0.664 | 0.672 | 0.656 | 0.680 | 0.664 |
| Snap bean | 3 | 0.612 | 0.713 | 0.553 | 0.610 | 0.706 | 0.655 |
| Rosaceae | 6 | 0.569 | 0.674 | 0.851 | 0.630 | 0.696 | 0.709 |
| Cucurbitaceae | 6 | 0.560 | 0.627 | 0.889 | 0.574 | 0.625 | 0.941 |
| Oleaceae | 4 | 0.558 | 0.675 | 0.786 | 0.632 | 0.727 | 0.629 |
| Cucumber | 5 | 0.549 | 0.632 | 0.894 | 0.572 | 0.632 | 0.908 |
| Wheat | 3 | 0.488 | 0.615 | 0.894 | 0.507 | 0.632 | 0.945 |
| Lilac | 3 | 0.473 | 0.625 | 0.912 | 0.558 | 0.695 | 0.832 |
T3SE profiles are based on 43 variable T3SES. VA gene profiles are based on 169 variable VA genes.
Number of strains isolated from each host.
Includes both kidney and snap beans.
These data permit us to ask more focused questions, such as, is there only one way to infect a specific host? And are similar VA or T3SE profiles the result of vertical descent from a common ancestor, convergent evolution, or horizontal gene transfer? The tomato isolates present an excellent opportunity to address these issues. Nine of 11 tomato isolates tightly cluster in P. syringae phylogroup 1, while the other two, PsyB76 and Pto2170, are highly divergent and found in phylogroup 2. The T3SE profile of Pto2170 is essentially indistinguishable from the 9 phylogroup 1 tomato isolates, as opposed to PsyB76, which has a highly divergent T3SE profile. Given that both the T3SE profile and evolutionary clustering of PsyB76 are different from other tomato pathogens, this strain has likely evolved the ability to infect tomato independently via convergent evolution. In contrast, Pto2170 shows evidence for the horizontal acquisition of T3SE from the phylogroup 1 tomato isolates, as it shares 36 variable T3SEs with the group 1 tomato isolates. The large-scale acquisition of T3SE via horizontal gene transfer is not entirely unlikely, given that many T3SE genes are clustered on genomic islands associated with mobile elements (Jackson et al. 2000). Twelve variable T3SEs were common to all tomato isolates, including: hopY1, hopL1, hopT1-2, hopX1 (C terminus), avrPto1, hopR1, CEL ORF4, hopB1, hopAK1, hopP1, hopAG:ISPssy, and hopO1-3. These may include those T3SEs that define the minimal set required for tomato specificity.
Genomic organization:
We extended our genomic analysis to address whether regions of the PtoDC3000 chromosome are highly variable with respect to their complement of VA genes. Clusters of high variation are putative genomic islands that may be more prone to gene acquisition and loss. Such genomic islands typically contain clusters of coordinately regulated loci required for niche specialization (Ilyina and Romanova 2002).
We mapped the variation observed for each VA gene onto the corresponding position in the PtoDC3000 genome (Figure 3). Variation was calculated as the percentage of strains that were scored as divergent for each gene. Regions of high variability include the hrp/hrc cluster, which encodes the T3SS apparatus, and the coronatine biosynthetic gene cluster. Significant clusters of variable loci were also found associated with T3SEs, flagellar synthesis genes, and yersiniabactin synthesis genes. Substantial numbers of T3SEs were observed to cluster near the hrp/hrc cluster, the coronatine biosynthetic genes, the largest plasmid, and at least two other genomic islands located at ∼0.55 and 0.95 Mb along the PtoDC3000 genome. The former island contains some highly variable genes encoding type IV pilus proteins, while the latter island contains four conserved type IV pilus biogenesis genes and putative virulence genes. The genomic regions flanking the conserved loci encoding the general secretory pathway include a number of highly variable genes, including genes encoding filamentous hemagglutinin proteins, tail tape measure proteins, and polysaccharide biosynthesis proteins.
