Abstract
Hedgehog proteins play critical roles in organizing the embryonic development of animals, largely through modulation of target gene expression. Little is currently known, however, about the kinds and numbers of genes whose expression is controlled, directly or indirectly, by Hedgehog activity. Using techniques to globally repress or activate Hedgehog signaling in zebrafish embryos followed by microarray-based expression profiling, we have discovered a cohort of genes whose expression responds significantly to loss or gain of Hedgehog function. We have confirmed the Hedgehog responsiveness of a representative set of these genes with whole-mount in situ hybridization as well as real time PCR. In addition, we show that the consensus Gli-binding motif is enriched within the putative regulatory elements of a sizeable proportion of genes that showed positive regulation in our assay, indicating that their expression is directly induced by Hedgehog. Finally, we provide evidence that the Hedgehog-dependent spatially restricted transcription of one such gene, nkx2.9, is indeed mediated by Gli1 through a single Gli recognition site located within an evolutionarily conserved enhancer fragment. Taken together, this study represents the first comprehensive survey of target genes regulated by the Hedgehog pathway during vertebrate development. Our data also demonstrate for the first time the functionality of the Gli-binding motif in the control of Hedgehog signaling-induced gene expression in the zebrafish embryo.
DURING animal development, cells communicate with each other to coordinate their proliferation and differentiation and to ensure that the right kind of tissues are assembled spatially and temporally within the embryo. Surprisingly, only a handful of intercellular signaling molecules have so far been identified and shown to participate in this process, their reiterative use in different cellular situations governing the generation of cellular diversity. The Hedgehog (Hh) family of lipid-modified secreted proteins are one such group of intercellular signals that have profound effects on the regulation of embryonic development of all animals (Ingham and McMahon 2001; Hooper and Scott 2005). Although the signaling pathway was first discovered in Drosophila, where it primarily acts to pattern the cuticle of the larva and the appendages of the adult fly, in vertebrates, a multitude of developmental processes in the embryo and the adult organism have now been shown to be regulated by Hh activity. These include effects on cell proliferation and cell survival as well as cell fate determination. Consistent with all of these influences of Hh on normal development and physiology, loss of Hh signaling in humans has been linked to a number of congenital abnormalities like holoprosencephaly, whereas excessive activity of the pathway appears to be the etiological factor in the initiation and growth of some of the most common forms of cancers (Altaba et al. 2002; McMahon et al. 2003).
Much of our understanding of the molecular details of the Hh pathway has come from genetic and biochemical experiments with components required for transduction of the signal in Drosophila. From all of these investigations it is evident that nuclear access of the transcriptional activator form of the zinc finger protein, Cubitus interruptus (Ci), and its activation of target gene transcription marks the culmination of events in the Hh transduction cascade (Ingham and McMahon 2001; McMahon et al. 2003; Hooper and Scott 2005). In vertebrates, three distinct Ci homologs, the Gli proteins, have subsumed the function of Ci in the regulation of target gene expression. Because Drosophila Ci and the Gli3, and possibly Gli2, proteins of vertebrates undergo phosphorylation and proteolytic processing in the absence of Hh to yield truncated transcriptional repressor forms, optimal induction of target gene expression is critically determined by the ratio of the activator (Gliact) vs. the repressor variants (Glirep) of these proteins within the nucleus. In addition, since Hh signaling can influence a wide diversity of developmental processes, it is clear that the kinds of genes that are activated in each of these circumstances varies with the cellular context and developmental time, and is likely to require cell-type-specific cofactors. For example, the gene encoding the Hh receptor protein Patched (Ptc) is a conserved and a direct target of Gliact in all cells that respond to Hh activity, irrespective of their lineage (Ingham and McMahon 2001; Hooper and Scott 2005). By contrast, in situations where Hh instigates cell proliferation, as among the precursors of the cerebellar granule cells within the mammalian brain, Gli proteins induce the expression of cyclin genes for stimulation of the cell cycle (Wechsler-Reya and Scott 2001; Roy and Ingham 2002). In the developing spinal cord, on the other hand, a concentration gradient of Hh is translated into graded Gli activity, which in turn activates the expression of a number of transcriptional regulators in a neuronal progenitor cell-type-specific manner (Jacob and Briscoe 2003). Thus, overall changes in gene expression in response to Hh can be accounted for by the summation of those whose expression is directly induced through the loss of Glirep and/or the activity of Gliact, together with others, whose expression is modulated (through activation or repression) by secondary and tertiary transcription factors acting downstream of Gli. Although it is clear that the endpoint in the Hh signal transduction cascade is the regulation of a diversity of target gene transcription leading to specific cellular responses, our knowledge on the number and the kinds of genes that are directly or indirectly regulated by Hh is quite inadequate.
We have been using genetic and cell biological analysis in the zebrafish to study the mechanism of Hh signaling in vertebrates and to understand how Hh specifies individual cell fates within specific lineages (Roy et al. 2001a,b; Wolff et al. 2003, 2004; Baxendale et al. 2004; Tay et al. 2005). As in amniotes, the primary source of Hh ligands within the early zebrafish embryo is the axial midline cells that compose the developing notochord and the floor plate (Krauss et al. 1993; Ekker et al. 1995; Currie and Ingham 1996). Hh activity that emanates from these restricted sources directs the specification of unique cell identities within the neural tube (Lewis and Eisen 2003). Hh also regulates specification of cell patterns within the myotome of vertebrate embryos, and in the zebrafish, it directs the induction of the slow-twitch muscle fiber progenitor fate (Blagden et al. 1997; Du et al. 1997; LEWIS et al. 1999b; ROY et al. 2001b; Baxendale et al. 2004). Mutational inactivation of key components within the signaling pathway such as Smoothened (Smo), the transmembrane protein that is activated when Hh binds Ptc and functions to transmit the signal intracellularly, completely inhibits development of ventral neuronal fates in the spinal cord and the acquisition of the slow-twitch muscle fiber identity in the myotome (Barresi et al. 2000; Varga et al. 2001). Conversely, ectopic activation of the Hh pathway by misexpression of Hh or a dominant negative (dn) version of protein kinase A (PKA), which antagonizes endogenous PKA-mediated phosphorylation and subsequent proteolysis of Gli into Glirep, results in the development of supernumerary ventral neural cell types at the expense of those in the dorsal neural tube (Krauss et al. 1993; Concordet et al. 1996; Hammerschmidt et al. 1996). Within the myotome, excessive numbers of slow-twitch fibers are specified, with a concomitant loss of the fast-twitch muscles—a cell type that represents the default fate of the muscle progenitors (Blagden et al. 1997; Du et al. 1997; Roy et al. 2001b; Baxendale et al. 2004).
Microarrays have been demonstrated to be a powerful technological platform for genomewide expression profiling of gene activity (Schena et al. 1998). The strikingly contrasting developmental effects of the loss and gain of Hh signaling in the zebrafish embryo prompted us to utilize this paradigm for a microarray-based screen, to discover new genes that are regulated by the Hh pathway during early vertebrate development. We have compared the transcriptional profiles of wild-type embryos together with those treated either with cyclopamine, an alkaloid drug that specifically inhibits Smo activity and completely abolishes Hh signaling, or dnPKA, which hyperactivates the pathway. This has led to the identification of a cluster of putative target genes whose expression is differentially regulated, either positively or negatively, by Hh. The list includes (a) genes that have been demonstrated in previous studies to be expressed in cells and tissues whose development is regulated by Hh, (b) several transcripts that have been studied in other developmental perspectives, but whose association with Hh signaling has not been implicated so far, and (c) many novel transcripts whose roles in Hh-regulated events during embryogenesis remain completely unexplored. A study of the developmental expression patterns and quantification of transcript abundance by real-time PCR of a small sample of these genes has revealed that the cell-type-specific distributions of their transcripts in wild-type embryos, as well as those with loss and gain of Hh signaling activity, are entirely commensurate with their expression profiles observed on the microarrays. Moreover, we have discovered that a significant number of the upregulated genes have one or more of the generic Gli-binding motif within their regulatory sequences, suggestive of their direct regulation by Hh. As proof of principle, we also demonstrate that a single perfect Gli-binding site present in an evolutionarily conserved enhancer fragment of nkx2.9, a new member of the zebrafish nk family of homeobox genes discovered in our screen, is sufficient for its spatially restricted expression within the ventral neural tube. We propose that the Hh signaling responsive target genes discovered in our screen will be a valuable resource for functional studies aimed at the elucidation of the mechanism by which the Hh pathway directs diverse cellular events during normal development, as well as under conditions of aberrant signaling that instigates a variety of disease states in humans.
MATERIALS AND METHODS
Zebrafish strains:
Wild-type and mutant strains of zebrafish were maintained under standard conditions of fish husbandry. The strain carrying a mutation in the zebrafish gli1 gene, detourts269, has been described previously.
Embryo samples and RNA extraction:
Zebrafish embryos were collected immediately after fertilization, maintained at 28.5°, and staged by developmental time (hours post-fertilization, hpf) and using morphological criteria (Kimmel et al. 1995). For RNA extraction, embryos (wild-type and experimental samples) were harvested at 24 hpf and stored at −80°. Total RNA was extracted from the frozen embryos using Trizol reagent (Gibco-BRL, Grand Island, NY), purified using Qiagen columns, and its quality was evaluated using gel electrophoresis. Reference RNA was prepared from stage-matched wild-type embryos. Sufficient amounts of reference RNA required for the entire project were prepared at one time and stored as aliquots at −80°.
Cyclopamine treatment, DNA and mRNA injection, in situ hybridization, and antibody labeling:
Cyclopamine samples were purchased from Toronto Research Chemicals. Fertilized eggs were soaked in medium containing 100 μm cyclopamine and grown to desired stages essentially as described previously (Barresi et al. 2001; Wolff et al. 2003). In vitro synthesized capped mRNA encoding dnPKA, Gli1 (approximately 0.1 μg/μl), or the plasmid containing the nkx2.9-gfp reporter transgene (approximately 25 ng/μl) was injected at the single cell stage and the embryos were allowed to develop until 12 or 24 hpf before harvesting. Whole-embryo in situ hybridization and antibody labeling with anti-GFP antibodies (from Abcam) were performed according to standard protocols.
