Abstract
The paleopolyploid soybean genome was investigated by sequencing homeologous BAC clones anchored by duplicate N-hydroxycinnamoyl/benzoyltransferase (HCBT) genes. The homeologous BACs were genetically mapped to linkage groups C1 and C2. Annotation of the 173,747- and 98,760-bp BACs showed that gene conservation in both order and orientation is high between homeologous regions with only a single gene insertion/deletion and local tandem duplications differing between the regions. The nucleotide sequence conservation extends into intergenic regions as well, probably due to conserved regulatory sequences. Most of the homeologs appear to have a role in either transcription/DNA binding or cellular signaling, suggesting a potential preference for retention of duplicate genes with these functions. Reverse transcriptase–PCR analysis of homeologs showed that in the tissues sampled, most homeologs have not diverged greatly in their transcription profiles. However, four cases of changes in transcription were identified, primarily in the HCBT gene cluster. Because a mapped locus corresponds to a soybean cyst nematode (SCN) QTL, the potential role of HCBT genes in response to SCN is discussed. These results are the first sequenced-based analysis of homeologous BACs in soybean, a diploidized paleopolyploid.
GENE duplication, arising from region-specific duplication or genomewide polyploidization, is a prominent feature of genome evolution. Gene and genome duplication have been shown to provide morphological and fitness advantages, create genetic redundancy, expand genome size, and provide a source for forming diverse/novel gene functions (Wendel 2000). Although found across most eukaryotic lineages, gene duplication appears to occur at an elevated rate in plants with up to 100% of all angiosperms having a polyploid or paleopolyploid history (Masterson 1994; Lockton and Gaut 2005). The high incidence of gene duplication in plants is probably due to its impact on genetic diversity and adaptation (Lawton-Rauh 2003).
Increased evidence of paleopolyploidy in plants once thought to be purely diploid has come to light in recent years. Comparative mapping studies as well as genome-sequencing efforts have revealed that both Arabidopsis and rice are paleopolyploids (Lynch and Conery 2000; Tagi 2000; Vision et al. 2000; Goff et al. 2002; Simillion et al. 2002; Blanc et al. 2003; Yu et al. 2003). Expressed sequence tag (EST)-based analyses of several plant genomes have also revealed evidence for large-scale genome duplications in a wide range of genera, including soybean (Blanc and Wolfe 2004a; Schlueter et al. 2004).
Further evidence of paleopolyploidy has been identified in soybean specifically. Soybean (Glycine max L. Merr.) is a member of the papilionoid Leguminosae tribe Phaseoleae. While most genera of the Phaseoleae have a genome complement of 2n = 22, soybean has a chromosome number of 2n = 40 (Hadley and Hymowitz 1973; Lackey 1980). Studies of soybean gene families have also suggested that soybean is a paleopolyploid (Lee and Verma 1984; Hightower and Meagher 1985; Grandbastien et al. 1986; Nielsen et al. 1989). Additionally, combined data from nine mapping populations uncovered extensive homeologous relationships among linkage groups, with 90% of soybean RFLP probes detecting more than two fragments (Shoemaker et al. 1996). In many cases nested duplications were observed, suggesting at least two rounds of duplication and diploidization (Shoemaker et al. 1996; Lee et al. 1999, 2001).
The most compelling evidence to date is from an analysis of duplicate genes identified from ESTs. Large numbers of conserved duplicate gene pairs with similar levels of divergence from one another allowed the identification of at least two major genome duplications in soybean (Blanc and Wolfe 2004a; Schlueter et al. 2004). These gene pairs are referred to as homeologs since they most likely resulted from a polyploidy event and not from single gene duplications. The coalescence estimates of these events are ∼14.5 and 41.6 million years ago (MYA) (Schlueter et al. 2004). BAC hybridization, BAC-end sequencing, and fingerprinting studies have suggested that the conserved homeologous regions within the soybean genome still retain upward of 46–86.5% structural identity (Marek et al. 2001; Foster-Hartnett et al. 2002; Yan et al. 2003, 2004). Recently, integration of the soybean genetic and physical map using FISH identified conserved homeologous regions between two chromosomes (Walling et al. 2006).
While these previous studies foreshadow the sequence-level conservation in homeologous regions in soybean, no studies to date have actually sequenced and characterized any of these duplicated regions. In maize, however, three separate homeologous regions have been studied: the Adh1 loci, the lg2/lrs1 loci, and the Orp loci (Ilic et al. 2003; Langham et al. 2004; Ma et al. 2005). Between the homeologous Adh1 regions, only four predicted genes/gene fragments were retained in both regions; for the lg2/lrs1 loci and the Orp loci, only the duplicated gene that anchored each region was retained. It appears from these analyses that the maize genome has undergone extensive rearrangements, transposable element insertions, and gene loss after duplication (Ilic et al. 2003; Langham et al. 2004; Ma et al. 2005). Conversely, an analysis of homologous CesA1 regions in cotton, a relatively recent allotetraploid, found extensive genic as well as intergenic sequence conservation with variation only in small insertions and deletions and transposable elements (Grover et al. 2004). Much as with maize and cotton, an analysis of homeologous regions in the soybean genome provides insights into the evolutionary forces that have shaped the genome after duplication. In this article, we report sequence-based analysis of homeologous regions in soybean.
MATERIALS AND METHODS
Duplicate BAC selection:
A BLASTN-based similarity search with default parameters (Altschul et al. 1990) was performed with previously identified duplicate gene pairs (Schlueter et al. 2004) against all genetically mapped RFLP sequences at NCBI. The duplicate pair corresponding to The Institute for Genome Research (TIGR) tentative contigs (TC) TC104546 and TC114014 (Quackenbush et al. 2000) showed similarity to the Phaseolus vulgaris RFLP probe pBng181 (AZ044940, AZ044941) with e-values of 1e−59 and 1e−43, respectively. These TCs correspond to N-hydroxycinnamoyl benzoyltransferase genes as annotated by Schlueter et al. (2004).
TC consensus sequence DNA alignments were done with virtual translation and the Clustalx method with default parameters (DNASTAR, Madison WI). Homeolog-specific PCR primers were designed using Oligo 6.82 (Molecular Biology Insights, Cascade, CO) The primers were tested against the G. max cultivar Williams 82 using a DNA Engine Gradient Cycler from MJ Research (Watertown, MA). PCR reactions were 10 μl in volume and contained 1.1× MasterAmp 2× PCR PreMix B (Epicentre, Madison, WI), 0.11 μm of each primer, 50 ng Williams 82 DNA, and 0.1375 unit of Taq DNA polymerase (Invitrogen, Carlsbad, CA). PCR cycling conditions were 94° for 2 min, 35 cycles of 94° for 45 sec, annealing temperature for 30 sec, 72° for 45 sec, followed by a final extension of 72° for 3 min. Products were gel purified and subsequently sequenced at the DNA Synthesis and Sequencing Facility (Iowa State University, Ames, IA) to verify homeolog specificity. The primer sequences for N-hydroxycinnamoyl/benzoyltransferase (HCBT) copy 1 (TC144014) were U, 5′-TGG TGC TGC AAT CTC TGA AGG T-3′ and L, 5′-GGA TTG GAC TTA GAA ACA GCA T-3′; for HCBT copy 2 (TC104546), primer sequences were U, 5′-CAA ACC ATA ATG CCA GTG CT-3′ and L, 5′-TTG TAT CCG GTG AAA GAC AG-3′.
Arrayed pools of the Williams 82 G. max BAC library (Marek and Shoemaker 1997) was PCR screened using the conditions described above. One BAC was identified for each primer pair, gmw1-74i13 for HCBT copy 1 (TC144014) and gmw1-52d3 for HCBT copy 2 (TC104546). BAC DNA was isolated using a plasmid midi kit (QIAGEN, Valencia, CA) and insert size was determined by NotI digest and CHEF gel electrophoresis. Digest conditions were 3 μl of BAC DNA, 1× NEBuffer 3, and 0.8 unit of NotI (New England Biolabs, Ipswich, MA). BAC-end sequences were obtained using M13 forward and reverse primers at the DNA Synthesis and Sequencing Facility (Iowa State University).