Conclusion:
We have provided the broadest survey of host-associated virulence factors from any pathogen by tapping into the natural genetic variation found in the P. syringae species complex. We have identified numerous candidate host-specificity genes, furthered our mechanistic understanding of how this remarkable bacterium adapts to diverse plant hosts, and provided a glimpse into the dynamic evolutionary processes that underlie these interactions. These data establish the basis for identifying mechanistic differences among evolutionarily distant pathogens that infect the same host and for determining why pathogens of divergent hosts have maintained similar complements of VA genes. We hope that this study will serve as a guide for focusing functional testing of host specificity and for framing hypotheses directed at understanding the complex mechanisms underlying host adaptation and pathogenesis.
Acknowledgments
We thank the many individuals who have provided strains to the Guttman lab and Sam Cartinhour for generously sharing DC3000 primer sequences. We acknowledge Nicholas Provart and Ryan Austin for microarray analysis advice; John Stinchcombe for statistical advice; Paul Debbie for assistance with microarray fabrication; and particularly John Stavrinides, Robyn Morgan, Wenbo Ma, and Pauline Wang for valuable input and critique. G.B.M. is partially supported by the National Science Foundation (DBI-0077622). S.F.S. was partially supported by an Ontario Graduate Scholarship. D.S.G. is a Canada Research Chair in Comparative Genomics and supported by grants from the Natural Science and Engineering Research Council of Canada and Performance Plants of Kingston, Ontario.
References
- Abramovitch, R. B., and G. B. Martin, 2004. Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7: 356–364. [DOI] [PubMed] [Google Scholar]
- Abramovitch, R. B., Y. J. Kim, S. Chen, M. B. Dickman and G. B. Martin, 2003. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22: 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J. R., and A. Collmer, 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42: 385–414. [DOI] [PubMed] [Google Scholar]
- Alfano, J. R., A. O. Charkowski, W. L. Deng, J. L. Badel, T. Petnicki-Ocwieja et al., 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97: 4856–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel, J. L., A. O. Charkowski, W. L. Deng and A. Collmer, 2002. A gene in the Pseudomonas syringae pv.tomato Hrp pathogenicity island conserved effector locus, hopPtoA1, contributes to efficient formation of bacterial colonies in planta and is duplicated elsewhere in the genome. Mol. Plant Microbe Interact. 15: 1014–1024. [DOI] [PubMed] [Google Scholar]
- Badel, J. L., K. Nomura, S. Bandyopadhyay, R. Shimizu, A. Collmer et al., 2003. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol Microbiol 49: 1239–1251. [DOI] [PubMed] [Google Scholar]
- Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik et al., 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Bender, C. L., F. Alarcon-Chaidez and D. C. Gross, 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63: 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beth Mudgett, M., 2005. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56: 509–531. [DOI] [PubMed] [Google Scholar]
- Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen et al., 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100: 10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan et al., 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S-enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185: 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J. H., J. M. Urbach, T. F. Law, L. W. Arnold, A. Hu et al., 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 102: 2549–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski, A. O., J. R. Alfano, G. Preston, J. Yuan, S. Y. He et al., 1998. The Pseudomonas syringae pv.tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180: 5211–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody, Y. S., and D. C. Gross, 1984. Fluorescent siderophore production and iron acquisition by Pseudomonas syringae pv syringae. Phytopathology 74: 826. [Google Scholar]
- Cornelis, G. R., 2002. The Yersinia Ysc-Yop virulence apparatus. Int. J. Med. Microbiol. 291: 455–462. [DOI] [PubMed] [Google Scholar]
- Cui, X., K. Kerr and G. Churchill, 2002. Transformations for cDNA microarray data. Stat. Appl. Genet. Mol. Biol. 2: 1–20. [DOI] [PubMed] [Google Scholar]