Microarray analysis:
Zebrafish microarrays (Compugen, San Jose, CA) containing 16,416 oligonucleotide probes of selected genes were used in this study. Details of array composition, putative annotation of the genes in the array, and other information are given elsewhere (Mathavan et al. 2005). The oligonucleotide probes were spotted onto poly-l-lysine coated microscope slides using a custom-built DNA microarrayer. Printed arrays were post-processed following the standard procedure described for cDNA arrays (Eisen and Brown 1999). Sample and reference RNA were reverse transcribed in the presence of Cy3–dUTP and Cy5–dUTP (Amersham), respectively, to fluorescently label the target cDNAs. The arrays were hybridized following the strategies described by Eisen and Brown (1999) with minor modifications. For each sample, a minimum of four hybridizations were performed. The signal intensities of Cy5 and Cy3 dyes in each spot and the local background were measured using the GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA) to calculate the net intensity of each spot for analysis.
Normalization of the two channels (sample and control) was performed for each slide using the intensity-based log ratio median method (Yang et al. 2003). To select genes that are differentially expressed in cyclopamine treatment and dnPKA mRNA injection experiments, a two-step filtering process was utilized. First, only those genes that were differentially expressed in cyclopamine treatment or dnPKA mRNA injection compared to their respective wild-type controls were selected. From this data set, the genes that are differentially expressed between cyclopamine treatment and dnPKA mRNA injection were then selected. For assessment of differential expression, a modified t-statistic (SAM) (Tusher et al. 2001) with the maximum standard deviation (among all genes) was used as the fudge constant; the critical P value was set at 0.01, and the critical median fold change was set at 2. Data sets extracted using above statistical analysis were clustered and visualized (Cluster and Tree View; Eisen et al. 1998). Putative annotations of the differentially expressed genes were obtained from the zebrafish chip annotation database (http://giscompute.gis.a-star.edu.sg/∼govind/zebrafish/version2/). Procedures for annotation retrievals and use of the database have been explained earlier (Mathavan et al. 2005).
Real-time PCR:
For transcript quantification by real-time PCR, the SYBR Green I (Roche Applied Science) RNA amplification kit was used on the LightCycler according to the manufacturer's instructions. The primers used for each of the genes are as follows:
AW777717: F- 5′ TCGCGTGTTATTCTCCAAAG 3′, R- 5′ AAGACTATGCGCAACACAGG 3′;
AW777561: F-5′ TCATTCCACTGGTTTGCTCT 3′, R-5′ TTGGAATGACCGAGTGGTTA 3′;
AW342624: F-5′ TCCAATGCAGGTTTCACATT 3′, R-5′ ACAACAATTGGACCCCTGTT 3′;
BG985673: F-5′ AGTCGCAGATCCAGGAGTTT 3′, R-5′ CTTCTCTCCGAACATGCTGA 3′;
AJ317957: F-5′ ACGTTCTTGAACCCCTTCAC 3′, R-5′ GCCGCACTCAACAATGATAC 3′;
AJ311846: F-5′ ACCATTCTTTGGCAAGGTTC 3′, R-5′ TACGGCTGTTCATCTTCTGC 3′;
BI866326: F-5′ CAACTGGAACGGAATGATTG 3′, R-5′ ATGGAGGTCAACAGGTAGGC 3′;
BI533161: F-5′ ATGATTTGGTTTCTGCCACA 3′, R-5′ TTGCCTAAAACCCTCAGCTT 3′.
Prior to quantification, the optimal concentrations of template, primers, and magnesium were determined. Serially diluted plasmid DNA samples were used to construct a standard curve to quantify the test samples as well as the efficiency of amplification.
Construction of the nkx2.9-gfp transgene:
A 1.75-kb fragment of genomic DNA immediately upstream of the nkx2.9 translation initiation site was amplified using the following pair of primers
Forward: 5′-CACGCAGATCTGCTCTTGGGTGTTTGAGAGC-3′
Reverse: 5′-CAGCTGTCGACTACACTGATGTCGCGGTGTT-3′ and cloned into the pEGFP1 plasmid (Clontech).
Microscopy and image processing:
Stained embryos were dissected from their yolk and mounted in 70% glycerol. All imaging was done using a Zeiss Axioplan 2 compound microscope equipped with a Nikon digital camera and image capture software. The images were subsequently arranged into montages using Adobe Photoshop 6.
Computational identification of Gli-binding motifs:
We looked for the presence of the perfect Gli-binding motif (GACCACCCA), or motifs with a single-base-pair mutation (Gli_m1), within the positive and negative strand of the flanking sequences (5 kb upstream of the translation start site and 5 kb downstream of the translation stop site) and within intragenic sequences (5 kb inside the gene, immediately downstream of the translation start site) of the genes that showed differential expression in response to Hh signaling. The 5′ and 3′ ESTs and complete coding sequences (cds) used for this analysis were obtained from the zebrafish UniGene database (Unigene build 89, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene). When available, the designated genomic sequences were extracted from the zebrafish Zv5 assembly, using the University of California, Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu). Intragenic sequences were extracted only for genes with full cds. Position weight matrices were constructed from the positive instances of the Gli_m1 motif found in sequences associated with upregulated genes and zebrafish unigene clusters using an expectation maximization algorithm. Sequence logos were generated using the standard program WebLogo (v. 2.8) (Crooks et al. 2004).
RESULTS
Strategy to globally repress or activate the Hh pathway in the zebrafish embryo:
We reasoned that a differential analysis of RNA expression profiles from embryos with complete absence vs. those with ectopic levels of Hh activity is likely to uncover genes that are specifically regulated by Hh signaling during development. To achieve this, we either treated batches of zebrafish embryos with cyclopamine, an antagonist of Smo, or injected them with mRNA-encoding dnPKA to hyperactivate the pathway. Using the expression of the zebrafish homolog of the ptc gene, ptc1, as a sensitive readout of Hh signaling levels as well as the expression patterns of several cell-type restricted markers as our assay, we and others have provided substantial evidence in earlier studies that cyclopamine is a specific inhibitor of Hh signaling, whereas loss of PKA activity results in the specific and global activation of the pathway in the zebrafish embryo (Concordet et al. 1996; Hammerschmidt et al. 1996; BARRESI et al. 2001; Wolff et al. 2003). Although zebrafish hh genes show dynamic expression in a variety of organizing centers throughout embryonic development (Krauss et al. 1993; Ekker et al. 1995; Currie and Ingham 1996), for the purpose of this screen, we selected a representative developmental stage—24 hpf—by which time the majority of the cell fates that depend upon Hh signaling from midline tissues become specified.
The relative mRNA abundance levels of embryos treated with cyclopamine or injected with dnPKA RNA were measured from genomewide expression profiling using zebrafish microarrays containing probes for more than 16,000 genes (Mathavan et al. 2005). The profiles of transcripts of a selected cohort of differentially expressed genes were grouped into two different clusters (Figure 1). A class of genes that did not significantly change their expression levels in the above treatments was also identified; these genes may not be involved directly or indirectly in the Hh pathway and were not considered for further investigation. For the rest of the analysis, we focused on genes that were differentially expressed due to loss or gain of Hh function. To reliably identify genes whose expression is critically dependent on Hh activity from the set of about 16,000 probes on the arrays, we have primarily focused on those whose transcript levels showed more than twofold change (increase or decrease) between the cyclopamine and dnPKA-treated embryos (P value of 0.01; see materials and methods).
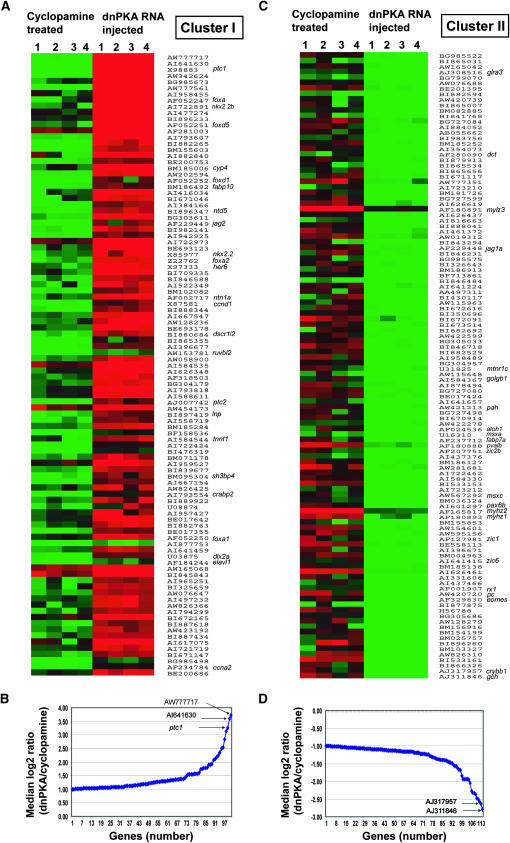
Figure 1.—
Expression profiles of significantly upregulated and downregulated genes in dnPKA mRNA-injected embryos compared to their expression in those treated with cyclopamine (minimum twofold change; P value > 0.01). (A) Upregulated genes (101 genes). (C) Downregulated genes (114 genes). For each treatment, four replicate hybridizations were performed and data obtained from each hybridization experiment are presented. (B, D) The pattern of differential expression (median log2 ratio; dnPKA mRNA/cyclopamine) of all the genes in cluster I (B) and cluster II (D) is graphically represented. The fold change in the differential expression ranged between 2 and 13 (median log2 ratio 1.0–3.7) for the genes that were upregulated and 2 and 8 (median log2 ratio 1.0–2.8) for the genes that were downregulated, respectively.