BAC sequencing and assembly:
BAC DNA was randomly sheared using a nebulizer (Invitrogen, San Diego) and size selected for 2–4 kbp on a 1% (w/v) low-melt agarose gel. Sheared DNA was phosphatase-treated, blunt-end repaired, and cloned into the vector pCR4Blunt-TOPO (TOPO shotgun subcloning kit, Invitrogen) The recombinant plasmids were transformed into TOP10 Escherichia coli cells by electroporation and selected on LB plates containing kanamycin. For gmw1-52d3, a second subclone library of 7- to 9-kbp fragments was also made as described above.
Subclones were sequenced at the USDA-ARS Mid-South-Area Genomics Laboratory using M13 forward and reverse primers on an ABI3730XL with BigDye3.1. Base calling, vector trimming, and contig assembly were done using SeqMan II, starting with a match size of 12 bp, a maximum gap of 70 bp, and a minimum match percentage of 95% and decreasing the minimum match percentage to 90% as necessary to merge contigs (DNAStar). The complete sequence of gmw1-52d3 was obtained through shotgun sequencing.
Closing of gaps on gmw1-74i13 was accomplished by two methods. In all cases but one, clone pairs spanned a gap and complete sequencing of that clone closed the gap. The last gap was closed using PCR primers designed from adjacent contigs. The PCR product was gel purified, subcloned into TOPO TA vector (Invitrogen), transformed into TOP10 E. coli cells (Invitrogen), and sequenced with M13 forward and reverse primers. All gap-closing sequencing was done at the DNA Synthesis and Sequencing Facility (Iowa State University).
Genetic mapping of BACs:
Each BAC was manually scanned for di- and trinucleotide repeats of at least 7 bp in length. Primer pairs flanking the simple sequence repeats (SSRs) were designed using Oligo 6.82 (Molecular Biology Insights) and tested against a variety of parental lines from mapping populations. PCR reactions were 10 μl in volume and contained 1× PCR buffer, 1.5 mm magnesium chloride, 5 mm dNTPs, 0.5 μm each primer, 50 ng parental DNA, and 0.025 unit of Taq DNA polymerase (Invitrogen). PCR cycling conditions were 94° for 2 min, 35 cycles of 94° for 45 sec, 60° for 30 sec, 72° for 45 sec, followed by a final extension of 72° for 3 min. Resulting bands were run on either a 3% (w/v) agarose 1× TAE (Tris, acetic acid, EDTA) gel for larger (>250 bp) products or a 6% (w/v) polyacrylamide 0.5× TBE (Tris, boric acid, EDTA) gel for smaller fragments.
The SSR from gw1-74i13 corresponds to a TAA/TAT repeat found from 99,536 to 99,593 bp with primer pair sequences of U, 5′-AGG AAG CTG CTT TAC AAC GTC-3′ and L, 5′-CAA AGC GTC CAT ACC AAA GTC A-3′ and a resulting PCR product of 682 bp in Williams 82. The SSR from gmw1-52d3 corresponds to a TA repeat found from 84195 to 84213 bp with primer pair sequences of U, 5′-AGT CAT CGA ATA AAC ATA G-3′ and L, 5′-AGT AAA AAC TTG AAA TTG G-3′ and a resulting PCR product of 150 bp. Genetic relationships between these SSRs and the established map were determined using MapMaker with a minimum lod score of 3.0 (Lander et al. 1987; Diers et al. 1992). Relative QTL positions were identified from the soybean composite map at Soybase (http://soybase.org).
Sequence analysis and annotation:
Gene prediction was done using a combination of ab initio and EST-alignment-based methods. For ab initio predictions, Genscan with Arabidopsis thaliana-based parameters (Burge and Karlin 1997), FgeneSH with Medicago truncatula-based parameters (http://www.softberry.com) and GeneMark.hmm with A. thaliana-based parameters (Lukashin and Borodovsky 1998) were run. For EST-verified structure prediction, GeneSeqer at PlantGDB (Schlueter et al. 2003) was used to align both soybean ESTs and other plant putatively unique transcripts (Dong et al. 2005) to the BAC sequences. These EST alignments and ab initio predictions can be viewed in an xGDB-based database at http://soybase.org/publication_data/Schlueter/GmaxGDB.html (S. D. Schlueter, M. Wilkerson and V. Brendel, unpublished results; http://xgdb.sourceforge.net). Each predicted gene was subjected to a BLASTP query of the NCBI nonredundant (nr) database with default parameters to assign a putative function. An e-value threshold of 1e−10 was used to assign putative function (supplemental Table 1 at http://www.genetics.org/supplemental/). Also, through BLASTP, any conserved motifs in predicted genes were identified.
AVID global pairwise alignments, with default parameters, were done between homeologous BACs to produce a VISTA plot to visualize nucleotide identity between sequences (Frazer et al. 2004). The percentage of identity and similarity between genes both intra- and interchromosomally was calculated using WATER (gap penalty of 10; extension penalty of 0.2; EMBOSS). Synonymous and nonsynonymous distances were calculated using PAML (Yang 1997) with the same parameters as those of Schlueter et al. (2004).
Putative retroelements were initially identified from ab initio gene predictions that were most similar by BLAST-based annotation to polyprotein sequences. Both BLASTN and TBLASTX were performed against the TIGR repeat databases (http://www.tigrblast.tigrorg/euk-blast/index.cgi?project=plant.repeats). Potential LTR retrotransposons were searched for using LTR_STRUC default parameters (McCarthy and McDonald 2003). Soybean-specific repetitive sequences identified from soybean BAC-end sequencing projects (Marek et al. 2001) were also searched using BLASTN. RepeatMasker was run utilizing Repbase (Smit, AFA & Green, P RepeatMasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html). A self-BLASTN of each BAC further identified putative repetitive regions as well as tandem duplications.
RT–PCR of homeologs:
Alignments between each gene pair, or gene family, were performed using ClustalX default parameters (DNASTAR). Primer pairs were designed for each gene to be specific for only one gene copy as described above for BAC identification (supplemental Table 2 at http://www.genetics.org/supplemental/). Additionally, when possible, each primer pair was designed to flank an intron as an internal control. Primer pairs were tested by PCR against G. max cultivar Williams 82 genomic DNA and subsequently sequenced to verify the homeolog specificity of each product.
Greenhouse-grown soybean tissue was collected from a range of organs and developmental stages of G. max cultivar Williams 82. For each time point, tissue was taken from at least three independent plants. Tissue for cotyledons, roots, and furled unifoliate was collected 3 days after emergence (DAE). Unfurled unifoliate tissue was collected 4 DAE. Another sample of cotyledons and roots was taken at 7 and 8 DAE, respectively. Furled trifoliolate was collected 11 DAE and unfurled trifoliolate at 15 DAE. Flowers and pods were taken at 60 and 76 DAE, respectively. All tissue was flash frozen with liquid nitrogen. mRNA was extracted and purified from frozen tissue using the RNeasy plant mini kit (QIAGEN), treated with a DNA-free DNase treatment and removal kit (Ambion, Austin, TX), and quantified using a NanoDrop ND-1000 spectrophotometer (Wilmington, DE).
Reverse transcriptase–PCR screens (RT–PCR) were conducted across all tissues with the above primers. Reactions were 25 μl in volume and used the SuperScript One-Step RT–PCR with platinum taq kit (Invitrogen) containing 1× reaction mix, 5 μm of each primer, 150 ng of RNA, and 1 μl of platinum taq DNA polymerase (Invitrogen). PCR cycling conditions were 42° for 45 min, 94° for 4 min, 35 cycles of 94° for 45 sec, annealing temperature for 30 sec, 72° for 45 sec, followed by a final extension of 72° for 5 min. Controls for RT–PCR reactions included a “minus” reverse transcriptase reaction to test for genomic DNA contamination, a water template reaction to test for reagent contamination, and a tubulin-positive control (Graham et al. 2002). All RT–PCR reactions were done with two to three independent biological replicates. These reactions provide a positive/negative screen for the presence of a transcript in a particular tissue.