- Cuppels, D. A., and T. Ainsworth, 1995. Molecular and physiological characterization of Pseudomonas syringae pv tomato and Pseudomonas syringae pv maculicola strains that produce the phytotoxin coronatine. Appl. Environ. Microbiol. 61: 3530–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt, U., and J. Hacker, 2001. Whole genome plasticity in pathogenic bacteria. Curr. Opin. Microbiol. 4: 550–557. [DOI] [PubMed] [Google Scholar]
- Dobrindt, U., U. Hentschel, J. B. Kaper and J. Hacker, 2002. Genome plasticity in pathogenic and nonpathogenic enterobacteria. Curr. Top. Microbiol. Immunol. 264: 157–175. [PubMed] [Google Scholar]
- Eisen, M. B., P. T. Spellman, P. O. Brown and D. Botstein, 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa, A., and J. R. Alfano, 2004. Disabling surveillance: bacterial type III secretion system effectors that suppress innate immunity. Cell. Microbiol. 6: 1027–1040. [DOI] [PubMed] [Google Scholar]
- Evertsz, E. M., J. Au-Young, M. V. Ruvolo, A. C. Lim and M. A. Reynolds, 2001. Hybridization cross-reactivity within homologous gene families on glass cDNA microarrays. Biotechniques 31: 1182, 1184, 1186 passim. [DOI] [PubMed] [Google Scholar]
- Expert, D., 1999. Withholding and exchanging iron: interactions between Erwinia spp. and their plant hosts. Annu. Rev. Phytopathol. 37: 307–334. [DOI] [PubMed] [Google Scholar]
- Feil, H., W. S. Feil, P. Chain, F. Larimer, G. Dibartolo et al., 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 102: 11064–11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G., J. D. Duran, S. Volko and T. Boller, 1999. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276. [DOI] [PubMed] [Google Scholar]
- Flor, H. H., 1971. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9: 275–296. [Google Scholar]
- Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell et al., 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 99: 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J. T., and B. A. Vinatzer, 2003. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 6: 20–28. [DOI] [PubMed] [Google Scholar]
- Guttman, D. S., and J. T. Greenberg, 2001. Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant Microbe Interact. 14: 145–155. [DOI] [PubMed] [Google Scholar]
- Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. Ranall and J. T. Greenberg, 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295: 1722–1726. [DOI] [PubMed] [Google Scholar]
- Guttman, D. S., S. J. Gropp, R. L. Morgan and P. W. Wang, 2006. Diversifying selection drives the evolution of the type III secretion system pilus of Pseudomonas syringae. Mol. Biol. Evol. 23 (in press). [DOI] [PubMed]
- Hacker, J., and E. Carniel, 2001. Ecological fitness, genomic islands and bacterial pathogenicity—a Darwinian view of the evolution of microbes. EMBO Rep. 2: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker, J., and J. B. Kaper, 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54: 641–679. [DOI] [PubMed] [Google Scholar]
- Hakenbeck, R., N. Balmelle, B. Weber, C. Gardes, W. Keck et al., 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 69: 2477–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, P. J., and J. D. Taylor, 2006. Patterns of interaction between isolates of three pathovars of Pseudomonas syringae and accessions of a range of host and nonhost legume species. Plant Pathol. 55: 46–53. [Google Scholar]
- Hwang, M. S. H., R. L. Morgan, S. F. Sarkar, P. W. Wang and D. S. Guttman, 2005. Phylogenetic characterization of virulence and resistance phenotypes in Pseudomonas syringae. Appl. Environ. Microbiol. 71: 5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina, T. S., and Y. M. Romanova, 2002. Bacterial genomic islands: organization, function, and evolutionary role. Mol. Biol. 36: 171–179. [PubMed] [Google Scholar]
- Israel, D. A., N. Salama, C. N. Arnold, S. F. Moss, T. Ando et al., 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Invest. 107: 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R. W., E. Athanassopoulos, G. Tsiamis, J. W. Mansfield, A. Sesma et al., 1999. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl. Acad. Sci. USA 96: 10875–10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R. W., J. W. Mansfield, D. L. Arnold, A. Sesma, C. D. Paynter et al., 2000. Excision from tRNA genes of a large chromosomal region, carrying avrPphB, associated with race change in the bean pathogen, Pseudomonas syringae pv. phaseolicola. Mol. Microbiol. 38: 186–197. [DOI] [PubMed] [Google Scholar]