Genes whose expression showed substantial positive regulation by Hh signaling:
The genes that were significantly (median log2 ratio > 1) upregulated in dnPKA mRNA-injected embryos (101 genes) compared to their transcript levels in the cyclopamine-treated group are presented in cluster I (Figure 1A; supplemental Figure 1 at http://www.genetics.org/supplemental/). A critical analysis of the expression profiles of the genes within this cluster, however, showed different degrees of modulations in the suppression or induction of transcript accumulation as a result of loss or gain of Hh function, respectively, with two readily apparent patterns.
Some genes showed a dramatic reduction of expression in cyclopamine-treated embryos and a strong upregulation in those injected with dnPKA mRNA (Figure 1A and Figure 2A, B; supplemental Figure 2 at http://www.genetics.org/supplemental/).
For others, the expression did not change appreciably on loss of Hh signaling, but the levels of their transcripts increased substantially on ectopic Hh pathway activity (Figure 1A and Figure 2C(i), D(i); supplemental Figure 2 at http://www.genetics.org/supplemental/).
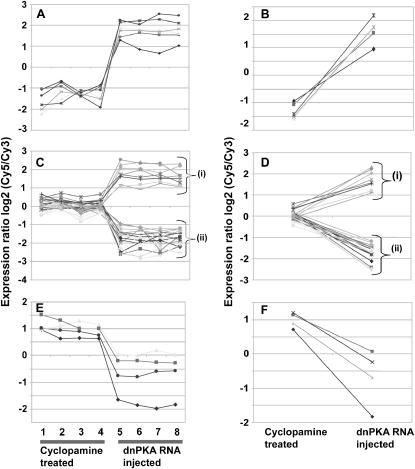
Figure 2.—
Three different patterns of gene expression in embryos treated with cyclopamine or injected with dnPKA mRNA. (A, B) Pattern 1. Genes in this group are significantly downregulated with cyclopamine treatment and upregulated on dnPKA mRNA injection. For each treatment, four hybridizations were performed and individual data points (A) and mean values (B) are presented. (C, D) Pattern 2. The genes in this group are not influenced dramatically by cyclopamine treatment, but they are significantly upregulated (C(i), D(i)) or downregulated (C(ii), D (ii)) in the dnPKA mRNA-injected batch (C, individual values; D mean values). (E, F) Pattern 3. Few genes represent this group and display increased levels of transcripts on cyclopamine treatment while they may or may not be influenced by dnPKA RNA treatment (E, individual values; F, mean values). Details of all the genes included in this figure are presented in supplemental Figure 2 at http://www.genetics.org/supplemental/.
On an average, the genes changed their levels of expression from a minimum of 2-fold to a maximum of about 13-fold (median log2 ratio 1.0–3.7), indicating that transcript abundance of some of the genes dramatically increased in dnPKA mRNA-injected embryos (Figure 1B).
Putative annotation of the genes in cluster I revealed that 26 of them have full annotation, while the rest are zebrafish gene collection (zgc) clones or ESTs (supplemental Figure 1 at http://www.genetics.org/supplemental/). Of the annotated clones, a number of them are known to be involved in the Hh signaling pathway from previous investigations, while the rest of the annotated genes and ESTs have so far not been linked, directly or indirectly, to Hh signaling. A classical example of an annotated gene involved in the pathway is ptc1, which is known to be upregulated in response to Hh in all cellular contexts that have been examined so far (Ingham and McMahon 2001; Hooper and Scott 2005). Expression profiles obtained in this study show that ptc1 is upregulated by about 10-fold in the dnPKA mRNA-injected embryos, compared to its expression in those treated with cyclopamine (Figure 1B). Besides ptc1, genes that have been known to be positively regulated by the Hh pathway in different developmental contexts that figured prominently in this cluster include cyclinD1 (ccnd1) (Kenney and Rowitch 2000; Duman-Scheel et al. 2002), ptc2 (Lewis et al. 1999a), nkx2.2a and 2.2b (Barth and Wilson 1995; Schafer et al. 2005), foxa (Odenthal et al. 2000), foxa2 (Norton et al. 2005), and netrin1a (net1a) (Lauderdale et al. 1998) among others. On the basis of the above results, we can conclude that the remainder of the genes in this cluster, which display expression profiles similar to that of the known Hh-regulated genes, can be expected to play a role in Hh-dependent developmental processes. For example, the EST clones with GenBank accession numbers AW777717 (UniGene id: Dr.2317; zebrafish UniGene build 85) and AI641630 (UniGene Id: Dr.27431; zebrafish UniGene build 85), increased their expression by about 13-fold in dnPKA mRNA-injected embryos compared to their levels in the cyclopamine-treated group (Figure 1B). The levels and profiles of transcript abundance of the above genes in response to loss or gain of Hh function are almost identical to the expression level of ptc1, implying that they are likely to be targets whose expression is critically dependent on Hh.
Genes that exhibited significant negative regulation by the Hh pathway:
Cluster II represents a collection of genes whose transcript abundance profile is somewhat diametrically opposite to the pattern observed in cluster I (Figure 1C). Following dnPKA mRNA injection, 114 genes displayed reduced amounts of transcript accumulation compared to their levels in cyclopamine-treated embryos (Figure 1C; supplemental Figure 3 at http://www.genetics.org/supplemental/). The differential expression of the genes between these groups ranged from two- to eightfold (Figure1D). For the vast majority of the genes, the levels were significantly downregulated in the dnPKA-injected embryos, while their expression was not that strongly influenced by cyclopamine treatment (Figure 2C(ii), D(ii); supplemental Figure 2 at http://www.genetics.org/supplemental/). Only a handful of genes showed a slight increase on cyclopamine treatment while their transcript levels either did or did not decrease substantially in the dnPKA mRNA-injected batch (Figure 2, E and F; supplemental Figure 2 at http://www.genetics.org/supplemental/). Of the total of 114 genes in cluster II, only 23 had full annotation with gene ontology (GO) terms; several of them appear to be related to the differentiation of the fast-twitch muscle cell type like fast muscle-specific myosin heavy chains (myhz1 and myhz2), fast myosin light chain (mylz3), and parvalbumin (pvalb)). Other genes have been previously linked to the development of eye structures (crystallins, pax6b, and retinal homeobox 1, rx1) and a number of them represent zinc finger proteins (zic1, zic2b, zic6) (see Figure 1C and supplemental Figure 3 at http://www.genetics.org/supplemental/). As mentioned previously, functional studies from our group and those of others have shown that midline-derived Hh activity promotes the specification of slow-twitch muscle fibers in the somite, and ectopic Hh signaling can convert the entire myotome to the slow-twitch fate at the expense of fast fibers (Blagden et al. 1997; Du et al. 1997; Roy et al. 2001b; Baxendale et al. 2004). On similar lines, cell fates along the proximo-distal axis of the developing eye are also regulated by Hh signaling, and high levels of Hh activity can suppress the specification of distal eye cell types(like the lens) at the expense of proximal fates (Ekker et al. 1995; MacDonald et al. 1995). Thus, detection of the downregulated genes in our microarray analysis is commensurate with their expression in tissues and cell types that are normally antagonized by Hh signaling.
Developmental expression patterns of selected novel target genes recapitulates their Hh signaling-dependent profiles on the microarrays:
To corroborate our microarray data, we have subjected a number of selected clones to whole-mount in situ hybridization on wild-type embryos at two distinct developmental stages, 12 hpf and 24 hpf, and compared their expression patterns with that observed in embryos treated with cyclopamine or injected with dnPKA mRNA.
- Genes that showed positive regulation in the microarray analysis (genes are referred to by their GenBank accession numbers)
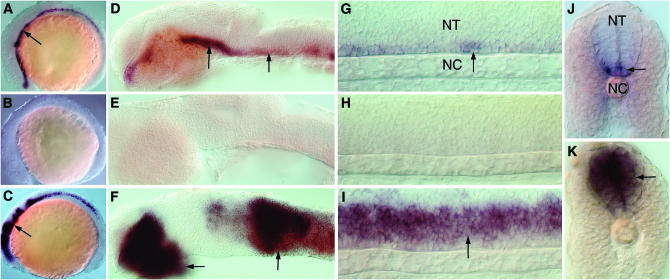
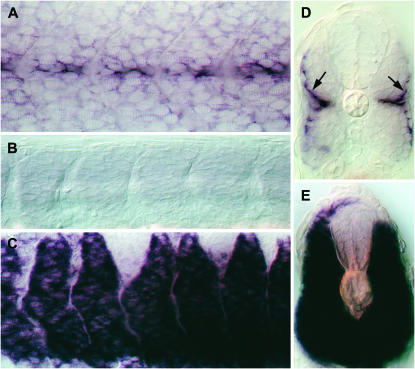
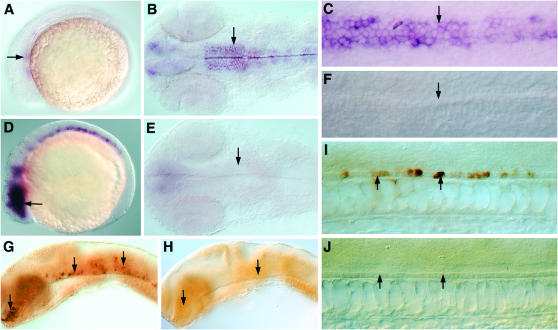
- AW777717: The expression of this gene is restricted exclusively to the developing central nervous system (CNS) in both of the developmental stages examined (Figure 3). Within the CNS, expression is observed in the ventral neuronal cells whose specification is dependent on Hh signals from the midline (Figure 3, A, D, G, and J). Consistent with this, the expression of this gene is completely lost in embryos exposed to cyclopamine (Figure 3, B, E, and H), while dnPKA mRNA injections resulted in its dramatic misexpression within the neural tube (Figure 3, C, F, I, and K). Consideration of sequence homology as well as expression profile indicates that this gene encodes the zebrafish ortholog of the mammalian homeodomain containing trancription factor Nkx2.9 (supplemental Figure 4A at http://www.genetics.org/supplemental/; Pabst et al. 1998). Previous studies have identified the zebrafish homologs of the nkx2.2 (nkx2.2a and nkx2.2b) (Barth and Wilson 1995; Schafer et al. 2005) and the nkx6 genes (Cheesman et al. 2004), which, like nkx2.9 described here, are definitive markers of ventral cell types in the neural tube and are induced by high levels of Hh activity. Our discovery of a zebrafish nkx2.9 gene ortholog further reinforces the view that the combinatorial activity of nkx family members in the patterning of the neural tube is well conserved in vertebrate embryos.
