Abstract
Sporulation is a well-studied process executed with varying efficiency by diverse yeast strains. We developed a high-throughput method to quantify yeast sporulation efficiency and used this technique to analyze a line cross between a high-efficiency oak tree isolate and a low-efficiency wine strain. We find that natural variation in sporulation efficiency mirrors natural variation in higher eukaryotes: it shows divergence between isolated populations, arises from loci of major effect, and exhibits epistasis. We show that the lower sporulation efficiency of the wine strain results from a failure to initiate sporulation, rather than from slower kinetics of meiosis and spore formation. The two strains differentially regulate many genes involved in aerobic respiration, an essential pathway for sporulation, such that the oak tree strain appears better poised to generate energy from this pathway. We also report that a polymorphism in RME1 that affects sporulation efficiency in laboratory strains also cosegregates with significant phenotypic differences in our cross of natural isolates. These results lay the groundwork for the study of variation in sporulation efficiency among natural isolates of yeast.
YEAST sporulation, the coupling of meiosis and spore formation in response to nutrient deprivation, provides a well-studied model of a cell fate decision in response to external cues (Kupiec et al. 1997). A variety of signaling mechanisms integrate environmental information to determine whether a yeast cell sporulates (Honigberg and Purnapatre 2003). The fraction of cells that sporulate in a culture, called the sporulation efficiency, varies heritably among yeast strains. For example, the best-studied laboratory strain, S288c, is a notoriously poor sporulator, so most studies of sporulation employ the efficient sporulator, SK1.
To accompany the wealth of genomic and loss-of-function analyses of sporulation, Deutschbauer and Davis (2005) began the genetic dissection of variation in sporulation efficiency by crossing S288c and SK1. They found three single-nucleotide polymorphisms that significantly affect sporulation efficiency in these laboratory strains, thus providing the first glimpse into how quantitative genetic variation governs phenotypic differences in this trait.
Sporulation efficiency also varies among natural isolates of yeast (Mortimer 2000). Many of these isolates are from wine fermentations, as Saccharomyces cerevisiae has long been known as a domesticated organism adapted to winemaking (Vaughan-Martini and Martini 1995). Researchers have also obtained yeast samples from other sources, including human infections and oak trees (McCusker et al. 1994; Naumov et al. 1998). A recent survey of genetic polymorphism demonstrates that the oak isolates of yeast represent a diverse, wild population that is genetically isolated from the wine strains (Fay and Benavides 2005). Oak strains therefore provide an untapped resource of wild genetic polymorphism resulting from natural selective pressures distinct from those experienced by the wine and laboratory strains.
To expand yeast sporulation efficiency as a model for the study of natural quantitative genetic variation, we set out to measure and characterize the sporulation efficiency of oak isolates and wine strains. Our first aim was to understand the nature of the genetic variance governing sporulation efficiency. Both the number of effective loci and the presence or absence of epistasis suggest how a trait may respond to selection (Falconer and Mackay 1996). In addition, if the genetic variance is governed by a small number of loci with large effect, the mapping of quantitative trait loci (QTL) provides an ideal experimental approach to identify contributing genetic factors (Lynch and Walsh 1998).
We also sought to identify pathways that potentially govern the variation of this trait. In addition to increasing our general understanding of sporulation, these data provide lists of candidate genes that aid in the dissection of QTL down to the level of individual genes and polymorphisms. Systematic analyses of sporulation in yeast have provided a large list of candidates (Chu et al. 1998; Primig et al. 2000; Deutschbauer et al. 2002; Enyenihi and Saunders 2003). These candidates are active at many different phases of sporulation, from the initiation of meiosis through formation of the spore wall. By identifying the specific stages from which phenotypic variation arises, we can narrow the list of candidates to genes expressed during those stages.
MATERIALS AND METHODS
Strains and growth conditions:
All strains were grown at 30° using standard media (Guthrie and Fink 1991). When needed for selection, the antibiotics nourseothricin, hygromycin, and G418 were filter sterilized and added to autoclaved media (Wach et al. 1994; Goldstein and McCusker 1999). Yeast strains used in this study are listed in Table 2. All are either isolates of S. cerevisiae from wine fermentations and oak trees or derivatives of such strains. The UCD wine strains originate from the University of California, Davis, stock center. Strains M5 and M13 are from the collection of R. Mortimer, and all YPS strains are from the collection of P. Sniegowski. Oak tree strains isolated by our laboratory (BC191, BC192, and BC193) were obtained by culturing soil samples from the base of trees using published methods (Sniegowski et al. 2002), modified as follows. To enrich for yeast as opposed other microorganisms, we twice subcultured, into SD medium with 8% glucose, each sample showing fermentation (bubbles) in the initial culture. We isolated a single colony from the second subculture of each sample. We determined whether this colony was S. cerevisiae by two methods. First, we used intergenic transcribed spacer PCR ribotyping (McCullough et al. 1998). Second, since all the oak yeast isolates sporulated, we mated our prototrophic natural isolates to a G418 resistant, ade2Δ haploid S. cerevisiae tester (Giaever et al. 2002) by mixing these haploid cells with spores from each natural isolate. We sporulated resulting hybrid ADE2+, G418-resistant colonies to test spore viability. Only strains with S. cerevisiae ribotyping patterns produced viable spores in this test cross, and we designated these strains as S. cerevisiae. BC192 and BC193 are original isolates, whereas BC191 is a monosporic derivative of a natural isolate.
TABLE 2.
Strains
| Strain | Origin | Location | Date | Genotype |
|---|---|---|---|---|
| UCD 51 | Vineyard | France | 1948 | |
| UCD 175 | Vineyard | Sicily, Italy | 1953 | |
| UCD 522 | Vineyard | France | Pre-1958 | |
| UCD 762 | Vineyard | Italy | Pre-1984 | |
| UCD 765 | Vineyard | Australia | ||
| UCD 820 | Vineyard | South Africa | Pre-1988 | |
| UCD 2120 | Vineyard | California | 1998 | |
| M5 | Vineyard | Italy | 1993 | |
| M13 | Vineyard | Italy | 1993 | |
| BC191 | Oak Tree | Illinois | 2003 | Monosporic isolate |
| BC192 | Oak Tree | Missouri | 2003 | |
| BC193 | Oak Tree | North Carolina | 2003 | |
| YPS606 | Oak Tree | Pennsylvania | 1999 | Monosporic isolate |
| YPS623 | Oak Tree | New Jersey | Monosporic isolate | |
| YPS2052 | YPS606 | Gift from P. Sniegowski | ho∷kanMX4 MATα | |
| YPS2056 | YPS606 | Gift from P. Sniegowski | ho∷kanMX4 MATa | |
| BC186 | YPS606 | Monosporic isolate | ||
| BC187 | UCD 2120 | Monosporic isolate | ||
| BC210 | BC186 × BC187 | |||
| BC216 | BC187 | ho∷natMX4 MATa | ||
| BC217 | BC187 | ho∷natMX4 MATα | ||
| BC233 | BC186 | SPS2:EGFP:kanMX4/SPS2:EGFP:kanMX4 | ||
| BC235 | BC187 | SPS2:EGFP:kanMX4/SPS2:EGFP:kanMX4 | ||
| BC240 | BC235 | SPS2:EGFP:natMX4/SPS2:EGFP:natMX4 | ||
| BC241 | BC235 | SPS2:EGFP:hygMX4/SPS2:EGFP:hygMX4 | ||
| BC248 | BC233 | SPS2:EGFP:hygMX4/SPS2:EGFP:hygMX4 | ||
| BC251 | BC233 × BC240 | SPS2:EGFP:kanMX4/SPS2:EGFP:natMX4 |
To generate spores during genetic manipulations, we grew oak isolates for 2 days in 1% yeast extract, 2% peptone, and 2% potassium acetate (YPA), and we grew wine isolates overnight in solid media containing 5% glucose, 3% nutrient broth, and 1% yeast extract and then placed a patch of these cells into 1% potassium acetate (sporulation medium) for 3 days.