Estimation of gene copy number:
Each annotated gene was searched against previously characterized Williams 82 specific EST-based gene family clusters (R. T. Nelson and R. C. Shoemaker, unpublished results) using BLASTN (Altschul et al. 1990). The number of family members identified corresponds to the number of unique ESTs in a family cluster. Additionally, Southern hybridizations were performed with each annotated gene (or homeologous pair/group) for a band-count/intensity-based copy number estimate. Williams 82 genomic DNA (15 μg) was digested with EcoRI, XhoI, and HindIII (2 units; New England Biolabs) run on 0.8% (w/v) agarose gel and transferred to Zeta-Probe blotting membranes (Bio-Rad, Hercules, CA). PCR primers (supplemental Table 2 at http://www.genetics.org/supplemental/) used in the RT–PCR analysis were used to generate Williams 82 genomic DNA probes for hybridizations. Additionally, primers for the six genes not common between BACs were designed (supplemental Table 3 at http://www.genetics.org/supplemental/). Labeling reactions were the same PCR conditions as for BAC identification except the dNTPs did not contain dCTP and 5 μl of [32P]dCTP was added to the reaction. Hybridizations were at 58° for 18 hr. Wash conditions were 10 min with 2× SSC (sodium chloride, sodium citrate), 0.1% SDS (sodium dodecyl sulfate), 15 min with 1× SSC, 0.1% SDS, and 15 min with 1× SSC, 0.1% SDS.
RESULTS
Identification, assembly, and mapping of soybean homeologous regions:
Shotgun sequencing of the BACs gmw1-74i13 and gmw1-52d3 yielded 5088 and 3896 sequence reads, respectively. Of the 3896 sequence reads from gmw1-52d3, 2072 were from the small fragment library and 1824 were from the large fragment library. For gmw1-74i13, sequence assembly yielded three major contigs. Sequencing across clone pairs closed two gaps; the final gap was closed by PCR amplification across the gap and subsequent sequencing. For gmw1-52d3, assembly was first done with the small-insert library. Addition of the larger-insert library to the assembly closed all gaps on gmw1-52d3. The overall assembled sequence length of gmw1-74i13 was 173,747 bp with an average coverage of 13× and of gmw1-52d3 was 98,760 bp with an average coverage of 16×.
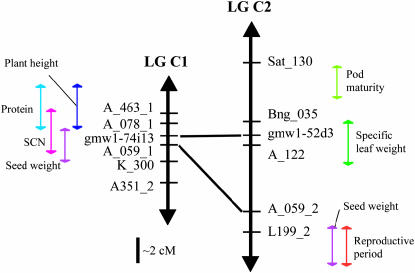
SSRs were identified from each BAC: 14 SSRs for gmw1-74i13 and 8 SSRs for gmw1-52d3. Genetic mapping placed gmw1-74i13 on linkage group C1 and gmw1-52d3 on linkage group C2 of the G. max A81-356022 × Glycine soja PI 468.916 mapping population (Diers et al. 1992; Shoemaker et al. 1996). Further orientation of these BACs on the composite maps (Song et al. 2003; http://soybase.org) shows that they map near the RFLP A_059 loci, another marker shared between C1 and C2 (Figure 1). The RFLP marker A_059, however, is not contained within these BAC sequences. A number of QTL are associated with markers in these regions of the linkage groups (Figure 1). Only one of the QTL appear to be retained between the linkage groups: a seed weight QTL (Orf et al. 1999). Gmw1-74i13 maps within the region of a soybean cyst nematode (SCN) resistance QTL as well (Yue et al. 2001). Other QTL that are close to gmw1-74i13 include a protein QTL (Lee et al. 1996a) and a plant height QTL (Lee et al. 1996b). Conversely, gmw1-52d3 falls within a specific leaf weight QTL (Mian et al. 1998). Other QTL in that region of linkage group C2 are QTL for pod maturity (Wang et al. 2003) and for reproductive period (Orf et al. 1999).
Figure 1.—
Map position of BACs gmw1-74i13 and gmw1-52d3 relative to the soybean composite map, linkage groups C1 and C2. Mapping of the BACs was based upon SSRs found in each BAC sequence. Lines show homeologous relationships between markers on these linkage groups. QTL are represented as colored arrows next to the linkage groups.
Sequence annotation of potential genes:
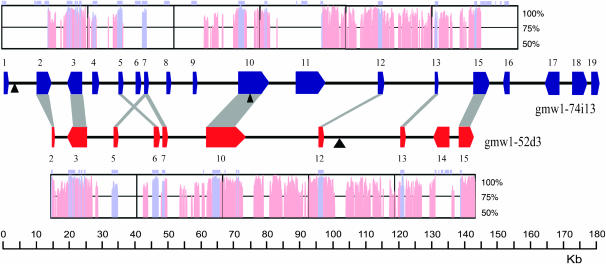
The three ab initio gene prediction programs used yielded somewhat similar gene structures, but varied enough that EST-alignment-based gene structure confirmation was warranted. On average, 65% of the predicted gene structures had exon coverage with a soybean EST and a total of 62% of the intron junctions were confirmed with EST support. A total of 28 genes was predicted between both BACs with gmw1-74i13 containing 19 genes and gmw1-52d3 containing 10 genes (Figure 2; http://soybase.org/publication_data/Schlueter/GmaxGDB.html). Between the two homeologous regions, 9 genes are mutually retained (Table 1; Figure 2).
Figure 2.—
Gene positions on homeologous soybean BACs gmw1-74i13 and gmw1-52d3. Each colored block arrow on the line represents a gene and gray boxes between genes show homeologs. All gene numbering corresponds to Table 1. The plots above and below the gene locations are VISTA plots showing the relative nucleotide identity between the two BACs. The light purple boxes on top of the VISTA plots correspond to exon positions. Degenerate polyprotein insertions are shown by black triangles.
TABLE 1.
Predicted gene features of homeologous BACs gmw1-74i13 and gmw1-52d3
| Total lengtha
|
Identity of exons | No. exons
|
Length of exons
|
No. of introns
|
Length of introns
|
Length aa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Putative function | 74i13 | 52d3 | 74i13 | 52d3 | 74i13 | 52d3 | 74i13 | 52d3 | 74i13 | 52d3 | 74i13 | 52d3 | Identity aa | Similarity aa | |
| 1 | WOX4 protein (homeobox–leucine zipper transcription factor protein) | 1016 | — | — | 3 | 567 | — | 2 | — | 449 | — | 189 | — | — | — | |
| 2 | Poly(A)-binding protein | 4128 | 225 | 96.0 | 9 | 1 | 1893 | 225 | 8 | — | 2235 | — | 630 | 75 | 97.3 | 98.6 |
| 3 | Membrane-like protein | 4104 | 4191 | 96.9 | 12 | 12 | 1422 | 1476 | 11 | 11 | 2682 | 2715 | 474 | 492 | 97.5 | 97.7 |
| 4 | N-hydroxycinnamoyl/benzoyl transferase-like 1 | 1226 | b | 4 | 807 | 3 | 419 | 269 | b | b | ||||||
| 5 | N-hydroxycinnamoyl/benzoyl transferase-like 2 | 1383 | 1368 | b | 1 | 1 | 1383 | 1368 | 0 | 0 | 0 | 0 | 461 | 456 | b | b |
| 6 | N-hydroxycinnamoyl/benzoyl transferase-like 3 | 1461 | 1374 | b | 1 | 1 | 1461 | 1374 | 0 | 0 | 0 | 0 | 487 | 458 | b | b |
| 7 | N-hydroxycinnamoyl/benzoyl transferase-like 4 | 1410 | 483 | b | 1 | 1 | 1410 | 483 | 0 | 0 | 0 | 0 | 470 | 161 | b | b |
| 8 | N-hydroxycinnamoyl/benzoyl transferase-like 5 | 1398 | — | b | 1 | — | 1398 | — | 0 | — | 0 | — | 466 | — | b | b |
| 9 | N-hydroxycinnamoyl/benzoyl transferase-like 6 | 1386 | — | b | 1 | — | 1386 | — | 0 | — | 0 | — | 462 | — | b | b |
| 10 | Zinc finger (C3HC4-type) 1 | 8157 | — | 92.5 | 4 | — | 2073 | — | 3 | — | 6084 | — | 691 | — | 88.3 | 90.8 |
| 11 | Zinc finger (C3HC4-type) 2 | 7685 | 6015 | 92.3 | 5 | 5 | 2043 | 2043 | 4 | 4 | 5642 | 3972 | 681 | 708 | 88.5 | 90.9 |
| 12 | Heat-shock transcription factor 31 | 1190 | 1174 | 95.3 | 2 | 2 | 1092 | 1092 | 1 | 1 | 98 | 82 | 364 | 364 | 94.5 | 97 |
| 13 | bHelix-loop-helix transcription factor | 795 | 777 | 80.7 | 1 | 1 | 795 | 777 | 0 | 0 | 0 | 0 | 264 | 259 | 77.9 | 82.4 |
| 14 | Remorin-like protein | — | 3885 | — | — | 7 | — | 1581 | — | 6 | — | 2304 | — | 527 | — | — |
| 15 | Arabidopsis-like expressed protein | 4684 | 3065 | 96.7 | 8 | 3 | 2406 | 1452 | 7 | 4 | 2278 | 1613 | 802 | 484 | 96.2 | 98.2 |
| 16 | bzip transcription factor | 3757 | — | — | 4 | — | 1254 | — | 3 | — | 2503 | — | 418 | — | — | — |
| 17 | DCL protein | 4452 | — | — | 3 | — | 636 | — | 2 | — | 3816 | — | 212 | — | — | — |
| 18 | Aromatic-rich family protein | 4088 | — | — | 4 | — | 756 | — | 3 | — | 3332 | — | 252 | — | — | — |
| 19 | PPR-repeat containing protein | 2092 | — | — | 4 | — | 1749 | — | 3 | — | 343 | — | 583 | — | — | — |
| Average | 92.9 | 91.5 | 93.7 | |||||||||||||
PPR, pentatricopeptide repeat. aa, amino acid.
