Abstract
A Japanese study reported that up to 16% of breast cancer samples harbor a sporadic mutation within the human Cav-1 gene, namely P132L. To date, however, no studies have examined the United States’ population. Here, we developed a novel allele-specific real-time PCR assay to detect the Cav-1 P132L mutation in mammary tumor cells isolated by laser capture microdissection from formalin-fixed paraffin-embedded breast cancer samples. We report that the Cav-1 P132L mutation is present in ∼19% of estrogen receptor α (ERα)-positive breast cancers but not in ERα-negative breast cancers. This is the first demonstration that the P132L mutation is exclusively associated with ERα-positive mammary tumors. We also identified six novel Cav-1 mutations associated with ERα-positive breast cancers (W128Stop, Y118H, S136R, I141T, Y148H, and Y148S). Thus, the overall incidence of Cav-1 mutations in ERα-positive breast cancers approaches 35% (greater than one-third). To mechanistically dissect the functional relationship between Cav-1 gene inactivation and ERα expression, we isolated primary mammary epithelial cells from wild-type and Cav-1−/− mice and cultured them in a three-dimensional system, allowing them to form mammary acinar-like structures. Under conditions of growth factor deprivation, Cav-1-deficient mammary acini displayed increased ERα levels and enhanced sensitivity toward estrogen-stimulated growth, with specific up-regulation of cyclin D1. Finally, we discuss the possibility that sporadic Cav-1 mutations may act as an initiating event in human breast cancer pathogenesis.
Multiple independent lines of experimental evidence suggest that Cav-1 functions as a mammary gland tumor suppressor gene.1,2 First, Cav-1 mRNA and protein levels are down-regulated in oncogene-transformed NIH 3T3 cells, in many human and mouse breast cancer cell lines, in primary human mammary gland tumors, and in transgenic breast cancer mouse models.3–6 Conversely, Cav-1 re-expression in breast cancer cell lines inhibits anchorage-dependent growth in soft agar and decreases their invasive potential.5,7 Cav-1 expression also reduces the migratory and invasive potential of MTLn3 cells, a metastatic mammary carcinoma line, by preventing epidermal growth factor (EGF)-induced lamellipodia formation and reducing cell migration.8 Forced expression of Cav-1 in the metastatic 4T1.2 mouse mammary carcinoma cell line also inhibits in vivo growth after orthotopic implantation into the mouse mammary gland.9
Interestingly, Cav-1−/− mice display several abnormal mammary-gland-specific phenotypes. For example, Cav-1−/− mammary glands exhibit signs of premalignant lesions, ie, ductal hyperplasia with wall thickening to three to four cell layers.10 Simultaneous loss of Cav-1 and of another tumor suppressor gene, INK4a, further perturbs mammary gland morphology, with increased ductal hyperplasia and lateral branching and the presence of fibrosis.11 Moreover, in the context of a mammary gland tumor-prone mouse model (MMTV-PyMT), genetic ablation of Cav-1 expression accelerates the appearance and growth of dysplastic lesions at the very early stages of mammary gland development, greatly facilitates mammary tumor formation at 14 weeks of age, and augments metastasis to distant sites, such as the lung.12,13
Genetic validation of the idea that Cav-1 functions as a tumor suppressor gene emerged from the observation that the human Cav-1 gene maps to the long arm of chromosome 7, in very close proximity to the D7S522 locus.14 This region includes a known fragile site (FRA7G) and is often associated with loss of heterozygosity in various human cancers, including breast, ovarian, and renal cell carcinomas.15–23 As such, a putative tumor suppressor gene is thought to be located within this chromosomal region. In support of this notion, a Japanese study detected a sporadic mutation in the Cav-1 gene, leading to a proline-to-leucine substitution at position 132 (P132L) in up to 16% of patients with primary breast tumors.24 Recombinant expression of the Cav-1 P132L mutant in NIH 3T3 cells induced cellular transformation, activation of the p42/44 mitogen-activated protein kinase signaling cascade, and promoted cellular invasion.24 Moreover, the Cav-1 P132L mutant was shown to act in a dominant-negative fashion, causing the mislocalization and intracellular retention of wild-type endogenous Cav-1 in a nontransformed human mammary epithelial cell line.10 As such, this heterozygous mutation leads to complete functional inactivation of the Cav-1 protein in the context of mammary epithelial cells. However, it remains unknown whether the Cav-1 P132L mutation or any other Cav-1 mutations are associated with human breast cancers in the United States.