Genetic manipulations:
We find that natural yeast isolates give much lower transformation efficiency than the standard laboratory strain (S288c) and that the efficiency varies with strain background. We carried out yeast transformations using the lithium acetate method, with modifications (Gietz and Woods 2002). Instead of a 40-min heat shock, we incubated cells at 30° for 30 min, then added DMSO to 8%, and incubated them at 42° for 15 min. Cells were then washed and resuspended in 800 μl of YPD and incubated overnight at room temperature before plating onto selective media. Transformants were streak purified, and those carrying the proper insertion were identified using colony PCR with primers both in the insertion cassette and flanking genomic loci.
Strain BC186 is a monosporic isolate from YPS606, which we obtained from Paul Sniegowski. YPS606 is itself a monosporic isolate from YPS142, a strain isolated from the bark of an oak tree in Pennsylvania (Sniegowski et al. 2002). We tagged SPS2 in BC186 with a yEGFP:kanMX4 construct using homologous recombination (Sheff and Thorn 2004) and sporulated the transformant to generate BC233 (primer sequences generated for this study are listed in Table 1). We replaced the kanMX4 marker in this strain with a hygMX4 marker and sporulated this transformant to generate BC248 (Goldstein and McCusker 1999).
TABLE 1.
Oligonucleotides generated for this study
| Name | Sequence 5′–3′ | Description |
|---|---|---|
| JG94 | TCAAAATGTCAGTTTATGCAGTTTTCACGGTATTGTTCTCGATAATTTTTGGTGACGGTGCTGGTTTA | Amplifies GFP:kanMX4 and targets to SPS2 |
| JG95 | TATCTCATAAGAACTAAATAAAATGGCTAGTATTTAAGTTAATCTTCTAACTCGATGAATTCGAGCTCG | Amplifies GFP:kanMX4 and targets to SPS2 |
| BC336 | GGATACAGTTCTCACATCACATCCGAACATAAACAACCATGG | Complementary to MX4 promoter |
| BC337 | CGTTAGTATCGAATCGACAGCAGTATAGCGACC | Complementary to MX4 terminator |
| JG111 | GTGGGAATAACAAGCGCGCTACTGGTAAGTG | Amplification of RME1 QTL |
| JG117 | CGGTTGCACCTCTTGTAAAACCTCTCTGTTCC | Amplification of RME1 QTL |
| JG120 | CGTAATAACCATTGAACTACATTAAAGAAATAACTGTCTCGC | Amplification of MKT1 QTL |
| JG124 | GGCATTGGTACCGCTGCTGG | Amplification of MKT1 QTL |
| JG128 | CCCAAACATTGGACGATGATTTAGAGTTGCC | Amplification of TAO3 QTL |
| JG133 | CCCTTGCCAAGAGAAATCCATAGCTGC | Amplification of TAO3 QTL |
| JG113 | GGGATAGTGGGTAAACGAGTAGC | Sequencing of RME1 QTL |
| JG114 | GCTTCGTCGTTGGTATAGTTTCGG | Sequencing of RME1 QTL |
| JG119 | GTCCATTAACTATTTTAGCGTGACTGAC | Sequencing of MKT1 QTL |
| JG123 | GCTGCAGTAAAATGGCCCG | Sequencing of MKT1 QTL |
| JG127 | CCTTCGGTAGCTGTTTTATATTACACTAC | Sequencing of TAO3 QTL |
| JG130 | CGAAAGGAAACAAATCAAGTATTCTAACC | Sequencing of TAO3 QTL |
BC187 is a monosporic isolate from UCD2120, a strain isolated from a wine barrel in California. We transformed this strain with the natMX4 cassette targeted to the HO locus to generate strains BC216 and BC217 (Goldstein and McCusker 1999). We tagged SPS2 in BC187 with a yEGFP:kanMX4 construct using homologous recombination and sporulated the transformant to generate BC235. We replaced the kanMX4 marker in this strain with a natMX4 resistance marker and sporulated this transformant to generate strain BC240. We replaced the kanMX4 marker in BC235 with a hygMX4 marker and sporulated this transformant to generate strain BC241.
To cross BC233 and BC240, we used a micro needle to place individual spores of each strain in direct contact. After 2 days, we selected for hybrids by replica plating the resulting colonies on media containing both nourseothricin and G418. Approximately 10% of colonies showed resistance to both antibiotics, indicating a mating between strain backgrounds. We picked one of these hybrids and designated it as BC251. We sporulated BC251 and isolated single spores to generate recombinant segregants. We picked only segregants from tetrads with 100% spore viability. These segregants were arrayed in 96-well plates with replicates of BC241, BC248, and BC251 and stored at −80° in YPD containing 20% glycerol.
Measurement of sporulation efficiency:
To measure the sporulation efficiencies of all the natural isolates (Figure 1), strains were grown overnight in YPD and then diluted 1:200 into YPA. After 16 hr, strains were washed and resuspended in sporulation medium to an optical density at 600 nm (OD600) of 1.0. Time points were recorded starting with the shift to sporulation medium. We counted at least 200 cells for each sample. To verify the SPS2:GFP reporter (Figure 4), we grew each strain in YPA and measured spore formation and fluorescence as each strain arrested growth and began to sporulate. We designated a cell as exhibiting GFP fluorescence if it displayed increased fluorescence—relative to a non-GFP-expressing population—at 510 nm using a Beckmann-Coulter Cytomics FC500 MPL with a 488-nm laser.
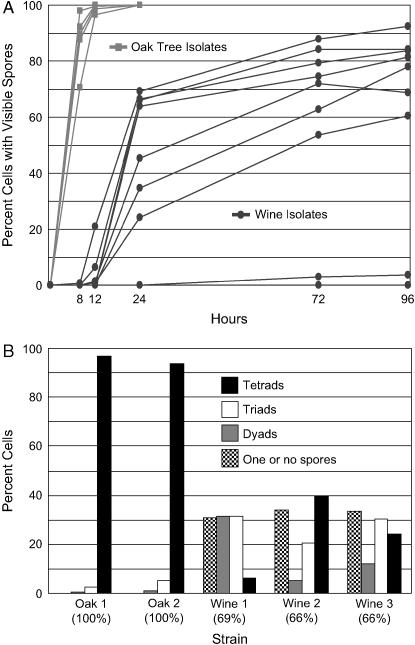
Figure 1.—
Sporulation efficiencies of natural isolates. (A) Sporulation time course of natural isolates in sporulation medium. (B) Frequencies of spores per cell for a subset of strains at the 24-hr time point. The 24-hr sporulation efficiencies are shown in parentheses. Cells with a single spore were rarely observed. Oak 1, BC186; Oak 2, BC191; Wine 1, M5; Wine 2, M13; and Wine 3, BC111.
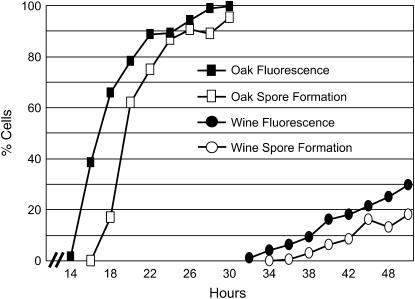
Figure 4.—
The SPS2:GFP reporter. A graph shows the fraction of cells in cultures of oak and wine strains that exhibit GFP fluorescence or visible spores during a sporulation time course.