Total length based on nucleotide exons plus introns from translation start to stop.
See Table 3 for identity and similarity information for the HCBT genes.
The average gene density of gmw1-74i13 is one gene every 9.1 kbp and of gmw1-52d3 is one gene every 9.9 kbp. This is less dense than previous estimates of one gene every 8 kbp (Young et al. 2003), one gene every 6 kbp (Triwitayakorn et al. 2005), or 5.8–6.7 kbp (Mudge et al. 2005). The average G/C content of the BACs is similar, with gmw1-74i13 being 32.27% and gmw1-52d3 being 31.85%.
The coding regions of predicted genes range in size from a partial 567-bp WOX4 homeobox–leucine zipper transcription factor protein to a 2454-bp gene similar to an A. thaliana expressed protein (Table 1; Figure 2). The intron/exon structure of genes on both BACs varied widely from a cluster of three to six single-exon HCBT genes to a membrane-like protein containing 12 exons. Details on the annotation of each predicted gene can be found in Table 1 and in supplemental Table 1 at http://www.genetics.org/supplemental/. Estimates on the copy number of each gene (or homeologous pair/group) were determined by EST-based methods as well as by Southern-based band counts (Table 2). In most cases, the EST-based count was an underestimate. On the occasion that Southern-based band counts are fewer than EST estimates, it is likely that a single hybridization signal may reflect more than one gene product of the same size.
TABLE 2.
Estimated gene family sizes of gmw1-74i13 and gmw1-52d3 predicted genes
| Gene | Putative function | EST-based family sizea | Southern-based band countb |
|---|---|---|---|
| 1 | WOX4 protein (homeobox–leucine zipper transcription factor protein) | 2 | 2 |
| 2 | Poly(A)-binding protein | 16 | 4+ |
| 3 | Membrane-like protein | 3–4 | 2 |
| 4 | N-hydroxycinnamoyl/benzoyl transferase-like 1 | 5 | 7 |
| 5 | N-hydroxycinnamoyl/benzoyl transferase-like 2 | 5 | 7 |
| 6 | N-hydroxycinnamoyl/benzoyl transferase-like 3 | 5 | 7 |
| 7 | N-hydroxycinnamoyl/benzoyl transferase-like 4 | 5 | 7 |
| 8 | N-hydroxycinnamoyl/benzoyl transferase-like 5 | 5 | 7 |
| 9 | N-hydroxycinnamoyl/benzoyl transferase-like 6 | 5 | 7 |
| 10 | Zinc finger (C3HC4-type) 1 | 2 | 2 |
| 11 | Zinc finger (C3HC4-type) 2 | 2 | 2 |
| 12 | Heat-shock transcription factor 31 | No ESTc | 3–4 |
| 13 | bHelix-loop-helix transcription factor | 2 | 2 |
| 14 | Remorin-like protein | 2–3 | 4 |
| 15 | Arabidopsis-like expressed protein | 2–4 | 7 |
| 16 | bzip transcription factor | 3–4 | 2 |
| 17 | DCL protein | 1–2 | 1 |
| 18 | Aromatic-rich family protein | 1 | 11+ |
| 19 | PPR-repeat containing protein | No EST | 4 |
EST count based on Williams and Williams 82 genotype only.
Band and band intensity count from Southern hybridizations to Williams 82 genomic DNA.
ESTs from other soybean cultivars suggest at least two copies.
Analysis of repetitive elements:
No full-length LTR-retrotransposon elements could be identified in either BAC sequence. However, three very degenerate polyprotein-like sequences were found from BLAST similarity searches of the nr database (Figure 2). Two degenerate polyproteins were found on gmw1-74i13; the first just 3′ of the WOX4-like gene and the second contained within the first intron of the first zinc-finger protein (Figure 2). The last degenerate polyprotein was found just 3′ of the heat-shock transcription factor on BAC gmw1-52d3 (Figure 2). None of these were conserved between the two BACs.
Comparison of homeologous soybean regions:
Nine genes shared between BACs gmw1-74i13 and gmw1-52d3 are conserved in both order and orientation. There are, however, several discernible differences between the homeologous soybean regions (Figure 2). BAC gmw1-52d3 contains one gene, remorin, which is not found in gmw1-74i13. Gmw1-74i13 contains an extra tandemly duplicated zinc-finger protein as well as three additional copies of HCBT. All full-length genes contain the same number of introns and exons. Gmw1-74i13 gene 5, the first HCBT gene, appears to be a fragmented copy with four exons whereas all other HCBT genes are single exon. A frameshift mutation is found in gmw1-52d3 gene 7, the last HCBT gene on this BAC, leading to a stop codon and truncation of the resulting protein. Additionally, the BAC ends of gmw1-52d3 are each in the middle of a gene; consequently, the 5′ PABP gene on gmw1-52d3 is truncated and likely missing eight exons and gene 14, and an Arabidopsis-like expressed protein is truncated and missing six exons (Table 1; Figure 2).
When we consider the composition of the genes shared between the homeologous regions, we find that the total length (exons plus introns) of each retained gene is similar. The average nucleotide identity between homeologous coding regions is 89.8% and the resulting amino acid identity and similarity is 88 and 90.7%, respectively, not including the HCBT genes. The HCBT genes have an average nucleotide identity of 75.1%, markedly less than that of the other homeologs.
Genic sequences are the most conserved between the BACs with upward of 95% nucleotide identity (Figure 2). What is striking is the conservation of noncoding sequence between the genes. Some of the conservation of intergenic sequence may be due to promoter elements and transcription-factor-binding sites, but conservation is greater than anticipated. The intergenic distance, however, is not as well conserved. Compared to gmw1-52d3, gmw1-74i13 contains 16,785 bp more DNA in the overlapping intergenic regions. This translates to 16.5% more noncoding DNA. However, the greater intergenic distance in gmw1-74i13 is due to a greater number of tandem duplications. As a result of more genes via tandem duplication, the average intergenic distance for gmw1-74i13 is 8486 bp and for gmw1-52d3 is 9450 bp.