It is believed that estrogen increases the proliferation rate of mammary epithelial cells and, thus, that estrogen exposure increases the risk of developing breast cancer. Estrogen binds to the estrogen receptor (ER), which belongs to a large family of nuclear receptors. ER functions as a transcription factor that, upon estrogen-induced ligand-activation, binds DNA and regulates the expression of estrogen-responsive genes. ERα is the primary mediator of estrogen responses during cell proliferation in the breast, whereas ERβ possesses antiproliferative properties.25 ERα is essential for mammary ductal growth, and ERα knockout (KO) mice lack duct formation. Despite this, in the normal adult mammary gland, ERα is found only in a small percentage (∼10 to 20%) of luminal epithelial cells. Interestingly, normal epithelial cells exhibit mutual exclusion of ERα expression and cell proliferation, as assessed by a lack of double immunostaining of ERα and the Ki-67 proliferation marker.26 However, ERα expression is elevated at the earliest stages of mammary tumorigenesis, such as ductal hyperplasia, and increases even further with increasing atypia.27,28 As such, the inverse correlation between ERα expression and proliferation is lost in some breast cancers, in which a large percentage of proliferating cells become ERα-positive. However, the molecular mechanisms for initiating increased steroid receptor expression in breast cancer cells remain largely unknown.
The aim of the present study was to evaluate the incidence of Cav-1 mutations in human breast cancers within the United States’ population. We found that ∼20% of primary breast cancers carry a Cav-1 mutation. Remarkably, Cav-1 mutations were exclusively found in ERα-positive breast tumors, with a relative incidence of 35%. Importantly, this is the first demonstration that Cav-1 mutations are associated with ERα-positive breast cancers. As such, we propose that Cav-1 loss-of-function may be one of the initiating mechanisms underlying ERα overexpression during early mammary tumorigenesis. To test this hypothesis directly, we reconstituted mammary acini formation in vitro using primary cultures of mammary epithelial cells derived from wild-type (WT) and Cav-1−/− mice. Interestingly, we demonstrate that, when cultured in the absence of a growth factor stimulus, Cav-1-null acini displayed ∼4-fold increased levels of ERα. In addition, in the absence of EGF, estrogen-stimulated Cav-1-deficient acini demonstrated enhanced growth rates and up-regulation of cyclin D1 levels.
Materials and Methods
Materials
Antibodies and their sources were as follows: ERα (H-184 and MC-20) and Cav-1 (N-20) from Santa Cruz Biotechnology (Santa Cruz, CA); E-cadherin was from BD Pharmingen (San Diego, CA); and cyclin D1 (Ab-3) from NeoMarkers (Fremont, CA). Other reagents were as follows: hydrocortisone, cholera toxin, insulin, β-estradiol, and gentamicin (from Sigma, St. Louis, MO), Collagenase type I, phenol red-free Dulbecco’s modified Eagle’s medium-F12 (from Gibco, Grand Island, NY); phenol-free reduced growth factor Matrigel (from Trevigen, Gaithersburg, MD); Lab-Tek II 8-well chamber slides (from Nalgene Nunc, Rochester, NY); and charcoal-stripped horse serum from Bioreclamation, Inc. (Hicksville, NY).