For analysis of sporulation efficiencies in the cross of BC233 and BC240 (Figures 5, 6, and 8 and Tables 4–6), we developed a set of conditions that yield more reproducible results and give greater disparity between the sporulation efficiencies of BC233 and BC251. Colonies were first inoculated into 0.5 ml of YPD in 2-ml 96-well blocks using a custom-made replica pinner and then grown at 30° with aeration and shaking at 325 rpm. After 14 hr, 8 μl of each culture were inoculated into 400 μl of sporulation medium (a 1:50 dilution) and incubated at 30° with aeration and shaking at 325 rpm. At the concentration of cells resulting from this dilution of a saturated culture, sporulation efficiency is insensitive to twofold changes in cellular concentration. We find that this is not the case in less dilute samples, in which sporulation efficiency becomes more concentration dependent. Time points were recorded starting at dilution into sporulation medium. The “early” time point was taken at 10 hr and the “late” time point at 30 hr. We defined sporulation efficiency as the percentage of cells exhibiting increased fluorescence at 510 nm using a Beckmann-Coulter Cytomics FC500 MPL with a 488-nm laser. At least 10,000 cells were analyzed from each sample. We routinely stored samples at −80° prior to analysis. For each intercross between high-efficiency segregants, 30 replicates of each parent were grown in a 96-well plate with 30 segregants. All phenotypic measurements and flow cytometer settings are available upon request.
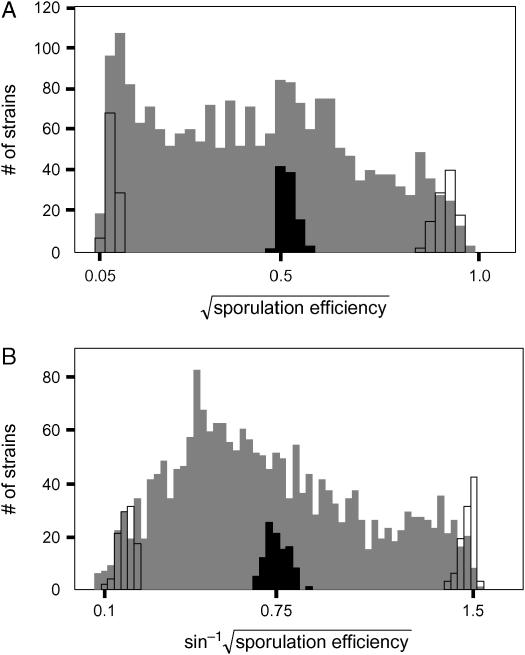
Figure 5.—
Segregation of sporulation efficiency. Histograms of sporulation efficiency at (A) 10 hr (early time point) and (B) 30 hr (late time point) after inoculation into sporulation medium are shown. We measured >2000 segregants at each time point. The raw data have been transformed to reduce the dependence of the sample variance upon the mean. Segregants, shaded bars; F1 hybrid, solid bars; parents, open bars.
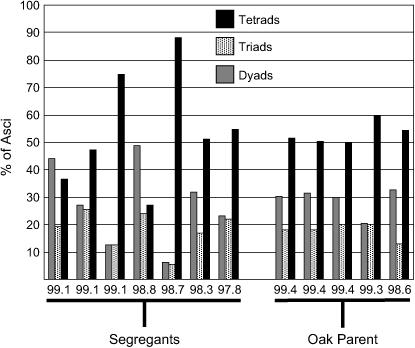
Figure 6.—
The fraction of asci producing two, three, our four spores in high-efficiency segregants. We quantified the fraction of tetrads, triads, and dyads in seven different high-efficiency segregants and in five replicates of the oak parent. Each strain or replicate is denoted on the x-axis by its percentage of sporulation efficiency at the late time point.
Figure 8.—
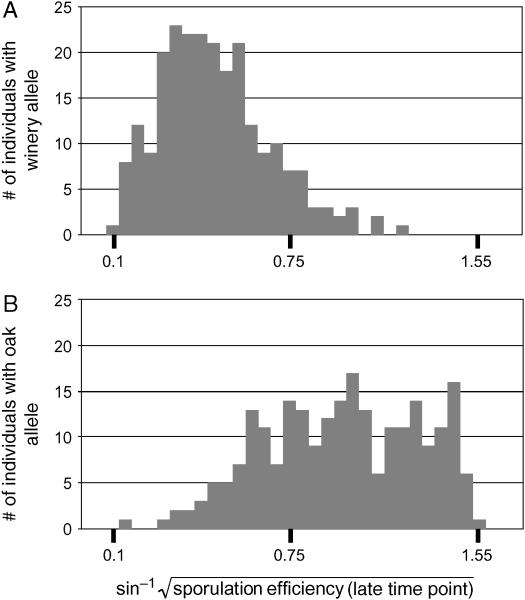
The effect of the RME1 locus on sporulation efficiency. Histograms of sporulation efficiency of segregants carrying (A) the wine-derived (n = 236) or (B) oak-derived (n = 234) allele at the RME1 locus are shown.
TABLE 4.
Sporulation efficiency at the early time point
| Strain type | Sample size | Mean | Variance | Segregational variance | Effective factors |
|---|---|---|---|---|---|
| Segregants | 2065 | 0.43 | 0.06 | 0.03 | 2.9 |
| Oak | 104 | 0.9 | 0.00058 | ||
| Hybrid | 102 | 0.51 | 0.00041 | ||
| Wine | 104 | 0.067 | 0.00015 |
The values are the result of a square-root transformation (Sokal and Rohlf 1995) of the raw values, which increases genetic additivity and reduces the dependence of the variance upon the mean.
TABLE 5.
Sporulation efficiency at the late time point
| Strain type | Sample size | Mean | Variance | Segregational variance | Effective factors |
|---|---|---|---|---|---|
| Segregants | 2069 | 0.72 | 0.12 | 0.06 | 3.5 |
| Oak | 104 | 1.46 | 0.00069 | ||
| Hybrid | 103 | 0.74 | 0.0017 | ||
| Wine | 104 | 0.17 | 0.00074 |
The values are the result of an arcsine transformation (Sokal and Rohlf 1995) of the raw values, which increases genetic additivity and reduces the dependence of the variance upon the mean.
TABLE 6.
Sporulation efficiency at the early time point in intercrosses between highly efficient recombinant progeny
| Parents | Parent means | Offspring mean | H2 | Segregational variance | Reduction in segregational variance (%) |
|---|---|---|---|---|---|
| A, B | 0.90, 0.90 | 0.89 | 0.87 | 0.00019 | 99 |
| C, B | 0.87, 0.89 | 0.89 | 0.86 | 0.00016 | 99 |
| A, D | 0.89, 0.89 | 0.88 | 0.95 | 0.00045 | 99 |
| D, C | 0.91, 0.88 | 0.87 | 0.97 | 0.00086 | 97 |
The values are the result of a square-root transformation (Sokal and Rohlf 1995) of the raw values, which increases genetic additivity and reduces the dependence of the variance upon the mean.
We calculated the segregational variance for doubled haploids as described elsewhere (Lynch and Walsh 1998). We calculated broad-sense heritability (H2) as
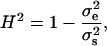 |
where 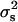 is the variance of the segregant population, and
is the variance of the segregant population, and 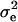 is the pooled variance of the parents (Moore and McCabe 2006). Our test for epistasis (deviation from additive inheritance) is an adaptation, identical to that used by Brem and Kruglyak (2005), of the Δ-statistic of Lynch and Walsh (1998). This test calculates a t-statistic representing the deviation of the offspring mean from the expected midparent value. Because all progeny are homozygous, any significant deviation of the observed mean from the midparent value must be due to epistasis.
is the pooled variance of the parents (Moore and McCabe 2006). Our test for epistasis (deviation from additive inheritance) is an adaptation, identical to that used by Brem and Kruglyak (2005), of the Δ-statistic of Lynch and Walsh (1998). This test calculates a t-statistic representing the deviation of the offspring mean from the expected midparent value. Because all progeny are homozygous, any significant deviation of the observed mean from the midparent value must be due to epistasis.