Synonymous and nonsynonymous distance measures were calculated using PAML between the retained duplicate genes. The average synonymous distance of the nontandemly duplicated homeologs (genes 2, 3, 10, 12, and 13) was 0.149, suggesting that this region was duplicated ∼12.2 million years ago. The tandemly duplicated genes were not considered in this estimate because their synonymous distances suggest tandem duplication after the polyploid event. On the basis of this evidence, these BACs likely represent homeologous segments that have been retained since the most recent (14 MYA) genome duplication in soybean (Schlueter et al. 2004).
Tandem duplication of N-hydroxycinnamoyl/benzoyltransferase:
Both gmw1-74i13 and gmw1-52d3 contain a conserved cluster of HCBT genes. These genes show a number of characteristics that set them apart from the surrounding genes. First, most of these genes are single-exon genes, with the exception of gmw1-74i13, gene 4 (HCBT 1), which contains four exons and is a fragmented copy. The gmw1-74i13 HCBT clustered genes are 60–80% identical at the nucleotide level and 58–88% similar at the amino acid level. Similarly, the gene cluster on gmw1-52d3 is 58–80% identical at the nucleotide level and 58–85% similar at the amino acid level (Table 3). Between gmw1-74i13 and gmw1-52d3 the nucleotide identity and amino acid similarity is slightly higher, ranging from 58 to 92% and 58 to 96%, respectively. Interestingly, gmw1-74i13, gene 5 (HCBT 2), and gmw1-52d3, gene 6 (HCBT 2), are 96% similar at the amino acid level (Table 3).
TABLE 3.
Identity, similarity, and synonymous and nonsynonymous distances of HCBT genes from gmw1-52d3 and gmw1-74i13
| 52d3-5 | 52d3-6 | 52d3-7 | 74i13-4 | 74i13-5 | 74i13-6 | 74i13-7 | 74i13-8 | 74i13-9 | |
|---|---|---|---|---|---|---|---|---|---|
| 52d3-5 | — | 80.3 | 59.1 | 63.8 | 80.3 | 80.3 | 83.5 | 79.3 | 81.9 |
| 78.3 | 53.0 | 55.4 | 78.0 | 79.5 | 80.5 | 76.2 | 79.9 | ||
| 85.7 | 59.1 | 61.2 | 85.3 | 86.6 | 88.2 | 84.2 | 86.6 | ||
| 52d3-6 | 0.134 | — | 58.2 | 65.2 | 91.4 | 82.3 | 83.1 | 76.8 | 80.7 |
| 0.461 | 52.4 | 56.5 | 91.1 | 79.4 | 78.5 | 71.0 | 76.8 | ||
| 58.4 | 61.4 | 96.2 | 87.1 | 86.7 | 82.0 | 85.9 | |||
| 52d3-7 | 0.168 | 0.149 | — | 62.9 | 60.8 | 59.0 | 65.0 | 58.1 | 59.4 |
| 0.523 | 0.466 | 52.6 | 52.6 | 50.9 | 59.7 | 51.1 | 52.0 | ||
| 59.6 | 58.7 | 57.0 | 62.8 | 56.2 | 58.6 | ||||
| 74i13-4 | 0.152 | 0.151 | 0.136 | — | 64.8 | 60.9 | 64.3 | 61.1 | 63.3 |
| 0.603 | 0.510 | 0.362 | 52.6 | 48.1 | 51.2 | 50.4 | 55.0 | ||
| 58.1 | 55.6 | 58.1 | 56.5 | 62.2 | |||||
| 74i13-5 | 0.122 | 0.044 | 0.137 | 0.244 | — | 81.7 | 81.3 | 76.9 | 79.9 |
| 0.449 | 0.218 | 0.428 | 0.643 | 80.4 | 77.7 | 70.8 | 78.0 | ||
| 88.7 | 85.4 | 81.4 | 87.1 | ||||||
| 74i13-6 | 0.123 | 0.125 | 0.184 | 0.303 | 0.116 | — | 82.4 | 76.9 | 79.2 |
| 0.522 | 0.374 | 0.469 | 0.829 | 0.384 | 78.8 | 71.7 | 75.9 | ||
| 86.4 | 81.8 | 84.4 | |||||||
| 74i13-7 | 0.104 | 0.128 | 0.095 | 0.198 | 0.119 | 0.124 | — | 76.5 | 80.9 |
| 0.396 | 0.362 | 0.244 | 0.542 | 0.395 | 0.357 | 71.8 | 76.9 | ||
| 81.1 | 86.6 | ||||||||
| 74i13-8 | 0.143 | 0.188 | 0.204 | 0.212 | 0.118 | 0.183 | 0.179 | — | 77.1 |
| 0.516 | 0.437 | 0.530 | 0.489 | 0.400 | 0.490 | 0.407 | 71.4 | ||
| 83.5 | |||||||||
| 74i13-9 | 0.119 | 0.143 | 0.170 | 0.163 | 0.129 | 0.146 | 0.136 | 0.180 | — |
| 0.409 | 0.415 | 0.450 | 0.586 | 0.470 | 0.489 | 0.454 | 0.544 |
Top italic numbers indicate percentage of nucleotide identity. Middle italic numbers indicate percentage of protein identity. Bottom italic numbers indicate percentage of protein similarity. Top nonitalic numbers indicate nonsynonymous distance. Bottom nonitalic numbers indicate synonymous distance.
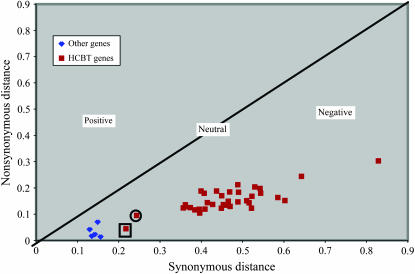
When synonymous and nonsynonymous distances are calculated between all HCBT copies, an intriguing trend appears (Table 3; Figure 3). Relative to the other homeologous genes, most of the HCBT genes have larger synonymous and nonsynonymous distances, both within BACs and between BACs. This suggests that the HCBT genes have evolved at a faster rate than the other genes in this region. Again, gmw1-74i13, gene 5 (HCBT 2), and gmw1-52d3, gene 6 (HCBT 2), as well as gmw1-74i13, gene 7 (HCBT 4), and gmw1-52d3, gene 7 (HCBT 3), differ from this trend and have much smaller synonymous and nonsynonymous distances (Figure 3).
Figure 3.—
A plot of synonymous distance vs. nonsynonymous distance for all soybean homeologous gene pairs. All red squares correspond to HCBT gene alignments while blue diamonds represent all other homeologs between the soybean BACs gmw1-74i13 and gmw1-52d3. The circled square corresponds to synonymous and nonsynonymous distances between gmw1-74i13, gene 7 (HCBT 4), and gmw1-52d3, gene 7 (HCBT 3). The blocked square similarly corresponds to gmw1-74i13, gene 5 (HCBT 2), and gmw1-52d3, gene 6 (HCBT 2).
RT–PCR analysis of homeologous genes:
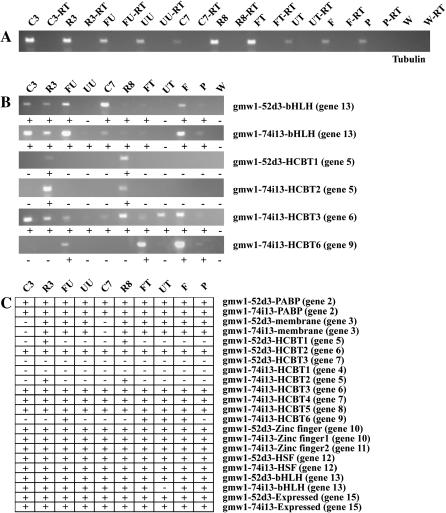
To better understand the functional evolution of these regions, 22 RT–PCR primer pairs that differentiated between each retained homeolog were designed (supplemental Table 3 at http://www.genetics.org/supplemental/). Ten different tissue types were chosen to look at a variety of organs and developmental stages. Negative controls confirmed that the mRNA samples were free of genomic DNA contamination. Tubulin was used as a positive control to verify the integrity of each mRNA sample.