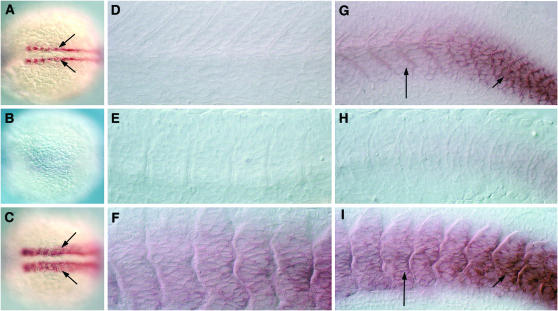
- AI416034: This gene is expressed predominantly in the adaxial cells, the precursors of the slow-twitch muscle fibers, in 12-hpf embryos (Figure 4A). This expression appears to be transient, since only remnants of the pattern are visible in the slow muscles in the tail end of embryos at 24 hpf (Figure 4, D and G). Because Hh activity is essential for the specification of the slow muscle cells, embryos with loss of Hh signaling exhibited little or no expression of this gene (Figure 4, B, E, and H), while those with ectopic Hh signaling resulting from overexpression of dnPKA showed upregulation of expression within the myotome (Figure 4, C, F, and I). Using 5′ and 3′ RACE PCR we amplified and assembled the full-length sequence for this gene that shows complete sequence identity to the zgc clone zgc:103659. BLAST searches revealed that this gene encodes a protein that shows the most significant homology to mammalian fibulins, extracellular matrix-associated proteins that contain epidermal growth factor (EGF)-like Ca2+ ion-binding domains (supplemental Figure 4B at http://www.genetics.org/supplemental/).
- AF281003: At 12 hpf, this gene is expressed in a zone of cells around the developing tail bud of wild-type embryos (data not shown). By 24 hpf, expression is observed prominently in all slow-twitch muscle cells (Figure 5A, D). In addition, this gene is expressed in the developing lens at this stage (data not shown). Consistent with its expression in slow muscle fibers, we did not observe any expression of the gene at 24 hpf in the myotome of embryos treated with cyclopamine (Figure 5B). On the contrary, in dnPKA mRNA-injected embryos there was ubiquitous expression of the gene throughout the myotome (Figure 5, C and E). We were able to identify a full-length clone of this gene in the zgc database, zgc:86932. Sequence analysis of the encoded protein indicates that it has strong homology to troponin C (supplemental Figure 4C at http://www.genetics.org/supplemental/). Troponins bind Ca2+ ions and are components of muscle sarcomere; this particular protein, therefore, is likely to represent the slow-twitch muscle-specific variant of troponin C.
- BM102082: In 12-hpf wild-type embryos, expression of this gene figures most prominently in the dorsal portion of the forebrain primordium and in the ventral region of the developing neural tube (Figure 6A). Subsequently, at 24 hpf, high levels of transcription can be observed all along the ventral region of the brain and the spinal cord, but in a much broader area than nkx2.9 (Figure 6, D and G). Embryos treated with cyclopamine showed a consistent loss of expression in the ventral neural tube at 12 hpf (Figure 6B). Marked loss of transcription is also apparent in the brain at 24 hpf, concomitant with a reduction in the overall levels of expression along the ventral spinal cord at this stage (Figure 6, E and H). In response to ectopic Hh signaling, there was a clear increase in the levels and an expansion in the domain of expression of the gene to the dorsal extent of the brain and spinal cord at 12 and 24 hpf (Figure 6, C, F, and I). This gene encodes a zebrafish homolog of the mammalian Heparan sulfate 6-o-endosulfatase, Sulfatase FP2 (supplemental Figure 4D at http://www.genetics.org/supplemental/), that is identical to a full-length clone, AY332607, available in the sequence database. In the quail embryo, the expression of a similar gene, sulfatase FP1 (also called qsulf1), is regulated by Shh signaling in the myotome (Dhoot et al. 2001). Although, expression of sulfatase FP2 has been examined in the developing CNS of the mammalian embryo (Nagamine et al. 2005), our analysis presents the first explicit connection of the regulation of expression of the gene by Hh activity.
- AI965251: We observed a rather ubiquitous expression of this gene in embryos at 12 hpf (data not shown). In 24-hpf embryos, however, a clear spatial pattern is apparent with high levels of expression all along the ventral region of the brain and spinal cord (Figure 7, A and D), a pattern quite similar to that of the sulfatase gene described above. In cyclopamine-treated embryos, there was a distinct reduction in the levels of expression in the spinal cord, and more prominently in the brain (Figure 7, B and E), whereas in those injected with dnPKA mRNA, strong upregulation of expression occurred dorsally in the brain and the spinal cord (Figure 7, C and F). The predicted protein encoded by the longest EST sequence available for this gene in the zebrafish unigene cluster exhibits significant homology to mammalian GREB1 (Gene regulated in breast cancer 1; Ghosh et al. 2000), a novel protein that is induced in response to estrogen in breast cancer cells (supplemental Figure 4E at http://www.genetics.org/supplemental/).
- Genes that showed downregulation in the microarray analysis
- AI722462: The expression of AI722462 localizes preferentially to the dorsal cells of the CNS of wild-type embryos at 12 as well as 24 hpf (Figure 8, A, D, and G and data not shown). This expression is distinctly expanded ventrally in embryos that lack Hh activity (Figure 8, B, E, and H). Conversely, in line with the ventralization of the neural tube on ectopic activation of Hh signaling, we found complete absence of expression of this gene from the dorsal neural cells in embryos overexpressing dnPKA (Figure 8, C and F). The protein encoded by this gene is the zebrafish homolog of Scube2, one member of a family of vertebrate extracellular matrix associated proteins that contain a signal peptide, CUB domain and EGF repeats (Yang et al. 2002). Recent functional studies with zebrafish embryos have suggested that Scube2 functions as a positive modulator of the Hh pathway (Kawakami et al. 2005; Woods and Talbot 2005; Hollway et al. 2006; our unpublished observations). The molecular mechanism of its action remains enigmatic, although it has been postulated that the protein functions by inhibiting the antagonizing effects of BMP-like molecules that are secreted from the dorsal neural tube, on Hh signaling (Kawakami et al. 2005). In light of this, our present analysis has uncovered an interesting negative regulation that exists between Hh activity and the expression of the scube2 gene in the dorsal neural tube.
- BI866326: The expression of this gene, like that of scube2, is confined to the dorsal neural tube in wild-type embryos at 24 hpf (Figure 9, A and D). Unlike scube2, however, we could not detect any specific pattern of expression at 12 hpf (data not shown). In line with the similarity in the domain of expression of this gene and scube2, we found that its expression expanded on cyclopamine treatment (Figure 9, B and E) and is completely repressed in embryos that were injected with dnPKA mRNA (Figure 9C). Database searches have revealed that this gene encodes a protein that is similar to glutamate ionotropic receptors of mammals (supplemental Figure 4F at http://www.genetics.org/supplemental/), which possibly explains the rather late onset of its expression in the differentiating dorsal neurons of the spinal cord.
Figure 3.—
In situ hybridization analysis of AW777717. (A–C) Expression in the developing CNS (arrows) of a 12-hpf wild-type embryo (A), after cyclopamine treatment (B), and after dnPKA mRNA injection (C). (D–F) Expression in the fore-, mid-, and hindbrain (arrows) of a 24-hpf wild-type (D), cyclopamine-treated (E), and a dnPKA mRNA-injected embryo (F). (G–I) Expression in the spinal cord (arrows) of a 24-hpf wild-type (G), a cyclopamine-treated (H), and a dnPKA mRNA-injected embryo (I). (J, K) Transverse sections of a 24-hpf wild-type (J) and a dnPKA-injected embryo (K), showing pattern of expression in the neural tube. NT, neural tube; NC, notochord. A–I are lateral views oriented anterior to the left and dorsal to the top; J and K represent frontal views of transverse sections.
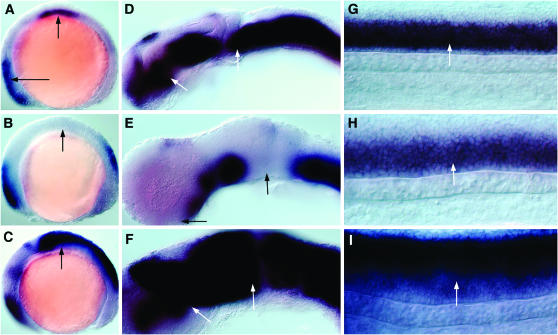
Figure 4.—
Expression pattern of AI416034. (A–C) Expression in the slow-twitch muscle precursors, the adaxial cells (arrows) of a 12-hpf wild-type embryo (A), after cyclopamine treatment (B) and on dnPKA mRNA injection (C). (D–F) Anterior somites of a 24-hpf wild-type embryo (D), after cyclopamine treatment (E), and after dnPKA mRNA injection (F). (G–I) Expression in the most posterior somites of a 24-hpf wild-type embryo (G), an embryo treated with cyclopamine (H), and an embryo injected with dnPKA mRNA (I). Anterior is to the left. A–C represent dorsal views; D–I represent lateral views, oriented dorsal to the top.