Analysis of mitochondrial DNA contributions to sporulation efficiency:
We generated petite haploid yeast strains by treating YPS2052, YPS2056, BC216, and BC217 with ethidium bromide (Guthrie and Fink 1991). We isolated multiple clones from each treated strain and confirmed respiratory deficiency by lack of growth on rich medium containing glycerol as the sole carbon source. For both the oak and the wine strain backgrounds, we crossed two independent petite derivatives of each mating type with a respiratory-proficient haploid from the opposite strain background to generate nuclear hybrids containing functional mtDNA from only one parent. The t-statistic does not reject the null hypothesis that hybrids sporulate equally using the mtDNA of either parent.
Flow-cytometric analysis of DNA content:
Overnight cultures grown in YPD were inoculated 1:200 into YPA. At each time point after inoculation, aliquots of 100 μl were examined for spore formation by microscopy, and 500-μl aliquots were fixed for SYTOX Green (Molecular Probes, Eugene, OR) staining and flow cytometry as described previously (Haase and Reed 2002). The interval of SYTOX Green fluorescence intensities displayed by cells in S-phase is clearly distinguishable in growing cultures. In stationary cultures, the beginning of meiotic DNA replication is easily marked by a sharp increase in the fraction of cells displaying this level of fluorescence.
Expression analysis:
For expression analysis, we first grew strains overnight in SD medium containing 2% glucose. Cells were then reinoculated in this medium to an OD600 of 0.2. After 4 hr, the cultures reached an OD600 of 1.0, and each was divided into two portions. The first was used to inoculate a new culture of SD glucose at an OD600 of 0.13. This subculture was grown an additional 3.5 hr to an OD600 of 0.6. We then washed this culture in water; resuspended it in 500 μl 10 mm Tris 7.5, 10 mm EDTA, 0.5% SDS (TES); and snap froze it in a dry ice and ethanol bath. The second portion of each culture was washed in SD acetate medium (2% potassium acetate) and resuspended to an OD600 of 0.6. Prior to freezing, this culture was incubated for 6 hr in SD acetate with no change in the OD600. We extracted RNA from frozen pellets using hot phenol and chloroform. We fluorescently labeled and hybridized RNA to 70-mer oligonucleotide microarrays using the 3DNA Array 350 kit (Genisphere, Hatfield, PA), according to the manufacturer's protocol. For RNA expression level comparison, we paired each experimental sample with a common reference pool composed of equal amounts of RNA from all samples.
We analyzed the results using the default settings in GenePix Pro 4.0 (Axon Instruments, Union City, CA). For each spot on the array, we calculated the log-transformed median of the pixel-by-pixel ratios of the two channel intensities (local background subtracted). The median of ratios is preferred over the mean because it follows a more Gaussian-like distribution, an important assumption for later analyses (Brody et al. 2002). We then applied a two-step mixed-model analysis of variance (ANOVA) to these log-transformed values. The first mixed ANOVA model removes the genomewide effect of different growing conditions and different genotype backgrounds on the expression levels,
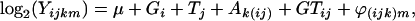 |
where Yijkm is the median of ratios for each spot, μ is the baseline expression on each spot independent of all other factors, Gi is the average genotype effect (BC186 vs. BC187), Tj is the average treatment effect (glucose vs. acetate medium), GTij is the average genotype-by-treatment interaction effect, Ak(ij) is the average array effect, and ϕ(ijk)m is the residual.
The second ANOVA model was applied to each gene separately using the residual ϕ(ijk)m from each spot as a response variable,
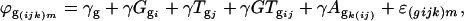 |
where 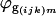 is the residual from the first ANOVA model for each spot, γg is the average gene expression for each gene g,
is the residual from the first ANOVA model for each spot, γg is the average gene expression for each gene g, 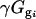 is the gene expression due to genotype i (BC186 vs. BC187),
is the gene expression due to genotype i (BC186 vs. BC187), 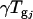 is the gene expression due to treatment j (glucose vs. acetate medium),
is the gene expression due to treatment j (glucose vs. acetate medium), 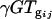 is the gene expression due to genotype i interacting with treatment j,
is the gene expression due to genotype i interacting with treatment j, 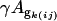 is the gene expression due to array effect, and ɛ(gijk)m is the residual.
is the gene expression due to array effect, and ɛ(gijk)m is the residual.
To avoid problems associated with multiple hypothesis testing, we did not use P-values calculated using the second ANOVA model to determine statistical significance. Instead, we calculated the q-value for each gene on the basis of the distribution of P-values across all genes. The q-value is a measure of the false discovery rate (FDR): the percentage of genes incorrectly deemed to have significant expression changes. The advantage of using q-values, rather than P-values, is that the calculation of q-values automatically takes into account that many hypotheses are being tested simultaneously. For each gene, we calculated the q-values for genotype effect, condition effect, and genotype-by-condition interaction effect using the program QVALUE (Storey and Tibshirani 2003). We considered a gene's expression to exhibit a significant effect (genotype or genotype-by-condition interaction) if its q-value for that effect is <0.05. A q-value of 0.05 for a genotype effect is equivalent to a P-value of 0.024. A q-value of 0.05 for a genotype-by-condition effect is equivalent to a P-value of 0.00245.
The ANOVA provided us with sets of genes displaying significant genotypic effects and/or interaction effects. We divided genes with significant genotypic effects into two sets on the basis of the strain that more highly expressed the gene. We tested for enrichment of functional categories in these subsets and the full interaction set, using the MIPS Functional Catalogue Database (http://mips.gsf.de/proj/funcatDB). P-values for this data set were calculated using the cumulative hypergeometric distribution (Sokal and Rohlf 1995). Microarray images, spot fluorescence ratios, and lists of differentially expressed genes are available upon request.
Genotyping of known sporulation QTL:
We sequenced published sporulation QTL (RME1, MKT1, and TAO3) in our parent strains using BigDye (Perkin-Elmer, Boston) termination sequencing of PCR products. We sequenced using primers ∼200 bp from the target nucleotide in both forward and reverse orientations. The primers used both for amplification and for sequencing are listed in Table 1. The PCR products generated from the parental alleles of the RME1 locus display a restriction fragment length polymorphism when digested with Tsp509I. To genotype the RME1 QTL in a large number of progeny, we digested PCR products amplified from colony PCR of the progeny and resolved the polymorphism by agarose gel electrophoresis. We determined statistical significance of marker–trait linkage using the Mann–Whitney test (Sokal and Rohlf 1995).
RESULTS
Sporulation efficiency varies among natural isolates:
To begin the genetic characterization of differences in sporulation efficiency among natural isolates of yeast, we measured the sporulation efficiencies of five strains originating from North American oak trees and nine strains from wine fermentations (strains are listed in Table 2). All of the oak tree strains sporulate more proficiently than the wine strains. At least 95% of the cells in all oak strain cultures form visible spores within 12 hr of inoculation into sporulation medium (Figure 1A). Far fewer cells form spores in wine cultures, and those that do take significantly longer than their oak tree counterparts. Oak strains predominantly form asci (sporulated cells) with four spores (tetrads), but many wine strains produce larger numbers of two-spored (dyads) and three-spored asci (triads) (Figure 1B). Wine strains produce tetrads and triads earlier in sporulation than dyads (data not shown).
The slow sporulation of a wine strain results from delayed entry into meiosis:
The delays in sporulation of wine strains could arise from a deficiency in detection or transduction of the signal to initiate meiosis or from defects in the machinery that executes meiosis and spore formation. The relative contribution of each mechanism bears upon the classes of genes we expect to carry polymorphisms affecting the trait. For example, many genes that function in spore formation are expressed only during sporulation, whereas genes that detect the signal to sporulate are expressed earlier.