These results demonstrate that 20 of the 22 predicted homeologs are transcribed. The first HCBT gene on gmw1-74i13 (gene 4) and the last HCBT gene on gmw1-52d3 (gene 7) showed no evidence of transcription. Only gene 13, homeologous bHLH proteins, and three HCBT genes show evidence for differential transcription between homeologs. Gene 13 on gmw1-74i13 shows no transcription in unfurled unifoliate whereas the gmw1-52d3 copy is transcribed in that tissue. Both the first HCBT gene on gmw1-52d3 (gene 4) and the second HCBT gene on gmw1-74i13 (gene 5) appear to be transcribed only in the below-ground portion of the plant. Gene 9 on gmw1-74i13 was detected in furled unifoliolate, furled trifoliolate, flowers, and pods (Figure 4). All other HCBT genes appeared to be transcribed in all tissues sampled.
Figure 4.—
RT–PCR reactions for the soybean homeolog-specific primers. (A) Control reactions to test for genomic DNA contamination of mRNA samples using Tubulin56 primers. (B) RT–PCR reactions for homeologs that show differential expression between BACs. (C) RT–PCR results for all homeologs between soybean BACs. The x-axis lists the tissue types and the y-axis lists the homeolog-specific primers. Genes that showed amplification (were expressed) are labeled with a plus sign. Genes that did not show amplification (no expression) are labeled with a minus sign. Tissue types are as follows: C3, cotyledons 3 DAE; R3, roots 3 DAE; FU, furled unifoliate 3 DAE; UU, unfurled unifoliate 4 DAE; C7, cotyledons 7 DAE; R8, roots 8 DAE; FT, furled trifoliolate 11 DAE; UT, unfurled trifoliolate 15 DAE; F, flowers 60 DAE; P, pods 76 DAE. All samples with a −RT are genomic DNA controls with no reverse transcriptase for mRNA amplification, but still contain Taq DNA polymerase.
DISCUSSION
Stability of homeologous soybean regions:
Previous detailed analyses of genomic sequences from homeologous regions in a paleopolyploid have been limited to maize (Illic et al. 2003; Langham et al. 2004; Ma et al. 2005). Their results found that gene content is relatively unstable between homeologous regions in the maize genome and that reciprocal deletions have led to the retention of only one copy in each homeologous region. This suggested that during diploidization, natural selection worked such that only one gene copy was retained. Further studies of BAC-end sequences in maize showed that while tandemly amplified genes are conserved, there is a surprising lack of retained homeologous genes, suggesting that during rediploidization maize has experienced significant gene loss through various means such as mutation and transposition (Messing et al. 2004). Our results are quite different from what has been observed in maize. We find that sequence and structure conservation of homeologous regions in soybean closely resemble those of homeologs in cotton, a relatively recent polyploid (Grover et al. 2004). We find an almost gene-for-gene retention between two soybean BACs as well as fairly high nucleotide identity in noncoding regions (Figure 2). These results support previous suggestions that the soybean genome has retained extensive conserved homeologous sequence (Shoemaker et al. 1996; Marek et al. 2001; Foster-Hartnett et al. 2002; Yan et al. 2003, 2004).
Both maize and soybean have been shown to be paleopolyploids with their most recent major genome duplication ∼11 and 14 MYA, respectively (Gaut and Doebley 1997; Schlueter et al. 2004). However, while the homeologous regions of maize have diverged greatly during rediploidization, those in soybean remain relatively stable. The evolutionary histories of maize and soybean need to be considered to explain these differences. The maize genome is well documented to have experienced a large amplification of transposable elements leading to shuffling of the rediploidizing genome (SanMiguel and Bennetzen 1998; Zhang and Peterson 1999; Lal and Hannah 2005). Conversely, there has been little to no evidence for a similar transposon explosion in the soybean genome. An analysis of soybean sequence around a cyst nematode resistance gene showed that only 3% of the predicted genes were transposable elements (Mudge et al. 2005). The lack of identifiable recent transposable element insertions in either gmw1-74i13 or gmw1-52d3 is in agreement with that observation. On the basis of a Poisson distribution of RFLP probes to soybean BAC pools, Mudge et al. (2004) proposed that the soybean gene space may be limited to as little as 24% of the genome. Similarly, using FISH, Lin et al. (2005) showed that some of the gene space in soybean might lack high-copy repetitive sequences. Our findings support these predictions; gene-rich regions may not be hotspots for recent retroelement insertions.
Four of the nine homeologs are similar to transcription factors: WOX4, the zinc-finger genes, heat-shock factor, and bHelix-loop-helix protein. Additionally, the HCBT genes are implicated with a role in disease response signaling and in the synthesis of phytoalexins. Blanc and Wolfe (2004b) found that, in Arabidopsis, duplicated genes encoding proteins involved in transcription or signal transduction are preferentially retained, while only one copy of genes involved in DNA repair are kept (more likely to be silenced). Our results with soybeans support their hypothesis. Most of the genes identified in the homeologous BACs are in some way involved in either signal transduction or binding of DNA. This could account for the greater-than-expected gene retention between these regions. Further, this suggests that there may be clustering of signaling genes or transcription factors within the soybean genome.
While there may be a bias toward the types of genes retained in the homeologous BACs analyzed, these findings show that retention of duplicate genes in homeologous regions in soybean may be common. Paleopolyploids are those species that have, over millions of years, undergone a switch from tetrasomic inheritance to disomic inheritance but still retain characteristics of a polyploid in the form of duplicate genes. Within a single species, there may be a mixture of diploid and tetraploid loci and, thus, rediploidization probably does not happen simultaneously for all chromosomes (Wolfe 2001). The high number of duplicate genes in homeologous regions suggests that, in soybean, the process of diploidization is a slow and ongoing process.
Sequence conservation was seen in the noncoding regions as well. Following a duplication event, genes may undergo several fates: retained function, subfunctionalization, neofunctionalization, or silencing (Force et al. 1999). After a period of relaxed selection, duplicates that have survived the birth and death process of duplication are under purifying negative selection and mutations in the coding regions tend to be silent (Kondrashov et al. 2002; Schlueter et al. 2004). However, many of the mutations that lead to duplicate gene retention and functional changes are not within the coding region, but rather upstream in regulatory sequences (Force et al. 1999). In this study we find that, while there have been some changes in the noncoding regions of these BACs, much of the noncoding sequence is retained. The ratio of synonymous-to-nonsynonymous distances for all of the homeologs shows that while some genes may be evolving faster, all genes are under negative/purifying selection (Figure 4). In other words, there is some selective constraint that is retaining both copies of homeologs.
Determination of potential copy numbers for each gene identified in these regions showed that in most cases each gene exists in at least two copies, and often more copies (Table 2). Some of the annotated genes are known to be members of large diverse gene families such as the poly(A)-binding proteins (Le and Gallie 2000) and heat-shock factors (Nover et al. 1998). However, copy number estimates from ESTs are usually low due to incomplete sampling of the transcriptome. Indeed, EST alignments to the predicted genes in these regions suggests that only 65% of the transcriptome is represented. Similarly, due to the high sequence similarity as well as the relatively similar gene sizes, even counts from Southern's may be skewed. However, these results also suggest that there may be more copies of these genes within the genome.
Homeologous gene transcription:
Although we expected to find evidence for transcriptional changes between homeologs, differences were quite limited. Only gene 13 (bHLH) and three HCBT genes showed evidence for differences in transcription between homeologs. These findings, along with the high sequence conservation in the noncoding regions between homeologs, suggest retention of transcription-factor-binding sites and thereby retention of transcription patterns. It is possible that changes were not observed because the transcription profiles are quantitative or that the tissues sampled may not be representative of those showing changes in transcription between homeologs.