Figure 5.—
Expression pattern of AF281003. (A–C) Expression in the slow-twitch muscle fibers of a 24-hpf wild-type embryo (A), after cyclopamine treatment (B), and on dnPKA injection (C). (D, E). Transverse sections showing expression in a 24-hpf wild-type embryo (D) and an embryo injected with dnPKA (E). The slow-twitch muscle fiber layer expressing the gene is indicated with arrows in D. A–C represent lateral views with anterior to the left and dorsal to the top; D and E are frontal views of transverse sections.
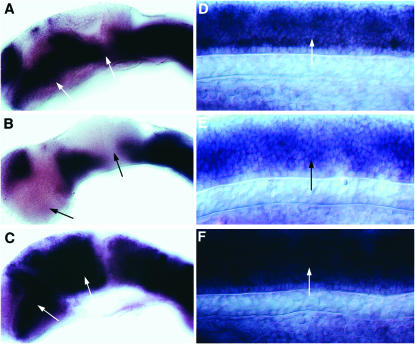
Figure 6.—
Expression pattern of BM102082. (A–C) Expression in the ventral neural tube (short arrows) of a 12-hpf wild-type embryo (A), after cyclopamine treatment (B), and after dnPKA mRNA injection (C). (A) Additional expression in the dorsal forebrain, that is independent of Hh regulation, is indicated (long arrow). (D–F) Expression in the fore-, mid-, and hindbrain (arrows) of a 24-hpf wild-type (D), cyclopamine-treated (E), and a dnPKA mRNA-injected embryo (F). (G–I) Expression in the spinal cord (arrow) of a 24-hpf wild-type (G), cyclopamine-treated (H), and a dnPKA mRNA-injected embryo (I). Lateral views are oriented anterior to the left and dorsal to the top.
Figure 7.—
Expression pattern of AI965251. (A–C) Expression in the fore-, mid-, and hindbrain (arrows) of a 24-hpf wild-type (A), cyclopamine-treated (B), and a dnPKA mRNA-injected embryo (C). (D–F) Expression in the spinal cord (arrow) of a 24-hpf wild-type (D), cyclopamine-treated (E), and a dnPKA mRNA-injected embryo (F). Lateral views are oriented anterior to the left and dorsal to the top.
Figure 8.—
In situ hybridization analysis of AI772462. (A–C) Expression pattern in the brain (arrows) of a 24-hpf wild-type embryo (A), an embryo treated with cyclopamine (B), and after dnPKA overexpression (C). Expression in the spinal cord (arrow) of a 24-hpf wild-type embryo (D), after cyclopamine treatment (E), and after dnPKA overexpression (F). (G, H) Transverse section of a 24-hpf wild-type embryo (G) and after cyclopamine treatment (H), showing expression in the spinal cord (arrow). A–F represent lateral views with anterior to the left and dorsal to the top; G and H are frontal views of transverse sections.
Figure 9.—
Expression pattern of BI866326. (A–C) Pattern of expression in the spinal cord (arrow) of a 24-hpf wild-type embryo (A), an embryo treated with cyclopamine (B), and after dnPKA mRNA injection (C). (D, E) Transverse sections showing expression in the spinal cord (arrow) of a 24-hpf wild-type embryo (D) and after cyclopamine treatment (E). A–C represent lateral views with anterior to the left and dorsal to the top; D and E are frontal views of transverse sections.
Analysis of real-time transcript accumulation of representative up- and downregulated genes further corroborates the Hh-dependent control of their expression:
In addition to in situ hybridizations, we have also performed real time PCR for eight clones that exhibited Hh-dependent expression profiles on the microarrays: four genes that are upregulated and another four that are downregulated by Hh signaling. Two of the clones that were tested by in situ hybridization were also subjected to real-time PCR and they displayed identical patterns of expression in both of these analyses (supplemental Figure 5 at http://www.genetics.org/supplemental/). Although the actual values obtained in the PCR vs. the array hybridization studies were not identical owing to variations in the sensitivity of the two methods, however, the overall patterns of expression were again highly compatible in both of these experiments (supplemental Figure 5 at http://www.genetics.org/supplemental/). Thus, the real-time PCR data provide an additional line of validation of the results obtained from the microarrays.
Gli protein-binding motif exists within the regulatory elements of zebrafish genes that are evolutionarily conserved direct Hh targets:
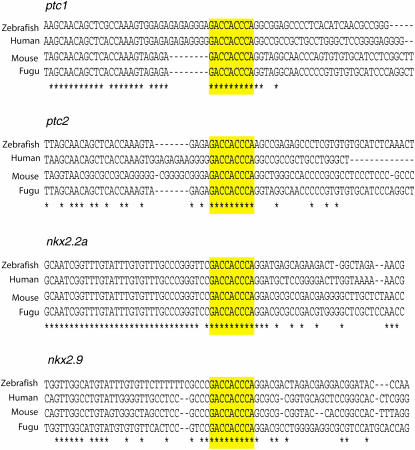
To understand how Hh activity controls the genes that showed differential expression in our screen, especially those that exhibited an upregulation of their expression, we have to determine whether they are directly activated by the Gli proteins or are induced by secondary transcriptional events downstream. The first line of investigation in this direction will involve the identification of the consensus Gli-binding motif within the regulatory sequences of these genes. In humans, the direct Hh target PTCH1, is controlled by the consensus Gli-binding motif GACCACCCA (hereafter referred to as Gli_0) in its promoter (Kinzler and Vogelstein 1990; Agren et al. 2004), and all of the three mammalian Gli proteins, Gli1, Gli2, and Gli3, are capable of binding to this consensus motif (Agren et al. 2004). In addition, there are several instances, such as for mammalian Gli1 and Myf5, in which Gli-dependent regulation of expression of Hh target genes is mediated through binding sites that are variations from the consensus (Dai et al. 1999; Gustafsson et al. 2002; Ikram et al. 2004). The zebrafish Gli1 and Gli2 proteins can induce reporter gene expression through such a variant Gli-binding motif, GAACACCCA (hereafter any variant Gli-binding motif with one mutation is referred to as Gli_m1), located within the 3′ enhancer region of the mouse Foxa2 (HNF3β) gene (Sasaki et al. 1997; Karlstrom et al. 2003), suggestive of a highly stringent level of conservation in Gli protein–DNA interaction. However, the presence of the Gli_0 or Gli_m1 sites within the genome of the zebrafish, even with respect to genes like ptc1 and patched2 (ptc2), that are thought to be evolutionarily conserved direct targets of Hh signaling, has not been analyzed to date. Using a bioinformatics approach, we were able to identify conserved enhancer elements with the perfect Gli-binding motif in the upstream sequences of zebrafish ptc1 and ptc2 implicating the existence of common mechanisms in the regulation of Hh target gene expression across vertebrate species (Figure 10).
Figure 10.—
Sequence alignments of the consensus Gli-binding motif and the flanking regions in the 5′ enhancers of the ptc1, ptc2, nkx2.2a, and nkx2.9 genes from zebrafish, fugu, mouse, and humans. The Gli-binding sites are highlighted in yellow.
Gli-binding sites are enriched in the regulatory elements of genes that showed positive induction in response to Hh:
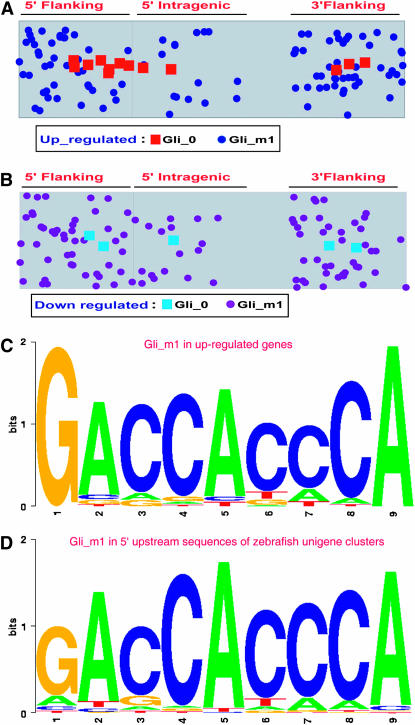
Prompted by the discovery of the Gli_0 motif in the upstream regulatory region of the zebrafish homologs of the evolutionarily conserved Hh target genes, we decided to look for the distribution of Gli_0 and/or the Gli_m1 motifs in the genes that displayed positive or negative regulation on the microarrays in response to Hh signaling. For the purpose of this search, we restricted our analysis to 5 kb of upstream and downstream (intragenic) sequences relative to the translation start site (since transcription start sites have not been accurately determined for the vast majority of the genes), as well as to 5 kb of sequence following the translation stop at the 3′ end of the genes (see materials and methods for further details of this analysis). Of the upregulated genes, 82 have been mapped to specific genomic locations. The Gli_0 motif was observed in 12 of these genes (8 sites in the 5′ upstream region, 2 in the intragenic, and 3 in the 3′ downstream region) (Figure 11A and supplemental Figure 6 at http://www.genetics.org/supplemental/). In addition to ptc1 (CK681296) and ptc2 (AJ007742), another well-established Hh target in this list is the hedgehog interacting protein gene (hhip; BM859918, DT078309; Chuang and McMahon 1999). Others, such as the nkx family members (nkx2.2a, BC076402; nkx2.2b, BC091555; nkx2.9, BC091676) and foxa (AF052247) have been recognized as targets of Hh signaling; however, it has so long been unclear whether they are directly induced by the Gli proteins. This study shows that these genes do contain the Gli_0 motif within their regulatory sequences, a finding that substantiates the view that their expression is indeed directly induced by Hh activity (Figure 10; Figure 11A; supplemental Figure 6 at http://www.genetics.org/supplemental/). The remainder of the genes with the Gli_0 motif are sulfatase fp2 (AY332608), foxd1 (BC075922), cellular retinoic acid binding protein 2 (crabp2, BC091960), a slow muscle-specific myosin essential light chain (BC062288), and a novel gene (CK678864) that shows homology to a new class of cysteine- and tyrosine-rich proteins from humans. Since we were able to detect the Gli_0 motif in only 10% of the upregulated genes, we scanned for its variant, the Gli_m1 site, and detected the motif in 31 additional genes that include distal-less homeobox 2a (dlx2a), cyclinD1 (cnnd1), foxa1, foxd5, and foxa2. While the majority of these have not been associated with Hh signaling previously, the expression of some like the dlx2a, ccnd1, and foxa2 has been documented to be controlled by the Hh pathway (Sasaki et al. 1997; Kenney and Rowitch 2000; Rallu et al. 2002; Duman-Scheel et al. 2002; Norton et al. 2005). In fact, the Gli_m1 site in the 3′ end of zebrafish foxa2 is identical to the Gli1-binding site present in the 3′ enhancer of the foxa2 gene of mammals. We found that a number of genes had more than one Gli_m1 motif, thus taking the total number of Gli-binding sites present among the 82 genes analyzed to 119 (50 in the 5′ region, 18 in intragenic, and 51 in the 3′ region) (Figure 11A; supplemental Figure 6 at http://www.genetics.org/supplemental/).