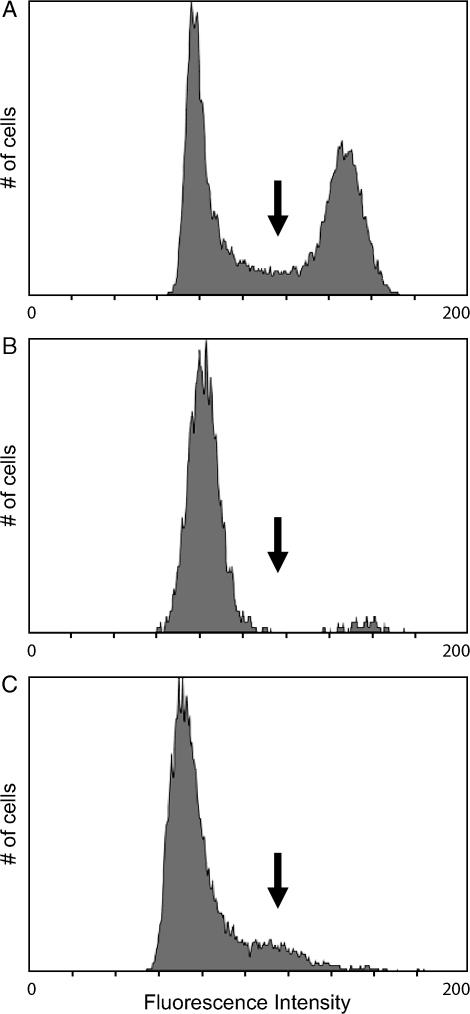
To assess the contributions of each type of delay to disparity in sporulation efficiency, we evaluated the sporulation kinetics of a wine strain, an oak strain, and their F1 hybrid (BC210). We chose the oak strain derivative BC186. BC186 is a single-spore descendant of YPS606, a highly efficient oak strain from Pennsylvania (Sniegowski et al. 2002). For our wine strain we chose BC187, a single-spore descendant of UCD2120. UCD2120 is a poorly sporulating strain isolated from a barrel fermentation unit in a California winery (L. Bisson, personal communication). Both BC186 (oak) and BC187 (wine) were derived from homothallic strains, so these single-spore isolates are homozygous diploids (Takano and Oshima 1970; Oshima and Takano 1971). After inoculation into rich medium supplemented with acetate, each strain grows to saturation before undergoing growth arrest, which is followed by meiosis and spore formation. Using DNA staining and flow cytometry, growth arrest is detectable by an absence of cells in S-phase. Meiotic DNA replication then returns cells to S-phase. The reemergence of S-phase cells in a saturated culture therefore allows us to mark the onset of sporulation (Figure 2).
Figure 2.—
Flow cytometry profiles of cultures at different stages of sporulation. Histograms of STYOX Green staining of cells in (A) a growing culture (2% acetate), (B) an arrested culture, and (C) an arrested culture in which cells have initiated meiotic DNA replication are shown. The arrows denote the fluorescence intensity of cells in S-phase.
For each strain, we compared the time points at which the first cells initiate meiosis to the appearance of the first visible spores. Regardless of strain background, ∼7–8 hr separate the initiation of DNA synthesis and the appearance of spores: cells of each strain require a similar amount of time to carry out sporulation once it begins (Table 3). In contrast, we observed differences in the time it takes for each strain to initiate sporulation. In oak cultures, most cells begin meiosis 2–4 hr after growth arrest. Few cells in a wine culture initiate meiosis, and do so only a full day after growth arrest. Therefore, the wine strain sporulates poorly because most cells of this genetic background do not initiate meiosis, and those that do begin later. The difference we observe in sporulation efficiency reflects the different times at which cells of each strain initiate meiosis, as opposed to differences in each strain's ability to execute meiosis and spore formation.
TABLE 3.
Sporulation kinetics of an oak strain, a wine strain, and their hybrid
| Strain | Growth arrest (hr) | Initiation of meiosis (hr) | Visible spores (hr) | Arrest → initiation (hr) | Initiation → visible spores (hr) |
|---|---|---|---|---|---|
| Oak | 11.5 | 13.5 | 20.5 | 2 | 7 |
| Hybrid | 14.75 | 19 | 26.5 | 4.25 | 7.5 |
| Wine | 17.75 | 36 | 44 | 18.25 | 8 |
All times denote hours since inoculation into acetate medium. These times are approximate, because we observed cells only at specific time points.
Development of a high-throughput measure of sporulation efficiency:
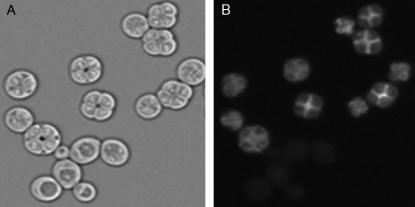
Quantitative traits such as sporulation efficiency usually display complex patterns of inheritance whose dissection requires phenotypic measurements of large sample sizes. We therefore developed an assay of sporulation efficiency by generating derivatives of the oak and wine strains (designated BC233 and BC240, respectively) that carry GFP fused to the C terminus of the coding region of SPS2. Yeast cells express SPS2 only during sporulation (Percival-Smith and Segall 1986), when it plays a role in spore wall formation (Coluccio et al. 2004), so strains carrying this fusion exhibit GFP fluorescence only during and after spore formation (Figure 3). In each strain, GFP fluorescence precedes visible spore formation by ∼2–4 hr (Figure 4). Thus, the fraction of cells expressing this reporter, measured by flow cytometry in 96-well plates, serves as an easily quantifiable measure of sporulation efficiency that does not require microscopy.
Figure 3.—
A fluorescent reporter of sporulation. Bright-field (A) and fluorescence (B) micrographs of a yeast strain carrying the SPS2:GFP reporter are shown. Only sporulating cells fluoresce.
Sporulation efficiency segregates as a complex trait:
We crossed the oak and wine strain derivatives carrying the SPS2:GFP reporter to generate an F1 hybrid (BC251). Because the strains are homothallic, sporulating the F1 produces homozygous diploid progeny. Using growth conditions that give reproducible values of sporulation efficiency (see materials and methods), we assayed the sporulation efficiency of the parents, F1, and >2000 recombinant progeny at two time points (early and late, see materials and methods) after inoculation into sporulation medium (Tables 4 and 5). The broad-sense heritability (H2) of both time points is >0.99 (see materials and methods), indicating that most of the phenotypic variation we observe is genetic. At both time points, the distribution of segregants contains multiple, distinct peaks, suggesting that loci of major effect segregate in this cross (Figure 5). The progeny modes occur between the wine parent and midparent values, indicating a deviation from additive inheritance. Because these segregants are completely homozygous, this deviation must be due to epistasis between segregating QTL. A t-test for epistasis is highly significant (P < 0.0001) at both time points (see materials and methods).
Our estimates indicate that a small number of loci account for most of the variance in this cross. For each 96-well plate we assayed, ∼1 segregant of 40 (51/2065) sporulates at higher efficiency than all four replicates of the oak parent, as measured at the early time point. The same proportion (54/2069) sporulates worse than the wine parent at the late time point. If these proportions represent the probability that an individual inherited all beneficial (or detrimental) alleles for sporulation, then five to six loci likely segregate in this cross (1/25.3 ≈ 1/40). We also computed the Castle–Wright estimator by comparing the segregational variance (variance due to the segregation of QTL) of the progeny to the variance of the parents (Lynch and Walsh 1998). This metric predicts that roughly three effective loci segregate in this cross at both time points. Although this method underestimates the actual number of segregating loci (Zeng et al. 1990), both metrics suggest a small number of factors, which is consistent with the bimodal distribution of progeny phenotypes. The phenotypes of a given strain at each time point are highly correlated (correlation coefficient = 0.945), suggesting (not surprisingly) that many of the same loci affect both time points. The tendency to make dyads vs. triads or tetrads also segregates in this cross. Some progeny produce a higher proportion of tetrads than the oak parent, whereas others produce mostly dyads. The most efficiently sporulating progeny vary widely for this phenotype (Figure 6). Some of these progeny sporulate significantly better than the oak parent at the early time point, indicating that some minor alleles beneficial for sporulation efficiency must be inherited from the poorly sporulating wine parent.