The duplication–degeneration–complementation model suggested by Force et al. (1999) proposed that the retention of duplicated genes after polyploidy is the result of changes allowing either new gene function or compartmentalized gene function. The HCBT genes seem to fit this model. Of particular interest is the putative root-specific expression of gmw1-74i13 HCBT 1 (gene 5) and gmw1-52d3 HCBT 2 (gene 5; Figure 4). Almost all other HCBT genes are expressed in all tissues sampled, except gmw1-74i13 HCBT 6 (gene 9). This suggests either that the two homeologs with root-specific expression have independently developed this expression through the loss of regulatory elements or that the other genes have become more broadly expressed relative to these copies.
Possible role for the tandemly duplicated HCBT genes:
Tandem duplications have been shown in plants to have a role in the evolution of large gene families (Tian et al. 2004; Schauser et al. 2005). In this study we identified a cluster of HCBT genes in both homeologous soybean BACs (Figure 2). HCBT functions as the first step of phytoalexin biosynthesis by catalyzing the reaction of anthranilate and benzoyl–CoA to N-benzoylanthranilate (Yang et al. 1997). The accumulation of these dianthramide phytoalexins has been associated with plant response to a pathogen attack (Yang et al. 1997). In particular, the phytoalexin glyceollin has been shown to accumulate at the infection site of SCN in roots of resistance cultivars (Huang and Barker 1991). Glyceollin is a product of the phenylpropanoid pathway of which the production of N-benzoylanthranilate is a precursor step (Hammerschmidt 1999). This association between phytoalexin accumulation during SCN infection and the identification of HCBT genes found within an SCN QTL is worth further investigation.
The HCBT genes have accumulated more mutations, both synonymous and nonsynonymous, than the other homeologs (Table 3). The larger HCBT synonymous and nonsynonymous distance measures both between BACs and within BACs suggest that these tandemly amplified genes are undergoing more rapid evolution than the surrounding genes. Although all the HCBT genes appear to be under negative or purifying selection, there might be a slight relaxation in this pressure that has allowed these genes to mutate more than the surrounding homeologs.
Genes involved in disease response can have regions that either are under positive selection or have accumulated a number of mutations allowing plasticity in response to various pathogens (Graham et al. 2002). Through tandem duplication, the sheer number of these genes allows more mutations to accumulate in both the coding and upstream promoter regions, allowing for a broader response to pathogen attack. HCBT proteins have conserved cystine residues that may allow the formation of disulfide bridges and thus the formation of dimers (Yang et al. 1997). By having multiple combinations of dimers, this also would permit a broader pathogen response.
This study provides us with our first glimpse at genic and intergenic sequence conservation in the paleopolyploid soybean genome. Not only was sequence conservation higher than expected, but also limited expression differences between homeologs were observed. Further studies of homeologous regions in soybean are warranted to better understand the evolutionary history of this paleopolyploid genome. On the basis of the G. max cultivar Forrest physical map, the portions of linkage groups C1 (adjacent to marker A_059_1) and C2 to which the homeologous BACs mapped are excellent candidates to study duplicated regions due to the higher-than-average number of BACs identified by A_059_1 on linkage group C1 (Wu et al. 2004; Schultz et al. 2006). Additionally, neither of the BACs mapped to the pBng_181 locus on A2 (http://www.soybase.org) although pBng181 showed a high similarity to the HCBT EST contig sequences. A region of A2 has previously been shown to have syntenic markers with C2 based on RFLP mapping (Shoemaker et al. 1996). Therefore, it is likely that an extensive network of homeologous segments may be available for further study in the soybean genome.
Acknowledgments
The authors thank Mary Duke and Xiaofen Liu for their sequencing expertise. We also thank Corrinne Grover for her knowledge and help annotating genes and identifying transposable elements and Jody Hayes for her skills in mapping SSR markers. Names are necessary to report factually on the available data; however, the U. S. Department of Agriculture (USDA) neither guarantees nor warrants the standard of the product, and the use of the name by the USDA implies no approval of the product to the exclusion of others that may also be suitable. This article is a contribution of the Corn Insect and Crop Genetics Research Unit (USDA-Agricultural Research Service). J. A. Schlueter was supported by a grant from the United Soybean Board.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and K. H. Wolfe, 2004. a Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., and K. H. Wolfe, 2004. b Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., K. Hokamp and K. H. Wolfe, 2003. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C., and S. Karlin, 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Diers, B. W., P. Keim, W. R. Fehr and R. C. Shoemaker, 1992. RFLP analysis of soybean seed protein and oil content. Theor. Appl. Genet. 83: 608–612. [DOI] [PubMed] [Google Scholar]
- Dong, Q., C. J. Lawrence, S. D. Schlueter, M. D. Wilkerson, S. Kurtz et al., 2005. Comparative plant genomics resources at PlantGDB. Plant Physiol. 139: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y.-L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster-Hartnett, D., J. Mudge, D. Danesh, H. Yan, D. Larsen et al., 2002. Comparative genomic analysis of sequences sampled from a small region on soybean molecular linkage group ‘G.’ Genome 45: 634–645. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., L. Pachter, A. Poliakov, E. M. Rubin and I. Dubchak, 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and J. F. Doebley, 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94: 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S. A., D. Ricke, T.-H. Lan, G. Presting, R. Wang et al., 2002. A draft sequence of the rice genome (Oryza sativa L. spp. Japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Graham, M. A., L. F. Marek and R. C. Shoemaker, 2002. Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics 162: 1961–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien, M. A., S. Berry-Lowe, B. W. Shirley and R. Meagher, 1986. Two soybean ribulose-1,5-bisphosphate carboxylase small subunit genes share extensive homology even in distant flanking sequences. Plant Mol. Biol. 7: 451–465. [DOI] [PubMed] [Google Scholar]
- Grover, C. E., H. Kim, R. A. Wing, A. H. Paterson and J. F. Wendel, 2004. Incongruent patterns of local and global genome size evolution in cotton. Genome Res. 14: 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley, H. H., and T. Hymowitz, 1973. Speciation and cytogenetics, pp. 96–116 in Soybeans: Improvement, Production and Uses, edited by B. E. Caldwell. American Society of Agronomy, Madison, WI.