Figure 11.—
The distribution of the Gli_0 and Gli_m1 motifs and the Gli_m1 position weight matrices in the up- and downregulated genes. (A) Plot showing the distribution of Gli_0 and Gli_m1 motifs in the genes that showed upregulation of expression due to dnPKA mRNA injection. Genomic locations of the motifs are presented in supplemental Figure 6 at http://www.genetics.org/supplemental/. (B) Plot showing the distribution of Gli_0 and Gli_m1 motif in the genes that showed downregulation of expression due to dnPKA mRNA injection. Genomic locations of the motifs are presented in supplemental Figure 7 at http://www.genetics.org/supplemental/. (C) Nucleotide variations within the Gli_m1 motif in the upregulated genes. The motif is described by differentially sized letters. The height of a letter at a particular position is directly proportional to the frequency of its occurrence. The first base (G) and the last base (A) are conserved in all of the Gli motifs present in the upregulated genes. (D) Similar analysis of the Gli_m1 motif in the downregulated genes. Unlike the upregulated genes, all the bases of the motif show mutations in the downregulated genes.
Using all of the Gli_m1 sequences observed in the upregulated genes, we constructed a position weight matrix in the form of sequence logos using the expectation-maximization algorithm to identify the Gli_m1 consensus sequence associated with upregulated genes (Figure 11C). It appears that of the nine bases in the Gli_0 consensus, the first and last base were not mutated in the upregulated genes while the other bases showed more variations, a finding that fully concurs with the efficacy of binding of mammalian Gli1, 2, and 3 to all possible base substitutions within the consensus motif (Hallikas et al. 2006).
A similar analysis for the downregulated genes revealed that only two genes, fast muscle-specific myhz2 (BC071279) and eomesodermin (eomes, AF287007), have the Gli_0 motif in their 5′ upstream regions. On the whole, 38 of the 114 downregulated genes have the Gli_0 or the Gli_m1 motifs in the 5′ upstream (48 sites), in the intragenic (18 sites), and in the 3′downstream regions (44 sites) (Figure 11B and supplemental Figure 7 at http://www.genetics.org/supplemental/). As indicated for the upregulated genes, we also found that some of the downregulated genes have more than one Gli_m1 motif in their sequence. A position weight matrix analysis for this motif in the 5′ sequences of 26,164 zebrafish unigenes (only those genes for which sufficient upstream sequence information are available have been considered) is presented in Figure 11D. It is noteworthy, that unlike the upregulated genes, the identified Gli_m1 motifs in this instance exhibited mutations in the flanking bases (Figure 11D). Overall, the Gli_m1 motif is marginally enriched (approximately 3%) in the upregulated genes compared to its distribution in the unigene clusters. This could indicate a possible functional relevance of the motif in the upregulated genes.
nkx2.9 is a direct target of Gli1 activity in the ventral neural tube of the zebrafish embryo:
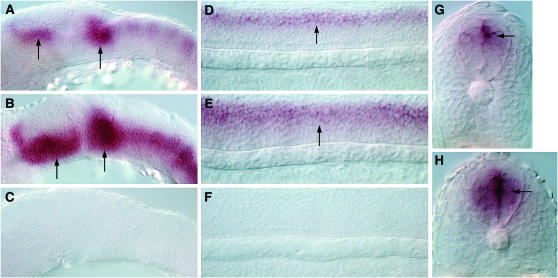
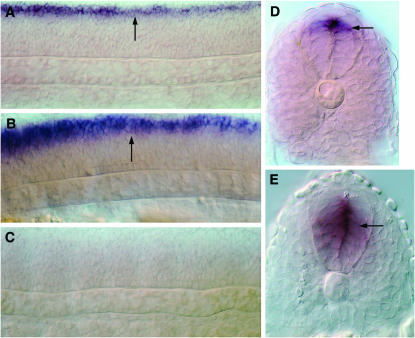
The final verification of the biological significance of the Gli-binding sites that we have been able to identify using bioinformatics will require the functional validation of their ability to direct the spatio-temporal transcription of the relevant genes in a Gli-dependent manner during the course of embryogenesis. As a starting point for this kind of analysis, we investigated whether the expression of nkx2.9, a gene whose transcription showed maximal positive responsiveness to Hh signaling in our microarray assay, is directly regulated by the Gli proteins through the Gli_0 motif in its enhancer. In contrast to mammals, where Gli2 and Gli3 are the primary mediators of Hh signaling in the neural tube, in the zebrafish embryo, the Gli1 protein appears to have assumed a more significant role. Mice that are homozygous for loss-of-function alleles of Gli1 are viable and are not associated with any developmental abnormalities (Park et al. 2000); however, gli1 mutant zebrafish are embryonic lethal and exhibit striking defects in the specification of Hh-dependent cell fates in the ventral neural tube (Karlstrom et al. 2003). In embryos homozygous for mutations in the detour (dtr) gene, which encodes zebrafish Gli1, there was a dramatic reduction in nkx2.9 expression at all levels along the antero-posterior axis of the developing neural tube (Figure 12, A, E, and F). On the contrary, injection of synthetic gli1 mRNA into wild-type embryos to induce high levels of Gli1 protein misexpression resulted in ectopic induction of nkx2.9 within the neural tube (Figure 12D). We next performed a transient transgenic assay by injecting a construct containing approximately 1.75 kb of nkx2.9 5′ enhancer sequence encompassing the Gli_0 motif fused upstream of the gfp reporter gene into newly fertilized zebrafish eggs. A previous analysis in the mouse embryo has indicated that the homologous enhancer fragment of the mouse Nkx2.9 gene is sufficient for directing reporter gene expression in the ventral neural tube (Santagati et al. 2003). In line with this mammalian study, we found that the injected zebrafish embryos displayed faithful expression of GFP in cells along the ventral region of the brain and spinal cord, recapitulating the endogenous expression pattern of nkx2.9 (Figure 12, G and I; 69 of 86 injected embryos showed this pattern). By contrast, injection of the same construct harboring a deletion of the Gli-binding site completely abolished GFP expression in the ventral neural tube (Figure 12, H and J; none of the 71 injected embryos showed GFP expression in the nkx2.9 domain). All of these findings not only provide incontrovertible evidence that the Hh-dependent induction of nkx2.9 expression is directly mediated by the activity of the Gli1 protein, but also validate, for the first time in the zebrafish, the functionality of the Gli_0 motif in Hh-dependent target gene regulation.
Figure 12.—
nkx2.9 expression in the ventral neural tube is directly regulated by Gli1 activity. (A) A 12-hpf gli1 mutant embryo, showing dramatic reduction in nkx2.9 expression (arrow). (B, C) Expression of nkx2.9 (arrow) along the ventral brain (B) and spinal cord (C) of a 24-hpf wild-type embryo. (D) A12-hpf, gli1 mRNA-injected embryo, showing ectopic nkx2.9 expression (arrow). (E, F) Loss of nkx2.9 expression (arrow) from the ventral brain (E) and spinal cord (F) of a 24-hpf gli1 mutant embryo. (G, I) Restricted expression of the nkx2.9-gfp transgene (arrows) exclusively in the ventral brain (G) and spinal cord (I) of a 24-hpf wild-type embryo, recapitulating the endogenous pattern of the gene. (H, J) Absence of GFP expression from the ventral brain (H) and spinal cord (J) of an embryo injected with a variant of the nkx2.9-gfp construct that lacks the Gli-binding site. A, D, G, H depict lateral views with anterior to the left and dorsal to the top; B, C, E, F show dorsal views with anterior oriented to the left.
DISCUSSION
Genetic studies, in Drosophila as well as vertebrates, have shown that the zinc finger containing Gli family of proteins is the primary mediator of Hh-dependent transcriptional regulation of target genes in responding cells (Ingham and McMahon 2001; Hooper and Scott 2005). It is now largely accepted that Hh modulates target gene transcription by inhibiting the formation of the proteolytically processed transcriptional repressor form of Gli, Glirep, and by promoting the accumulation of the transcriptional activator variant, Gliact, in the nucleus (Ingham and McMahon 2001; Hooper and Scott 2005). Therefore, genes that are transcriptionally activated due to the loss of Glirep and/or the generation of Gliact can be classified as direct targets of the Hh pathway. If a direct target gene encodes a transcriptional regulator it can, in turn, activate or repress many other downstream target genes. These downstream genes are indirect or secondary targets of Hh signaling. In our microarray-based screen for Hh pathway targets, the expression pattern of several genes mimicked that of ptc1 with strong decline in levels of expression in cyclopamine-treated embryos and prominent levels of ectopic induction in embryos overexpressing dnPKA, implicating that Hh signaling plays a central role in positively regulating their expression during development. Other genes were not affected as dramatically on loss of Hh signaling, but were clearly induced to high levels on dnPKA overexpression. The expression of these genes is possibly regulated by multiple signals that include Hh, or Hh activity is required for inducing their expression only in a subset among all of the cell types within which the genes are normally expressed. Consequently, loss of Hh signaling does not dramatically affect their overall levels of transcript accumulation, but ectopic Hh activity can result in ectopic induction of their expression. Among the genes that were strongly repressed in response to Hh, only a handful seemed to show some degree of ectopic induction in cyclopamine-treated embryos, while the vast majority did not exhibit any significant degree of upregulation on the loss of Hh activity. The expression of none of these genes is likely to be regulated through direct repression since Hh signaling does not induce the formation of Glirep. For example, ventralization of the neural tube in embryos with ectopic Hh signaling occurs through the ectopic dorsal activation of the ventrally expressed genes like the nkx family members. The Nkx proteins, in turn, repress genes like pax6 and dbx2, which are normally expressed in the dorsal region within the neural tube (Jacob and Briscoe 2003). This paradigm is likely to be true in many other instances where alternative fates are adopted by cells in response to Hh activity. The expression of genes that showed negative regulation in our assay could be inhibited by Hh through such indirect repressive effects.