Although the best progeny sporulated with almost identical efficiencies, they showed variation in the shape of the flow-cytometer histograms that measure GFP fluorescence intensity. They also produced curves of slightly different shape in a detailed time-course experiment (data not shown). This suggested that each recombinant strain may have reached high sporulation efficiency through the inheritance of different alleles. We therefore sought to determine if any loci of major effect segregated in our high-efficiency progeny. We performed intercrosses between four segregants that rank in the top 2% of sporulation efficiencies at both time points and make at least 50% tetrads. Progeny from these crosses exhibit dramatically reduced (∼99%) segregational variance relative to the original cross of the oak and wine parents (Table 6). Therefore, these strains share in common any alleles with large effect on sporulation efficiency. Yet even in these crosses of strains with extremely similar phenotypes, most of the variance we observe is genetic; H2 is still >0.85. This indicates the segregation of minor alleles between these strains.
Gene expression profiles indicate higher respiratory activity in the oak strain:
Our results demonstrate that the genetic factors causing differences in sporulation efficiency between these oak and wine strains act upstream of meiotic DNA synthesis. This is consistent with causative polymorphisms affecting genes expressed before sporulation begins. With this in mind, we used oligonucleotide arrays to determine if nonsporulating oak (BC186) and wine (BC187) cells differentially express genes or pathways that may influence sporulation efficiency. We analyzed samples of each strain grown in a defined medium containing nitrogen base and either glucose or acetate (see materials and methods). In the glucose cultures, each strain grows exponentially, whereas the acetate cultures remain static. The oak strain eventually sporulates after 24 hr in acetate, but we sampled the strains after only 6 hr in this medium. Therefore, comparison of gene expression in these conditions illuminates how each strain responds to sporulation-inducing conditions before the transcriptional program of sporulation begins.
We isolated RNA from each sample and implemented an experimental design in which each sample was fluorescently labeled and hybridized competitively with a common reference. The data were analyzed using an analysis of variance to detect genes with significant genotype and genotype-by-carbon source components to their expression variance. A total of 1174 genes possess a significant genotypic variance component (are differentially expressed between the two strains regardless of carbon source) at a FDR (see materials and methods) of 0.05. The set of genes expressed higher in the oak strain displays significant enrichment for genes in functional categories (Ruepp et al. 2004) connected to the mitochondrion and respiration (Table 7). Components of the mitochondrial ATP synthase, the electron transport chain, mitochondrial protein import machinery, and the mitochondrial ribosome express higher in the oak strain, regardless of the carbon source. Furthermore, many of these genes also exhibit a significant genotype-by-carbon source interaction: in acetate, both the oak and wine strains upregulate the expression of respiratory genes, but induction in the oak strain is significantly greater.
TABLE 7.
A subset of functional categories showing significant enrichment for genes with significant genotypic effects
| Functional categorya | Strain with higher expression | No. of genes with effects | No. of genes in functional category | P-value |
|---|---|---|---|---|
| Mitochondrion, 42.16 | Oak | 69 | 166 | 2.53E-28 |
| Respiration, 02.13 | Oak | 45 | 135 | 1.77E-14 |
| Mitochondrial transport, 20.09.04 | Oak | 30 | 101 | 1.02E-08 |
| Transposable elements, 38 | Wine | 48 | 123 | 4.98E-22 |
| Metabolism of methionine, 01.01.06.05 | Wine | 9 | 34 | 0.0011 |
Numbers for each category represent their designation in the MIPS Functional Catalogue (Ruepp et al. 2004).
In total, an interaction between genotype and carbon source played a significant role for 183 genes at an FDR of 0.05. For these genes, the relative expression in each strain differs in glucose and acetate. In addition to the respiration genes, this list showed functional enrichment for the metabolism of energy reserves (P = 0.004), specifically enzymes in the glycogen synthesis pathway, which consumes ATP to build up energy stores (Francois and Parrou 2001). Both strains increase expression of the glycogen synthesis pathway when grown in acetate, but the wine strain induces all components to a greater degree than the oak.
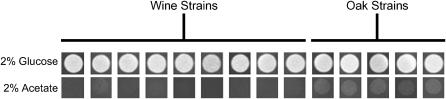
Oak and wine strains show different growth rates on nonfermentable carbon sources:
Our expression analysis suggests that the oak strain is better able to generate energy from nonfermentable carbon sources. To test this hypothesis, we plated dilutions of all our wine and oak strains onto plates supplemented with glucose or acetate (Figure 7). On acetate, all oak strains form larger visible colonies prior to any of the wine strains. We noted no corresponding differences in growth using glucose as the carbon source. Similar to sporulation efficiency, the ability to grow using acetate is greater in the oak isolates than in the wine strains.
Figure 7.—
Comparison of the growth of oak and wine strains on acetate. Photographs were taken 24 hr after plating ∼7500 cells of each strain on rich agar medium containing 2% glucose or 2% potassium acetate.
Oak–wine hybrids sporulate equally using the mitochondrial genome of either parent:
A functioning mitochondrial genome is an essential prerequisite for sporulation, and genes encoded in the mitochondrial DNA (mtDNA) are differentially expressed between the oak and wine strains. We tested whether polymorphisms in mtDNA cause differences in sporulation efficiency between these strains. We generated diploid hybrid strains containing the haploid genomes of both the oak (BC186) and wine (BC187) backgrounds, but with mtDNA from only one parent. If inheritance of the mtDNA affects sporulation efficiency, then we expect that strains inheriting mtDNA from the oak strain sporulate more proficiently than those with mtDNA from the wine strain. Instead, all the hybrid strains sporulate equally (∼60% after 12 hr). Furthermore, efficiently sporulating progeny from the cross of the oak and wine strains possess mtDNA markers identical by descent with the wine parent (data not shown). Therefore, both in the hybrid and in recombinant genetic backgrounds, inheritance of mtDNA from the oak strain parent is not necessary for high sporulation efficiency.
Polymorphism in RME1 cosegregates with variation in sporulation efficiency:
Three single-nucleotide polymorphisms (SNPs) cause differences in sporulation efficiency between SK1, an efficiently sporulating laboratory strain, and S288c, a poorly sporulating laboratory strain (Deutschbauer and Davis 2005). We sequenced these three QTL in our oak and wine parents. Two of the SNPs, coding polymorphisms in MKT1 and TAO3, do not segregate between our two strains, which carry the more common allele for each SNP (Deutschbauer and Davis 2005). The third SNP, a noncoding polymorphism upstream of RME1, segregates in our cross. The oak-derived allele shares a single-base-pair insertion with SK1 that increases sporulation efficiency when placed in the S288c background. In fact, all oak isolates in our collection (Table 2) carry the allele at this locus known to increase sporulation efficiency in the cross of laboratory strains, whereas all winery isolates carry the allele known to decrease sporulation efficiency.