Patients and Tumor Tissue Procurement
All patients included in the study were female, with the histopathological diagnosis of invasive ductal carcinoma of the breast, under an Institutional Review Board-approved protocol at Montefiore Medical Center. No subpopulations were excluded. The clinical and pathological information regarding age at diagnosis, histology, stage according to the fifth version of the American Joint Committee on Cancer, status of the ER, time to first relapse or time to progression, and overall survival were summarized and recorded in a breast cancer database. Of the formalin-fixed, paraffin-embedded tissue blocks from >150 patients examined, analyzable DNA was obtained from only 55 patients who were included in this study. The study materials were coded to protect confidentiality. The tumor areas for microdissection were identified by two expert surgical pathologists using hematoxylin and eosin-stained slides.
Quality of the Genomic DNA
The quality of the resulting genomic DNA was stringently assessed by low percentage agarose gel electrophoresis and by conventional polymerase chain reaction (PCR) using primer set 1 to amplify the sequence of Cav-1. Only the 55 Cav-1 PCR-positive patient samples were selected for further mutational analysis.
Isolation of Breast Tumor Cells by Laser Capture Microdissection (LCM)
Sections (5-μm thickness) from formalin-fixed, paraffin-embedded human breast cancer blocks were placed onto standard glass slides (Fisher Scientific, Pittsburgh, PA), deparaffinized, rehydrated, and stained with hematoxylin and eosin according to standard procedures. A PixCell IIe LCM system (Arcturus, Mountain View, CA) was used to isolate breast cancer cell areas from normal cells and place them onto a thin polymer film (CapSure LCM Caps, Arcturus), using a laser beam of 7.5-μm diameter. About 3,000 to 10,000 laser shots were needed to obtain analyzable DNA from each tissue specimen. As normal controls, normal mammary epithelial cells from the same sample were isolated either by LCM or by macrodissection (if normal tissue was predominant in the sample).
Conventional PCR
After lysis at 55°C overnight, genomic DNA was extracted from LCM-isolated cells using a DNeasy tissue kit (Qiagen, Valencia, CA), according to the manufacturer’s recommendations, and eluted with 20 to 30 μl of distilled water. Genomic DNA (5 to 10 μl) was used for each conventional or allele-specific PCR analysis. For conventional PCR, the forward primer (5′-CCAGCTTCACCACCTTCACT-3′) and reverse primer (5′-CACAGACGGTGTGGACGTAG-3′) were used to amplify a 210-bp DNA fragment corresponding to a 70-amino acid region (amino acids 88 to 156), which includes the entire transmembrane domain (amino acids 102 to 134) of the Cav-1 gene (GenBank accession number: NM001753, See also Table 1). Each PCR reaction was performed in a 50-μl final volume containing ∼20 to 100 ng of genomic DNA, 10 mmol/L Tris-HCl, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 10 mg of gelatin, 10 pmol/L of each primer, 200 μmol/L of each dNTP, and 0.2 U of TaqDNA polymerase (Promega, Madison, WI). PCR was performed in a thermal cycler (model 9600; PerkinElmer-Cetus, Boston, MA) using the following program: denaturation at 95°C for 5 minutes, followed by 35–40 amplification cycles (denaturation at 95°C for 60 seconds, annealing at 56°C for 60 seconds, and extension at 72°C for 60 seconds), and final extension at 72°C for 10 minutes. Both positive and negative controls were performed in parallel for each PCR reaction. The template for the positive control was genomic DNA extracted from the human breast cancer cell line MCF-7. Negative control reactions were performed without DNA template to exclude nonspecific amplification.
Table 1.