- Jin, Q., R. Thilmony, J. Zwiesler-Vollick and S. Y. He, 2003. Type III protein secretion in Pseudomonas syringae. Microbes Infect. 5: 301–310. [DOI] [PubMed] [Google Scholar]
- Kim, J., K. Ahn, S. Min, J. Jia, U. Ha et al., 2005. Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology 151: 3575–3587. [DOI] [PubMed] [Google Scholar]
- King, E. O., M. K. Ward and D. E. Raney, 1954. Two simple media for the demonstration of phycocyanin and fluorescin. J. Lab. Clin. Med. 44: 301–307. [PubMed] [Google Scholar]
- Lavie, M., B. Seunes, P. Prior and C. Boucher, 2004. Distribution and sequence analysis of a family of type III-dependent effectors correlate with the phylogeny of Ralstonia solanacearum strains. Mol. Plant Microbe Interact. 17: 931–940. [DOI] [PubMed] [Google Scholar]
- Levin, B. R., 1996. The evolution and maintenance of virulence in microparasites. Emerg. Infect. Dis. 2: 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, M. T. S., and B. N. Kunkel, 2005. The Pseudomonas syringae avrRpt2 gene contributes to virulence on tomato. Molecular Plant-Microbe Interactions 18: 626–633. [DOI] [PubMed] [Google Scholar]
- Lin, N. C., and G. B. Martin, 2005. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol. Plant Microbe Interact. 18: 43–51. [DOI] [PubMed] [Google Scholar]
- Lindeberg, M., J. Stavrinides, J. H. Chang, J. R. Alfano, A. Collmer et al., 2005. Unified nomenclature and phylogenetic analysis of extracellular proteins delivered by the type III secretion system of the plant pathogenic bacterium Pseudomonas syringae. Mol. Plant Microbe Interact. 18: 275–282. [DOI] [PubMed] [Google Scholar]
- Lindroos, H. L., A. Mira, D. Repsilber, O. Vinnere, K. Naslund et al., 2005. Characterization of the genome composition of Bartonella koehlerae by microarray comparative genomic hybridization profiling. J. Bacteriol. 187: 6155–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang, J. M., and N. T. Keen, 1995. Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol. Plant Microbe Interact. 8: 49–57. [DOI] [PubMed] [Google Scholar]
- Losada, L., T. Sussan, K. Pak, S. Zeyad, I. Rozenbaum et al., 2004. Identification of a novel Pseudomonas syringae Psy61 effector with virulence and avirulence functions by a HrpL-dependent promoter-trap assay. Mol. Plant Microbe Interact. 17: 254–262. [DOI] [PubMed] [Google Scholar]
- Ma, W., F. F. T. Dong, J. Stavrinides and D. S. Guttman, 2006. Diversification of a type III effector family via both pathoadaptation and horizontal gene transfer in response to a coevolutionary arms race. PLoS Genetics (in press). [DOI] [PMC free article] [PubMed]
- Malloff, C. A., R. C. Fernandez and W. L. Lam, 2001. Bacterial comparative genomic hybridization: a method for directly identifying lateral gene transfer. J. Mol. Biol. 312: 1–5. [DOI] [PubMed] [Google Scholar]
- McDermott, P. F., R. D. Walker and D. G. White, 2003. Antimicrobials: modes of action and mechanisms of resistance. Int. J. Toxicol. 22: 135–143. [DOI] [PubMed] [Google Scholar]
- Morris, C. E., C. Glaux, X. Latour, L. Gardan, R. Samson et al., 2000. The relationship of host range, physiology, and genotype to virulence on cantaloupe in Pseudomonas syringae from cantaloupe blight epidemics in France. Phytopathology 90: 636–646. [DOI] [PubMed] [Google Scholar]
- Murray, A. E., D. Lies, G. Li, K. Nealson, J. Zhou et al., 2001. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc. Natl. Acad. Sci. USA 98: 9853–9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, K., M. Melotto and S. Y. He, 2005. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8: 361–368. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T., and F. Brunner, 2002. Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5: 318–324. [DOI] [PubMed] [Google Scholar]
- Perrin, A., S. Bonacorsi, E. Carbonnelle, D. Talibi, P. Dessen et al., 2002. Comparative genomics identifies the genetic islands that distinguish Neisseria meningitidis, the agent of cerebrospinal meningitis, from other Neisseria species. Infect. Immun. 70: 7063–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnicki-Ocwieja, T., D. J. Schneider, V. C. Tam, S. T. Chancey, L. Shan et al., 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 99: 7652–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman, A. R., R. W. Jackson, J. W. Mansfield, V. Kaitell, R. Thwaites et al., 2005. Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr. Biol. 15: 2230–2235. [DOI] [PubMed] [Google Scholar]