The RME1 locus is linked to a QTL exerting a major effect upon sporulation efficiency in this cross (Figure 8). Inheritance of the oak-derived allele increases sporulation efficiency, on average, by 28% at the early time point and 42% at the late time point (n = 470, P ≪ 0.001). In a linear regression on genotype, inheritance of the RME1 polymorphism gives an r2 of 0.41 at the early time point and 0.47 at the late time point. The polymorphism occurs 309 bp upstream of the RME1 open reading frame, suggesting a regulatory effect in S288c. We therefore checked our microarray data to see whether the RME1 message is expressed differentially between our parent strains. RME1 displays a significant genotype effect: the wine strain overexpresses RME1 relative to the oak strain in both glucose and acetate (P = 0.013) (Figure 9).
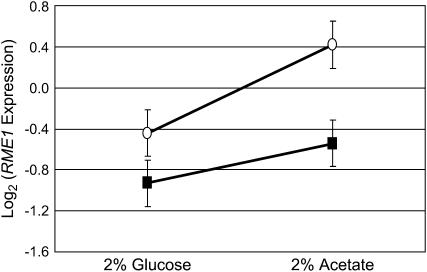
Figure 9.—
mRNA expression of RME1 in (▪) oak (BC186) and (○) wine (BC187) strains. Expression is measured as the Log2-transformed ratio of expression of the sample compared to a common reference. The error bars represent the standard error of the measurements in the ANOVA model (see materials and methods). BC187 expresses RME1 significantly greater under both conditions (P = 0.013).
DISCUSSION
The quantitative genetics of sporulation efficiency in natural isolates:
Using laboratory strains, Deutschbauer and Davis (2005) demonstrated the tractability of yeast sporulation efficiency as a quantitative trait. To expand the usefulness of this model in quantitative and evolutionary genetics, we have measured sporulation efficiency and its genetic variance in natural strains. We find that wild isolates from oak trees sporulate more efficiently than strains obtained from wine fermentations. In a cross between strains from these two populations, the genetic sources of variation lie in the nuclear genome, as opposed to the mitochondrial DNA.
We found that the difference in phenotype lies in the decision to initiate meiosis and that the speed of sporulation in both strain backgrounds is equal once the process has begun. This allowed us to use SPS2, a gene expressed late in sporulation, as a reporter gene to calculate sporulation efficiency in an automated, high-throughput assay. This assay enabled the rapid analysis of a large sample of recombinant progeny, providing a general picture of the genetic variation governing sporulation efficiency between two natural isolates.
A large fraction of the variance can be explained by a small number of loci exerting large effects upon the trait. This conclusion promises that genetic mapping will be successful in identifying QTL accounting for most of the variation in sporulation efficiency (Lynch and Walsh 1998). Intercrosses of our progeny with the highest sporulation efficiencies (within the top 2%) contained only 1–3% of the segregational variance observed in the initial cross of the oak and wine strains. QTL with large effect do not segregate between these individuals. These QTL could therefore be identified by techniques that selectively genotype extreme progeny, such as bulk segregant analysis (Brauer et al. 2006).
Despite the reduction in segregational variance, the broad-sense heritability of sporulation efficiency in these intercrosses was still high, revealing the segregation of minor-effect loci that might be missed by a bulk analysis. The intercrosses produced transgressive segregants with sporulation efficiencies slightly above that of either recombinant parent. The occurrence of transgressive segregants points to the wine strain contributing some of the minor alleles promoting increased sporulation efficiency. The identification of minor loci will likely require QTL detection strategies that genotype a large sample of progeny.
The distribution of phenotypes among natural isolates opposes their pattern of sequence polymorphism. Although oak strains show uniformly high sporulation efficiencies, they compose a more genetically variable population (J. Gerke and B. Cohen, unpublished results) than the relatively homogeneous wine strains (Fay and Benavides 2005), which vary widely in their sporulation efficiencies. This observation suggests stronger selection for efficient sporulation—or a correlated trait—in oak tree populations. The phenotypic means of the progeny in our cross are significantly shifted away from the midparent—toward the wine parent—at both time points. This indicates that, collectively, the wine alleles (negative with respect to sporulation efficiency) are epistatic to the oak alleles. Given selection for sporulation efficiency in the oak isolates, the negative epistasis of wine alleles could serve as a barrier to gene flow between oak and wine populations, since the inheritance of a wine allele would be detrimental to the fitness of a strain in the oak habitat. Accordingly, we hypothesize that the major loci affecting sporulation efficiency do not segregate in the oak population. Selection against sporulation efficiency or correlated traits in the wine isolates remains an open question. The lower sporulation efficiency of wine strains could be the result of genetic drift rather than selection. For example, a relaxation of selection on high sporulation efficiency would allow this phenotype to diverge. Whatever the case, the wider variation of sporulation efficiency among wine isolates raises the question of whether major alleles segregate in this population.
This is not the case with one known QTL governing yeast sporulation efficiency. In our sample of isolates, the genotype of a polymorphism at the RME1 locus is fixed between strains from the two populations. We find that this single-base-pair change, which directly affects sporulation efficiency in laboratory isolates, also shows linkage as a major-effect QTL in our cross. From a panel of 13 strains, Deutschbauer and Davis conclude that the allele promoting sporulation efficiency is rare (Deutschbauer and Davis 2005). Although it was rare in their sample of strains, our results show that it is common—if not fixed—in oak strains. The inclusion of oak strains in future yeast studies promises to provide additional genetic diversity that may not be present in other populations. In fact, it is possible that the large effect of the RME1 QTL in our cross is due, in part, to linked polymorphisms not present in laboratory isolates.
In addition to absolute differences in sporulation efficiency, we observed that sporulated cells from different wine strains produce distinct fractions of asci containing dyads, triads, or tetrads. Dyads form when only two nonsister haploid genomes are enclosed in spores (Davidow et al. 1980). This decision is regulated by the availability of spindle pole body components and nutrient flux through the glyoxylate pathway (Nickas et al. 2004). Strains probably produce tetrads earlier than dyads in a sporulation time course because nutrient availability decreases over time (Esposito et al. 1969; Fast 1973). Since our wine strains sporulate slowly, we expect them to produce dyads with greater frequency. Our cross of wine and oak tree isolates also reveals the presence of natural genetic variation in tetrad vs. dyad formation; even rapidly sporulating progeny vary with respect to the frequency of dyad formation. Crosses of highly efficient progeny with disparate frequencies of dyad formation will determine the genetic variance of this phenotype and reveal if efficiently sporulating dyad makers result from a different allelic combination than efficient tetrad makers.
Our study of sporulation efficiency indicates that loci of large effect account for much of the genetic variation observed between lines drawn from phenotypically diverged populations and reveals epistasis between the loci segregating in these lines. In addition, minor alleles with opposite effects in each strain generate transgressive segregants. Loci of large effect, epistasis, and transgressive segregation are commonly observed in crosses of phenotypically diverged lines in higher eukaryotes (Lynch and Walsh 1998). Despite their single-cell lifestyle, natural isolates of yeast promise to provide a useful model for eukaryotic quantitative genetics.
The functional basis of variation in sporulation efficiency:
A first step in unraveling the genetic architecture of a trait is to gain an understanding of the genes that could potentially carry polymorphisms affecting the phenotype (Mackay 2001). Through a prioritized candidate gene approach, this knowledge can expedite the narrowing of QTL to one or a few genes. In this respect, yeast sporulation provides an attractive trait, as it is a well-studied developmental program. Multiple transcriptional analyses show that >1000 genes are differentially expressed during meiosis and sporulation (Chu et al. 1998; Primig et al. 2000). Loss-of-function genetic studies have designated >400 genes that positively or negatively affect sporulation (Kupiec et al. 1997; Deutschbauer et al. 2002; Enyenihi and Saunders 2003). The genes implicated in these studies provide the most likely candidates to carry polymorphisms affecting sporulation efficiency.