Genomewide computational search for enhancer modules containing the Gli-binding motif represents a strategy that is complementary to our functional screen for the identification of Hh target genes (for example, see Hallikas et al. 2006). While the former is restricted exclusively to the discovery of the direct targets, a functional approach such as the one described here has the merit of sampling, in an unbiased way, for all kinds of target genes—both direct and indirect. This additional benefit is significant when one considers that for many biological outcomes of Hh signaling, the central determinant of a specific phenotype is the activity of an indirect target gene. To cite an example, expression of Tbx1, which has a critical role in craniofacial development in mammals, is indirectly regulated by Hh signaling. The primary targets for Hh in this context are genes encoding the Fox proteins, which in turn, directly activate Tbx1 transcription through a Fox protein-binding motif in the enhancer of the Tbx1 gene (Yamagishi et al. 2003). Nevertheless, through in silico analysis of the distribution of the consensus Gli-binding motif and its variations in the neighborhood sequences of the upregulated genes, we have made an attempt to distinguish those that could be direct targets. Genetic studies in Drosophila have shown that the repressor, as well as the activator form of the Gli ortholog Ci, associates with the same Gli consensus site within the enhancer of the decapentaplegic (dpp) gene (Methot and Basler 1999; Muller and Basler 2000). Cirep switches the expression of dpp off in the absence of Hh signaling, while the loss of Cirep and generation of Ciact that result from Hh pathway activation induce high levels of dpp expression in the same cells. In the ptc promoter, however, the Gli motif is targeted exclusively by Ciact (Methot and Basler 1999; Muller and Basler 2000), indicating that differential access of Glirep and Gliact to the Gli-binding site imposes another dimension in the regulation of Hh signaling-dependent gene expression. The Gli sites that we have identified in the positively regulated genes from our screen can be envisaged to function through similar mechanisms—their biological relevance, however, will have to await functional characterization through procedures exemplified by our analysis of the nkx2.9 enhancer. We also recognize that the limited amount of sequence information that has been examined for the presence of these motifs must have resulted in the underestimation of the precise numbers of direct target genes. It is worth noting though, that for the majority of the well-established direct target genes, we were able to identify the Gli-binding motif(s) within the 5 kb of flanking sequences in the proximity of the translation start and stop sites.
Strikingly, many genes among those that showed downregulation in response to Hh also contain the Gli_0 or the Gli_m1 motifs within their flanking and/or intragenic sequences. Although the functionality of these binding sites must again be confirmed before we can attribute any biological importance, assuming that some of these are indeed targeted by Gli, we can envisage at least two possible reasons for their presence in this class of genes. First, the Gli recognition sites in many of these genes could be exclusively recognized by the repressor, and not the activator forms of the Gli proteins. In Drosophila, the hh gene itself is a target of Ci activity, but the Gli-binding site in hh appears to be accessible only to Cirep. hh transcription is induced when Ci activity, both repressor and activator, is completely abolished, but the gene is never activated even in cells that are exposed to the highest levels of Hh (Methot and Basler 1999; Muller and Basler 2000). Thus, even small amounts of Cirep are sufficient to prevent hh transcription, and the preponderance of Ciact in these maximally responding cells cannot overcome this repression and promote hh induction. Such a mechanism of control could be operative for several of the downregulated genes that have the Gli-binding motif. Second, Hh affects morphogenesis through several temporally distinct inductive interactions, sometimes within the same cell or tissue type, during the course of development (McMahon et al. 2003). We have shown in a previous report that the timing of exposure of the zebrafish somitic muscle precursors to Hh determines the kinds of fates that they will ultimately adopt (Wolff et al. 2003). In the early embryo, Hh promotes slow-twitch muscle specification in myoblasts in proximity to the midline, at the expense of the fast-twitch fate. However, later in embryogenesis, Hh activity in fast muscle precursors is necessary for the generation of diversity within the fast lineage itself, as exemplified by the Hh-dependent expression of the engrailed (eng) genes in a subset of fast fibers. Thus, Hh can antagonize the development of one cell type at a particular time and have a completely different effect on the same lineage at a later point in development. Given this scenario, genes that are downregulated in response to Hh in one particular cellular context or developmental time could be positively regulated in a different tissue or even within the same cell type, but at a different time point in embryogenesis. Our screen was confined to the assessment of transcriptional changes ensuing from the global loss or activation of Hh signaling in early embryogenesis. Taking this limitation into account, it will be of interest to perform similar screens in the future with regulated administration of cyclopamine at specific developmental stages, as well as the use of conditional transgenes to temporally or spatially activate the pathway (Wolff et al. 2003).
The novel target genes of the Hh pathway that have been identified from this study should be of considerable significance in helping us build a more comprehensive picture of how an Hh-induced network of gene activity translates into the regulation of cell fates during development. Determination of the expression patterns, effects of loss of function, and analyses of the regulatory elements of these genes will clarify their tissue-specific requirements and provide insights into whether their expression is influenced directly or indirectly by Hh activity. Given the remarkable conservation in the molecular details of vertebrate development, it can be expected that homologs of many of the genes that our assay has recognized as Hh pathway targets in the zebrafish will also be involved in mediating Hh-dependent responses within the developing mammalian embryo. Moreover, several studies have now underscored a dramatic and direct association of aberrations in the Hh pathway, with a wide spectrum of congenital abnormalities and malignancies in humans (Altaba et al. 2002). In spite of this connection, we currently have strikingly little appreciation of the cellular and molecular basis for these Hh signaling-related anomalies. It is conceivable that misregulated expression of the human homologs of some of the target genes that we have discovered in the zebrafish could be the critical determinants of certain Hh signaling associated pathologies.
Acknowledgments
We thank R. Karlstrom for providing us with the zebrafish gli1 cDNA and S. Choksi for his insightful comments on the manuscript. This work was funded by the Institute of Molecular and Cell Biology, the Genome Institute of Singapore, the Bioinformatics Institute, the Agency for Science, Technology and Research, and the Biomedical Research Council of Singapore.
References
- Agren, M., P. Kogerman, M. I. Kleman, M. Wessling and R. Toftgard, 2004. Expression of the PTCH1 tumor suppressor gene is regulated by alternative promoters and a single functional Gli-binding site. Gene 330: 101–114. [DOI] [PubMed] [Google Scholar]
- Altaba, A., P. Sanchez and N. Dahmane, 2002. Gli and hedgehog in cancer: tumors, embryos and stem cells. Nat. Rev. Cancer. 2: 361–372. [DOI] [PubMed] [Google Scholar]
- Barresi, M. J., H. L. Stickney and S. H. Devoto, 2000. The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development 127: 2189–2199. [DOI] [PubMed] [Google Scholar]
- Barresi, M. J., J. A. D'angelo, L. P. Hernandez and S. H. Devoto, 2001. Distinct mechanisms regulate slow-muscle development. Curr. Biol. 11: 1432–1438. [DOI] [PubMed] [Google Scholar]
- Barth, K. A., and S. W. Wilson, 1995. Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121: 1755–1768. [DOI] [PubMed] [Google Scholar]
- Baxendale, S., C. Davison, C. Muxworthy, C. Wolff, P. W. Ingham et al., 2004. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat. Genet. 36: 88–93. [DOI] [PubMed] [Google Scholar]
- Blagden, C. S., P. D. Currie, P. W. Ingham and S. M. Hughes, 1997. Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes. Dev. 11: 2163–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman, S. E., M. J. Layden, T. Von Ohlen, C. Q. Doe and J. S. Eisen, 2004. Zebrafish and fly Nkx6 proteins have similar CNS expression patterns and regulate motoneuron formation. Development 131: 5221–5232. [DOI] [PubMed] [Google Scholar]
- Chuang, P. T., and A. P. McMahon, 1999. Vertebrate Hedgehog signaling modulated by induction of a Hedgehog-binding protein. Nature 397: 617–621. [DOI] [PubMed] [Google Scholar]
- Concordet, J. P., K. E. Lewis, J. W. Moore, L. V. Goodrich, R. L. Johnson et al., 1996. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122: 2835–2846. [DOI] [PubMed] [Google Scholar]
- Crooks, G. E., G. Hon, J. M. Chandonia and S. E. Brenner, 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie, P. D., and P. W. Ingham, 1996. Induction of a specific muscle cell type by a hedgehog-like protein in zebrafish. Nature 382: 452–455. [DOI] [PubMed] [Google Scholar]
- Dai, P., H. Akimaru, Y. Tanaka, T. Maekawa, M. Nakafuku et al., 1999. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274: 8143–8152. [DOI] [PubMed] [Google Scholar]