Regular and Allele-Specific Primer Sets for Cav-1 WT and CAV-1 (P132L)
| Specificity | Forward primer | Reverse primer | PCR product size (bp) |
|---|---|---|---|
| Membrane spanning domain and flanking sequences | 5′-ccagcttcaccaccttcact-3′ | 5′-cacagacggtgtggacgtag-3′ | 210 (Primer set 1) |
| WT allele-specific (P132) | 5′-cacatctgggcagttgtacc-3′ | 5′-cacagacggtgtggacgtag-3′ | 93 (Primer set 2) |
| Mutant allele-specific (P132L) | 5′-cacatctgggcagttgtact-3′ | 5′-cacagacggtgtggacgtag-3′ | 93 (Primer set 3) |
| Mutant allele-specific (P132L), with degeneracy | 5′-cacatctgggcagttgtrct-3′ | 5′-cacagacggtgtggacgtag-3′ | 93 (Primer set 4) |
Four sets of primers are shown. Primer set 1 was designed to amplify the entire region (amino acids 87 to 156), and to subject genomic DNA to direct sequencing. Note that primers sets 2, 3, and 4 are allele-specific, recognizing the WT or P132L allele, and were employed in real-time PCR screening. The reverse primers are identical in all four sets. Also, primer set 4 was designed with degeneracy, due to a polymorphism at valine 131. Sequence differences are indicated in bold and are underlined. r = a or g degeneracy.
Sequencing
PCR products were separated by electrophoresis on 1.6 to 1.8% agarose gels before visualization via UV light. The PCR products were gel-extracted using a Gel Extraction Kit (Qiagen). To detect Cav-1 mutations, direct automated sequencing of PCR products was performed using the forward PCR primer by the dye-terminator fluorescence sequencing method on a fluorescent sequencer (model 3700; Applied Biosystems, Foster City, CA) at the DNA Sequencing Facility (Albert Einstein College of Medicine). All Cav-1 mutations were confirmed by direct sequencing using the reverse PCR primer. In addition, Cav-1 mutations were confirmed on independently LCM-isolated normal and breast tumor cells from the same tissue block or from different tissue blocks from the same patient, if available.
Allele-Specific Real-Time-PCR Amplification
A strategy to quickly detect the P132L mutation was designed using allele-specific real-time PCR. The template for allele-specific real-time PCR was either genomic DNA (50 to 100 ng) or the 210-bp PCR product from the conventional PCR (1 to 10 ng), described above. The allele-specific primers were designed to distinguish the P132L mutant from its wild-type counterpart. Because of a naturally occurring polymorphism in the third nucleotide of amino acid codon 131, the forward primer was designed with degeneracy (see Table 1). Amplification was performed using the allele-specific forward primer and a common reverse primer (50 to 900 nmol/L), using a SYBR Green master mixture (containing heat-activated AmpliTaq Gold DNA polymerase, dNTPs, buffer, SYBR Green (Applied Biosystems, Foster City, CA), and a reference dye). An ABI PRISM 7900HT (Applied Biosystems) was used for real-time PCR amplification and fluorescence melting curve analysis. Amplification consisted of a 2-minute AmpErase UNG incubation at 50°C, a 10-minute preincubation at 95°C to activate the TaqDNA polymerase, followed by 35 to 45 cycles (denaturation at 95°C for 15 seconds, primer annealing, and extension for 1 minute at 60°C) in 96-well plates. The fluorescence melting curve was analyzed immediately after amplification by measuring the fluorescence intensity of the PCR product from 60 to 95°C at a slope of 2%. The maximum rate of fluorescence change occurred at the Tm of the PCR product. The relative quantification of the target gene was acquired and analyzed using SDS 2.0 software (Applied Biosystems). The size of the expected PCR products was confirmed by agarose gel electrophoresis, and the candidate mutation was validated by direct sequencing.
Primer Specificity
We checked our primer sequences very carefully, and they do not co-amplify other caveolins, such as Cav-2 or Cav-3. The DNA and protein sequences of the caveolins are actually quite divergent. If Cav-2 or Cav-3 sequences were co-amplified, we would have detected them, because they are easily distinguished based on their divergent DNA sequences.