Our results further narrow the search space for natural variation governing this trait. Many genes needed for sporulation are expressed only during meiosis and spore formation (Primig et al. 2000). Since the phenotypic difference between the oak and wine strains we study is due to delays prior to meiosis, that class of genes can be ruled out as candidates. Instead, the causal sequence differences must affect genes expressed before sporulation begins.
Our analysis of gene expression suggests that genes regulating respiration and starvation response may affect sporulation efficiency. Many respiration genes are expressed significantly higher by the oak strain. In addition, the oak strain shows stronger upregulation of these genes in acetate. These results suggest that the oak strain is capable of greater respiratory activity, especially in conditions that promote sporulation. Our demonstration that the oak isolates grow faster than wine strains on nonfermentable carbon sources supports the hypothesis that the expression differences are functional and may extend to the rest of our isolates. Sporulation requires respiratory activity, so increased respiration could conceivably lead to higher sporulation efficiency. Furthermore, in acetate, the wine strain upregulates genes in the glycogen synthesis pathway to a greater degree than the oak strain. These genes, considered part of a starvation response (Lillie and Pringle 1980; Gasch et al. 2000), are used to store energy as glycogen for later use. Collectively, our data fit a model in which the oak strain produces more energy from acetate to drive growth and sporulation, but the wine strain—presumably adapted for fermentative alcohol production—has less respiratory activity and instead stores more glycogen, a hallmark of the starvation response. This model echoes a hypothesis proposed to explain differences in sporulation efficiency between SK1 and W303, a poor sporulator related to S288c (Williams et al. 2002; Winzeler et al. 2003). In addition, a quantitative reverse-genetics study revealed that respiration and starvation response genes were significantly enriched in the set of genes affecting sporulation efficiency (Deutschbauer et al. 2002). Together, these data suggest that genes regulating respiration and carbon utilization are likely candidate genes governing sporulation efficiency.
In contrast, the three functional polymorphisms known to affect sporulation efficiency do not lie in genes involved in respiration or carbon metabolism. In fact, only RME1, a negative transcriptional regulator of meiosis, is an obvious candidate gene (Mitchell and Herskowitz 1986). Thus, although candidate genes within sporulation QTL may provide a starting point for fine mapping, an unbiased, genomewide approach still provides the best starting point for genetic dissection (Deutschbauer and Davis 2005). As more QTL are identified, it will be interesting to see what portion of the genetic variation in this trait fits with the models generated by functional genomics.
Acknowledgments
We thank James Falvo and Erin O'Shea for providing GFP fusion strains, Tim Schedl and Rob Mitra for access to Nomarski and fluorescent microscopes, Shoshana Goldberg for technical assistance, Paul Sniegowski and Justin Fay for providing strains, Jim Cheverud for advice on statistical analysis of progeny distributions, and Justin Fay and members of the Cohen Lab for advice and discussions.
References
- Brauer, M. J., C. M. Christianson, D. A. Pai and M. J. Dunham, 2006. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics 173: 1813–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R. B., and L. Kruglyak, 2005. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl. Acad. Sci. USA 102(5): 1572–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody, J. P., B. A. Williams, B. J. Wold and S. R. Quake, 2002. Significance and statistical errors in the analysis of DNA microarray data. Proc. Natl. Acad. Sci. USA 99(20): 12975–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein et al., 1998. The transcriptional program of sporulation in budding yeast. Science 282(5389): 699–705. [DOI] [PubMed] [Google Scholar]
- Coluccio, A., E. Bogengruber, M. N. Conrad, M. E. Dresser, P. Briza et al., 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 3(6): 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidow, L. S., L. Goetsch and B. Byers, 1980. Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics 94: 581–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer, A. M., and R. W. Davis, 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Deutschbauer, A. M., R. M. Williams, A. M. Chu and R. W. Davis, 2002. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99(24): 15530–15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi, A. H., and W. S. Saunders, 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, M. S., R. E. Esposito, M. Arnaud and H. O. Halvorson, 1969. Acetate utilization and macromolecular synthesis during sporulation of yeast. J. Bacteriol. 100(1): 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics. Longman, Essex, England.
- Fast, D., 1973. Sporulation synchrony of Saccharomyces cerevisiae grown in various carbon sources. J. Bacteriol. 116(2): 925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., and J. A. Benavides, 2005. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 1(1): 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois, J., and J. L. Parrou, 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25(1): 125–145. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen et al., 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11(12): 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896): 387–391. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15(14): 1541–1553. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Haase, S. B., and S. I. Reed, 2002. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1(2): 132–136. [PubMed] [Google Scholar]
- Honigberg, S. M., and K. Purnapatre, 2003. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116(11): 2137–2147. [DOI] [PubMed] [Google Scholar]
- Kupiec, M., B. Byers, R. E. Esposito and A. P. Mitchell, 1997. Meiosis and sporulation in Saccharomyces cerevisiae, pp. 889–1036 in The Molecular and Cellular Biology of the Yeast Saccharomyces Cell Cycle and Cell Biology, edited by J. R. Pringle, J. R. Broach and E. W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Lillie, S. H., and J. R. Pringle, 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143(3): 1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mackay, T. F., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- McCullough, M. J., K. V. Clemons, J. H. McCusker and D. A. Stevens, 1998. Intergenic transcribed spacer PCR ribotyping for differentiation of Saccharomyces species and interspecific hybrids. J. Clin. Microbiol. 36(4): 1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker, J. H., K. V. Clemons, D. A. Stevens and R. W. Davis, 1994. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics 136: 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, A. P., and I. Herskowitz, 1986. Activation of meiosis and sporulation by repression of the RME1 product in yeast. Nature 319(6056): 738–742. [DOI] [PubMed] [Google Scholar]
- Moore, D. S., and G. P. McCabe, 2006. Introduction to the Practice of Statistics. W. H. Freeman, New York.
- Mortimer, R. K., 2000. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 10(4): 403–409. [DOI] [PubMed] [Google Scholar]
- Naumov, G. I., E. S. Naumova and P. D. Sniegowski, 1998. Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of North American oaks. Can. J. Microbiol. 44(11): 1045–1050. [PubMed] [Google Scholar]
- Nickas, M. E., A. E. Diamond, M. J. Yang and A. M. Neiman, 2004. Regulation of spindle pole function by an intermediary metabolite. Mol. Biol. Cell 15(6): 2606–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima, Y., and I. Takano, 1971. Mating types in Saccharomyces: their convertibility and homothallism. Genetics 67: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith, A., and J. Segall, 1986. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 6(7): 2443–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway et al., 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26(4): 415–423. [DOI] [PubMed] [Google Scholar]
- Ruepp, A., A. Zollner, D. Maier, K. Albermann, J. Hani et al., 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32(18): 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff, M. A., and K. S. Thorn, 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21(8): 661–670. [DOI] [PubMed] [Google Scholar]
- Sniegowski, P. D., P. G. Dombrowski and E. Fingerman, 2002. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 1(4): 299–306. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995. Biometry: The Principles and Practice of Statistics in Biological Research. W. H. Freeman, New York.
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100(16): 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, I., and Y. Oshima, 1970. Allelism tests among various homothallism-controlling genes and gene systems in Saccharomyces. Genetics 64: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan-Martini, A., and A. Martini, 1995. Facts, myths and legends on the prime industrial microorganism. J. Ind. Microbiol. 14(6): 514–522. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10(13): 1793–1808. [DOI] [PubMed] [Google Scholar]
- Williams, R. M., M. Primig, B. K. Washburn, E. A. Winzeler, M. Bellis et al., 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99(21): 13431–13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., C. I. Castillo-Davis, G. Oshiro, D. Liang, D. R. Richards et al., 2003. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. B., D. Houle and C. C. Cockerham, 1990. How informative is Wright's estimator of the number of genes affecting a quantitative character? Genetics 126: 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]