- Dhoot, G. K., M. K. Gustafsson, X. Ai, W. Sun, D. M. Standiford et al., 2001. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science 293: 1663–1666. [DOI] [PubMed] [Google Scholar]
- Du, S. J., S. H. Devoto, M. Westerfield and R. T. Moon, 1997. Positive and negative regulation of muscle cell identity by members of the hedgehog and TGF-beta gene families. J. Cell. Biol. 139: 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman-Scheel, M., L. Weng, S. Xin and W. Du, 2002. Hedgehog regulates cell growth and proliferation by inducing cyclin D and cyclin E. Nature 417: 299–304. [DOI] [PubMed] [Google Scholar]
- Eisen, M. B., and P. O. Brown, 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303: 179–205. [DOI] [PubMed] [Google Scholar]
- Eisen, M. B., P. T. Spellman, P. O. Brown and D. Botstein, 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker, S. C., A. R. Ungar, P. Greenstein, D. P. Von Kessler, J. A. Porter et al., 1995. Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr. Biol. 5: 944–955. [DOI] [PubMed] [Google Scholar]
- Ghosh, M. G., D. A. Thompson and R. J. Weigel, 2000. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60: 6367–6375. [PubMed] [Google Scholar]
- Gustafsson, M. K., H. Pan, D. F. Pinney, Y. Liu, A. Lewandowski et al., 2002. Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification. Genes. Dev. 16: 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallikas, O., K. Palin, N. Sinjushina, R. Rautiainen, J. Partanen et al., 2006. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124: 47–59. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt, M., M. J. Bitgood and A. P. Mcmahon, 1996. Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes. Dev. 10: 647–658. [DOI] [PubMed] [Google Scholar]
- Hollway, G. E., J. Maule, P. Gautier, T. M. Evans, D. G. Keenan et al., 2006. Scube2 mediates Hedgehog signaling in the zebrafish embryo. Dev. Biol. 294: 104–118. [DOI] [PubMed] [Google Scholar]
- Hooper, J. E., and M. P. Scott, 2005. Communicating with Hedgehogs. Nat. Rev. Mol. Cell. Biol. 6: 306–317. [DOI] [PubMed] [Google Scholar]
- Ikram, M. S., G. W. Neill, G. Regl, T. Eichberger, A. M. Frischauf et al., 2004. GLI2 is expressed in normal human epidermis and BCC and induces GLI1 expression by binding to its promoter. J. Invest. Dermatol. 122: 1503–1509. [DOI] [PubMed] [Google Scholar]
- Ingham, P. W., and A. P. Mcmahon, 2001. Hedgehog signaling in animal development: paradigms and principles. Genes. Dev. 15: 3059–3087. [DOI] [PubMed] [Google Scholar]
- Jacob, J., and J. Briscoe, 2003. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 4: 761–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstrom, R. O., O. V. Tyurina, A. Kawakami, N. Nishioka, W. S. Talbot et al., 2003. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development 130: 1549–1564. [DOI] [PubMed] [Google Scholar]
- Kawakami, A., Y. Nojima, A. Toyoda, M. Takahoko, M. Satoh et al., 2005. The zebrafish-secreted matrix protein you/scube2 is implicated in long-range regulation of hedgehog signaling. Curr. Biol. 15: 480–488. [DOI] [PubMed] [Google Scholar]
- Kenney, A. M., and D. H. Rowitch, 2000. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 20: 9055–9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, C. B., W. W. Ballard, S. R. Kimmel, B. Ullmann and T. F. Schilling, 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203: 253–310. [DOI] [PubMed] [Google Scholar]
- Kinzler, K. W., and B. Vogelstein, 1990. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 10: 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss, S., J. P. Concordet and P. W. Ingham, 1993. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75: 1431–1444. [DOI] [PubMed] [Google Scholar]
- Lauderdale, J. D., S. K. Pasquali, R. Fazel, F. J. Van Eeden, H. E. Schauerte et al., 1998. Regulation of netrin-1a expression by hedgehog proteins. Mol. Cell. Neurosci. 11: 194–205. [DOI] [PubMed] [Google Scholar]
- Lewis, K. E., and J. S. Eisen, 2003. From cells to circuits: development of the zebrafish spinal cord. Prog. Neurobiol. 69: 419–449. [DOI] [PubMed] [Google Scholar]
- Lewis, K. E., J. P. Concordet and P. W. Ingham, 1999. a Characterization of a second patched gene in the zebrafish Danio rerio and the differential response of patched genes to Hedgehog signaling. Dev. Biol. 208: 14–29. [DOI] [PubMed] [Google Scholar]
- Lewis, K. E., P. D. Currie, S. Roy, H. Schauerte, P. Haffter et al., 1999. b Control of muscle cell-type specification in the zebrafish embryo by Hedgehog signaling. Dev. Biol. 216: 469–480. [DOI] [PubMed] [Google Scholar]
- MacDonald, R., K. A. Barth, Q. Xu, N. Holder, I. Mikkola et al., 1995. Midline signaling is required for Pax gene regulation and patterning of the eyes. Development 121: 3267–3278. [DOI] [PubMed] [Google Scholar]
- Mathavan, S., S. G. Lee, A. Mak, L. D. Miller, K. R. Murthy et al., 2005. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS. Genet. 1: 260–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, A. P., P. W. Ingham and C. J. Tabin, 2003. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53: 1–114. [DOI] [PubMed] [Google Scholar]
- Methot, N., and K. Basler, 1999. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96: 819–831. [DOI] [PubMed] [Google Scholar]
- Muller, B., and K. Basler, 2000. The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic gli-binding sites. Development 127: 2999–3007. [DOI] [PubMed] [Google Scholar]
- Nagamine, S., S. Koike, K. Keino-Masu and M. Masu, 2005. Expression of a heparan sulfate remodeling enzyme, heparan sulfate 6-O-endosulfatase sulfatase FP2, in the rat nervous system. Dev. Brain. Res. 159: 135–143. [DOI] [PubMed] [Google Scholar]
- Norton, W. H., M. Mangoli, Z. Lele, H. M. Pogoda, B. Diamond et al., 2005. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphe neurones and cranial motoneurones. Development 132: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal, J., F. J. Van Eeden, P. Haffter, P. W. Ingham and C. Nusslein-Volhard, 2000. Two distinct cell populations in the floor plate of the zebrafish are induced by different pathways. Dev. Biol. 219: 350–363. [DOI] [PubMed] [Google Scholar]
- Pabst, O., H. Herbrand and H. H. Arnold, 1998. Nkx2–9 is a novel homeobox transcription factor which demarcates ventral domains in the developing mouse CNS. Mech. Dev. 73: 85–93. [DOI] [PubMed] [Google Scholar]
- Park, H. L., C. Bai, K. A. Platt, M. P. Matise, A. Beeghly et al., 2000. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127: 1593–1605. [DOI] [PubMed] [Google Scholar]
- Rallu, M., R. Machold, N. Gaiano, J. G. Corbin, A. P. McMahon et al., 2002. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development 129: 4963–4974. [DOI] [PubMed] [Google Scholar]
- Roy, S., and P. W. Ingham, 2002. Hedgehogs tryst with the cell cycle. J. Cell. Sci. 115: 4393–4397. [DOI] [PubMed] [Google Scholar]
- Roy, S., T. Qiao, C. Wolff and P. W. Ingham, 2001. a Hedgehog signaling pathway is essential for pancreas specification in the zebrafish embryo. Curr. Biol. 11: 1358–1363. [DOI] [PubMed] [Google Scholar]
- Roy, S., C. Wolff and P. W. Ingham, 2001. b The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes. Dev. 15: 1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati, F., K. Abe, V. Schmidt, T. Schmitt-John, M. Suzuki et al., 2003. Identification of cis-regulatory elements in the mouse Pax9/Nkx2–9 genomic region: implication for evolutionary conserved synteny. Genetics 165: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, H., C. Hui, M. Nakafuku and H. Kondoh, 1997. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124: 1313–1322. [DOI] [PubMed] [Google Scholar]
- Schafer, M., D. Kinzel, C. Neuner, M. Schartl, J. N. Volff et al., 2005. Hedgehog and retinoid signaling confines nkx2.2b expression to the lateral floor plate of the zebrafish trunk. Mech. Dev. 122: 43–56. [DOI] [PubMed] [Google Scholar]
- Schena, M., R. A. Heller, T. P. Theriault, K. Konrad, E. Lachenmeier et al., 1998. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 16: 301–306. [DOI] [PubMed] [Google Scholar]
- Tay, S. Y., P. W. Ingham and S. Roy, 2005. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development 132: 625–634. [DOI] [PubMed] [Google Scholar]
- Tusher, V. G., R. Tibshirani and G. Chu, 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, Z. M., A. Amores, K. E. Lewis, Y. L. Yan, J. H. Postlethwait et al., 2001. Zebrafish smoothened functions in ventral neural tube specification and axon tract formation. Development 128: 3497–3509. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya, R., and M. P. Scott, 2001. The developmental biology of brain tumors. Annu. Rev. Neurosci. 24: 385–428. [DOI] [PubMed] [Google Scholar]
- Wolff, C., S. Roy and P. W. Ingham, 2003. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 13: 1169–1181. [DOI] [PubMed] [Google Scholar]
- Wolff, C., S. Roy, K. E. Lewis, H. Schauerte, G. Joerg-Rauch et al., 2004. iguana encodes a novel zinc-finger protein with coiled-coil domains essential for Hedgehog signal transduction in the zebrafish embryo. Genes Dev. 18: 1565–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, I. G., and W. S. Talbot, 2005. The you gene encodes an EGF-CUB protein essential for Hedgehog signaling in zebrafish. PLoS. Biol. 3: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, H., J. Maeda, T. Hu, J. McAnally, S. J. Conway et al., 2003. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 17: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., H. Haddad, C. Tomas, K. Alsaker and E. T. Papoutsakis, 2003. A segmental nearest neighbor normalization and gene identification method gives superior results for DNA-array analysis. Proc. Natl. Acad. Sci. USA 100: 1122–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R. B., C. K. Ng, S. M. Wasserman, S. D. Colman, S. Shenoy et al., 2002. Identification of a novel family of cell-surface proteins expressed in human vascular endothelium. J. Biol. Chem. 277: 46364–46373. [DOI] [PubMed] [Google Scholar]