- Hammerschmidt, R., 1999. Phytoalexins: What have we learned after 60 years? Annu. Rev. Phytopathol. 37: 285–306. [DOI] [PubMed] [Google Scholar]
- Hightower, R., and R. Meagher, 1985. Divergence and differential expression of soybean actin genes. EMBO J. 4: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.-S., and K. R. Barker, 1991. Glyceollin I in soybean-cyst nematode interactions: spatial and temporal distribution in roots of resistant and susceptible soybeans. Plant Physiol. 96: 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, K., P. J. SanMiguel and J. L. Bennetzen, 2003. A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc. Natl. Acad. Sci. USA 100: 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F. A., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin, 2002. Selection in the evolution of gene duplications. Genome Biol. 3: 0008.1–0008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey, J. A., 1980. Chromosome numbers in the Phaseoleae (Fabaceae:Faboideae) and their relation to taxonomy. Am. J. Bot. 67: 595–602. [Google Scholar]
- Lal, S. K., and L. C. Hannah, 2005. Plant genomes: massive changes of the maize genome are caused by helitrons. Heredity 95: 421–422. [DOI] [PubMed] [Google Scholar]
- Lander, E., P. Green, J. Abrahamson, A. Barlow, M. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Langham, R. J., J. Walsh, M. Dunn, C. Ko, S. A. Goff et al., 2004. Genome duplication, fractionation and the origin of regulatory novelty. Genetics 166: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton-Rauh, A., 2003. Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenet. Evol. 29: 396–409. [DOI] [PubMed] [Google Scholar]
- Le, H., and D. R. Gallie, 2000. Sequence diversity and conservation of the poly(A)-binding protein in plants. Plant Sci. 152: 101–114. [Google Scholar]
- Lee, J. S., and D. P. S. Verma, 1984. Chromosomal arrangement of leghemoglobin genes in soybean. Nucleic Acids Res. 11: 5541–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. M., A. Bush, J. E. Specht and R. C. Shoemaker, 1999. Mapping duplicate genes in soybean. Genome 42: 829–836. [Google Scholar]
- Lee, J. M., D. Grant, C. E. Vallejos and R. C. Shoemaker, 2001. Genome organization in dicots. II. Arabidopsis as a ‘bridging species’ to resolve genome evolution events among legumes. Theor. Appl. Genet. 103: 765–773. [Google Scholar]
- Lee, S. H., M. A. Bailey, M. A. R. Mian, T. E. Carter, Jr., E. R. Shipe et al., 1996a. RFLP loci associated with soybean seed protein and oil content across populations and locations. Theor. Appl. Genet. 93: 649–657. [DOI] [PubMed] [Google Scholar]
- Lee, S. H., M. A. Bailey, M. A. R. Mian, T. E. Carter, Jr., D. A. Ashley et al., 1996b. Molecular markers associated with soybean plant height, lodging, and maturity across locations. Crop Sci. 36: 728–735. [Google Scholar]
- Lin, J.-Y., B. H. Jacobus, P. SanMiguel, J. G. Walling, Y. Yuan et al., 2005. Pericentromeric regions of soybean (L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile. Genetics 170: 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton, S., and B. S. Gaut, 2005. Plant conserved non-coding sequences and paralogue evolution. Trends Genet. 21: 60–65. [DOI] [PubMed] [Google Scholar]
- Lukashin, A., and M. Borodovsky, 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Ma, J., P. SanMiguel, J. Lai, J. Messing and J. L. Bennetzen, 2005. DNA rearrangement in orthologous Orp regions of the maize, rice and sorghum genomes. Genetics 170: 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek, L. F., and R. C. Shoemaker, 1997. BAC contig development by fingerprint analysis in soybean. Genome 40: 420–427. [DOI] [PubMed] [Google Scholar]
- Marek, L. F., J. Mudge, L. Darnielle, D. Grant, N. Hanson et al., 2001. Soybean genomic survey: BAC-end sequences near RFLP and SSR markers. Genome 44: 572–581. [PubMed] [Google Scholar]
- Masterson, J., 1994. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264: 421–424. [DOI] [PubMed] [Google Scholar]
- McCarthy, E. M., and J. F. McDonald, 2003. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19: 362–367. [DOI] [PubMed] [Google Scholar]
- Messing, J., A. K. Bharti, W. M. Karlowski, H. Gundlach, H. R. Kim et al., 2004. Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 101: 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian, M. A. R., D. A. Ashley and H. R. Boerma, 1998. An additional QTL for water use efficiency in soybean. Crop Sci. 38: 390–393. [Google Scholar]
- Mudge, J., Y. Huihuang, R. L. Denny, D. K. Howe, D. Danesh et al., 2004. Soybean bacterial artificial chromosome contigs anchored with RFLPs: insights into genome duplication and gene clustering. Genome 47: 361–372. [DOI] [PubMed] [Google Scholar]
- Mudge, J., S. B. Cannon, P. Kalo, G. E. D. Oldroyd, B. A. Roe et al., 2005. Highly syntenic regions in the genomes of soybean, Medicago truncatula and Arabidopsis thaliana. BMC Plant Biol. 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, N. C., C. D. Dickinson, T. J. Cho, V. H. Thanh, B. J. Scallon et al., 1989. Characterization of glycinin gene family in soybean. Plant Cell 1: 313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover, L., K.-D. Scharf, D. Galiardi, P. Vergne, E. Czarnecka-Vernet et al., 1998. The hsf work: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones 1: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orf, J. H., K. Chase, T. Jarvik, L. M. Mansur, P. B. Cregan et al., 1999. Genetics of soybean agronomic traits: I. comparison of three related recombinant inbred populations. Crop Sci. 39: 1642–1651. [Google Scholar]
- Quackenbush, J., F. Liang, L. Holt, G. Pertea and J. Upton, 2000. The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 28: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., and J. L. Bennetzen, 1998. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82: 37–44. [Google Scholar]
- Schauser, L., W. Wieloch and J. Stougaard, 2005. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60: 229–237. [DOI] [PubMed] [Google Scholar]
- Schlueter, J. A., P. Dixon, C. Granger, D. Grant, L. Clark et al., 2004. Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876. [DOI] [PubMed] [Google Scholar]
- Schlueter, S. D., Q. Dong and V. Brendel, 2003. GeneSeqer@PlantGDB: gene structure predication in plant genomes. Nucleic Acids Res. 31: 3597–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J. L., D. Kurunam, K. Shopinski, M. J. Iqbal, K. Samreen et al., 2006. The soybean genome database (SoyGD): a browser for display of duplicated, polyploid, regions and sequence tagged sites on the integrated physical and genetic maps of Glycine max. Nucleic Acids Res. 34: D758–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, R., K. Polzin, J. Labate, J. Specht, E. C. Brummer et al., 1996. Genome duplication in soybean (Glycine subgenus soja). Genetics 144: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion, C., K. Vandepoele, M. C. E. Van Montagu, M. Zabeau and Y. Van de Peer, 2002. The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99: 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Q. J., L. F. Marek, R. C. Shoemaker, K. G. Lark, V. C. Concibido et al., 2004. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109: 122–128. [DOI] [PubMed] [Google Scholar]
- Tian, C., P. Wan, S. Sun, L. Jiayang and C. Mingsheng, 2004. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54: 519–532. [DOI] [PubMed] [Google Scholar]
- Triwitayakorn, K., V. N. Njiti, M. J. Iqbal, S. Yaegashi, C. Town et al., 2005. Genomic analysis of a region encompassing Qrfs1 and Qrfs2: genes that underlie soybean resistance to sudden death syndrome. Genome 48: 125–138. [DOI] [PubMed] [Google Scholar]
- Vision, T. J., D. G. Brown and S. D. Tanksley, 2000. The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117. [DOI] [PubMed] [Google Scholar]
- Walling, J. G., R. Shoemaker, N. Young, J. Mudge and S. Jackson, 2006. Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 172: 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., G. L. Graef, A. M. Procopiuk and B. W. Diers, 2003. Identification of putative QTL that underlie yield in interspecific soybean backcross populations. Theor. Appl. Genet. 108: 458–467. [DOI] [PubMed] [Google Scholar]
- Wendel, J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]
- Wolfe, K. H., 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2: 333–341. [DOI] [PubMed] [Google Scholar]
- Wu, C., S. Sun, P. Nimmakayala, F. A. Santos, K. Meksem et al., 2004. A BAC- and BIBAC-based physical map of the soybean genome. Genome Res. 14: 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. H., J. Mudge, D. J. Kim, D. Larsen, R. C. Shoemaker et al., 2003. Estimates on conserved microsynteny among the genomes of Glycine max, Medicago truncatula and Arabidopsis thaliana. Theor. Appl. Genet. 106: 1256–1265. [DOI] [PubMed] [Google Scholar]
- Yan, H. H., J. Mudge, D. J. Kim, R. C. Shoemaker, D. R. Cook et al., 2004. Comparative physical mapping reveals features of microsynteny between Glycine max, Medicago truncatula, and Arabidopsis thaliana. Genome 47: 141–155. [DOI] [PubMed] [Google Scholar]
- Yang, Q., K. Reinhard, E. Schiltz and U. Matern, 1997. Characterization and heterologous expression of hycroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol. Biol. 35: 777–789. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 15: 555–556. [DOI] [PubMed] [Google Scholar]
- Young, N. D., J. Mudge and Y. N. Ellis, 2003. Legume genomes: more than peas in a pod. Curr. Opin. Plant Biol. 6: 199–204. [DOI] [PubMed] [Google Scholar]
- Yu, Y. S., T. Rambo, J. Currie, C. Saski, H. R. Kim et al., 2003. In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300: 1566–1569. [DOI] [PubMed] [Google Scholar]
- Yue, P., P. R. Arelli and D. A. Sleper, 2001. Molecular characterization of resistance to Heterodera glycines in soybean PI 438439B. Theor. Appl. Genet. 102: 921–928. [Google Scholar]
- Zhang, J., and T. Peterson, 1999. Genome rearrangements by nonlinear transposons in maize. Genetics 153: 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]