- Porwollik, S., C. A. Santiviago, P. Cheng, L. Florea, S. Jackson et al., 2005. Differences in gene content between Salmonella enterica serovar enteritidis isolates and comparison to closely related serovars gallinarum and Dublin. J. Bacteriol. 187: 6545–6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager, R., S. Mirold, E. Tietze, U. Strutz, B. Knuppel et al., 2000. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical isolates of Salmonella enterica. Int. J. Med. Microbiol. 290: 605–617. [DOI] [PubMed] [Google Scholar]
- Rohmer, L., D. S. Guttman and J. L. Dangl, 2004. Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in plant pathogenic Pseudomonas syringae. Genetics 167: 1341–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins et al., 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97: 14668–14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, S. F., and D. S. Guttman, 2004. The evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, N. A., A. Underwood, A. M. Kearns and G. Hallas, 2004. A virulence-associated gene microarray: a tool for investigation of the evolution and pathogenic potential of Staphylococcus aureus. Microbiology 150: 3763–3771. [DOI] [PubMed] [Google Scholar]
- Sawada, H., F. Suzuki, I. Matsuda and N. Saitou, 1999. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 49: 627–644. [DOI] [PubMed] [Google Scholar]
- Sokurenko, E. V., D. L. Hasty and D. E. Dykhuizen, 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7: 191–195. [DOI] [PubMed] [Google Scholar]
- Stavrinides, J., W. Ma and D. S. Guttman, 2006. Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathogens 2(10): e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboada, E. N., R. R. Acedillo, C. D. Carrillo, W. A. Findlay, D. T. Medeiros et al., 2004. Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity. J. Clin. Microbiol. 42: 4566–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboada, E. N., R. R. Acedillo, C. C. Luebbert, W. A. Findlay and J. H. E. Nash, 2005. A new approach for the analysis of bacterial microarray-based comparative genomic hybridization: insights from an empirical study. BMC Genomics 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauschek, M., R. J. Gorrell, R. A. Strugnell and R. M. Robins-Browne, 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA 99: 7066–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiamis, G., J. W. Mansfield, R. Hockenhull, R. W. Jackson, A. Sesma et al., 2000. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 19: 3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong et al., 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101: 4865–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, K., V. C. Tam, A. R. Records, T. Petnicki-Ocwieja and J. R. Alfano, 2002. The ShcA protein is a molecular chaperone that assists in the secretion of the HopPsyA effector from the type III (Hrp) protein secretion system of Pseudomonas syringae. Mol. Microbiol. 44: 1469–1481. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, W. B., D. C. Melles, A. Alaidan, M. Al-Ahdal, H. A. M. Boelens et al., 2005. Host- and tissue-specific pathogenic traits of Staphylococcus aureus. J. Bacteriol. 187: 4584–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bodman, S. B., W. D. Bauer and D. L. Coplin, 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 29: 29. [DOI] [PubMed] [Google Scholar]
- Wiebe, W. L., and R. N. Campbell, 1993. Characterization of Pseudomonas syringae pv maculicola and comparison with P.s. tomato. Plant Dis. 77: 414–419. [Google Scholar]
- Yao, Y. F., D. E. Sturdevant, A. Villaruz, L. Xu, Q. Gao et al., 2005. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect. Immun. 73: 1856–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura, K., S. Maki-Yonekura and K. Namba, 2002. Growth mechanism of the bacterial flagellar filament. Res. Microbiol. 153: 191–197. [DOI] [PubMed] [Google Scholar]
- Yu, J., A. Penaloza-Vazquez, A. M. Chakrabarty and C. L. Bender, 1999. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol. Microbiol. 33: 712–720. [DOI] [PubMed] [Google Scholar]
- Zhong, S. B., A. Khodursky, D. E. Dykhuizen and A. M. Dean, 2004. Evolutionary genomics of ecological specialization. Proc. Natl. Acad. Sci. USA 101: 11719–11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiesler-Vollick, J., A. E. Plovanich-Jones, K. Nomura, S. Bandyopadhyay, V. Joardar et al., 2002. Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv. tomato DC3000 genome. Mol. Microbiol. 45: 1207–1218. [DOI] [PubMed] [Google Scholar]