Synonymous Nucleotide Polymorphisms
Several synonymous nucleotide polymorphisms in the Cav-1 gene were identified in our study, eg, the third nucleotide of P132P (CCA → CCA/G) and S136S (AGC → AGT/C). We discussed the chromatogram sequencing results with the Director of the Sequencing Facility at our institution: Although the A and G of the P132 were not completely lined up, its location and surrounding nucleotide sequences exclude the possibility of an insertion, and it thus should be considered as a polymorphism. This was further supported by its absence in the normal tissues from the same archival tissue blocks and in the tumors that did not have P132L mutations. All of the caveolin-1 mutations and polymorphisms found in the genomic DNA of tumor cells were not detected in the genomic DNA of corresponding normal cells. Interestingly, Lièvre et al29 also observed the presence of three synonymous polymorphisms in tumor tissue DNA but not in matched normal tissue DNA. We do not understand the biological significance of these synonymous polymorphisms in the tumor cells. The results were reproducible by repeated PCR and sequencing analyses using genomic DNA isolated from different LCM isolations of the same tissue block or using a different tumor block whenever it was available.
Statistical Analysis
The P values for age, stage, and time to first relapse were calculated using the paired or unpaired Student’s t-test. Frequency comparisons were analyzed using Fisher’s exact test. The 95% confidence interval was calculated using the relevant 2 × 2 contingency tables. Differences with P < 0.05 were considered statistically significant.
Immunohistochemistry
Sections (5-μm thickness) from archived paraffin-embedded human breast tissues were deparaffinized, rehydrated, and quenched with 1.5% H2O2. For ER staining, slides were treated with DakoCytomation Target Retrieval Solution (Dako, Carpinteria, CA) in a steam bath at 95°C for 45 minutes. After equilibration in phosphate-buffered saline for 15 minutes, slides were placed in an autostainer apparatus (Dako) and stained with antibodies to ERα (1:50 dilution; monoclonal antibody clone 1D5; Dako). Immunoreactivity was detected using the Dako EnVision method, according to the manufacturer’s recommended procedures.
It is important to note that the antibody used initially for the clinical screening of ERα positivity (by the Department of Pathology) was a mouse monoclonal. However, all further immunohistochemistry experiments performed in the Lisanti laboratory used rabbit polyclonal antibodies directed against ERα (H-184 and MC-20, from Santa Cruz Biotechnology). This approach provided independent validation of the ER positivity of a given clinical sample. Similarly, sections were also immunostained with a rabbit polyclonal antibody directed against cyclin D1 (Ab-3, from NeoMarkers).
For negative controls, slides were subjected to the same procedures, including antigen retrieval, except for 1) omitting the primary antibody or 2) treating samples with nonimmune rabbit IgG. Both of these critical negative controls clearly demonstrated the specificity of the immunostaining that we observed.
Animal Studies
All animals were housed and maintained in a pathogen-free environment/barrier facility at the Institute for Animal Studies at the Albert Einstein College of Medicine under National Institute of Health guidelines. Mice were kept on a 12-hour light/dark cycle with ad libitum access to chow and water. Cav-1 KO mice were generated as previously described.30 All WT and Cav-1 KO mice used in this study were in the FVB/N genetic background.12,13
Isolation of Mammary Epithelial Cells
Primary mammary gland organoids (ie, freshly isolated intact acini) were isolated from 2-month-old virgin mice, as previously described,31 with minor modifications. The fourth and fifth mammary glands were removed aseptically, minced with a surgical razor blade, incubated with agitation (for 2 to 3 hours at 37°C) in 30 to 35 ml of Growth Media (Dulbecco’s modified Eagle’s medium/F12, 5% horse serum, 20 ng/ml EGF, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, Pen/Strep), containing 2 mg/ml collagenase type I, and 50 μg/ml gentamicin. Then, cell suspensions were spun 10 minutes at 1000 rpm to eliminate the floating fat cells. Cell pellets were resuspended in 10 ml of Assay Media (Dulbecco’s modified Eagle’s medium/F12, 2% horse serum, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, Pen/Strep). To separate single cells (mainly fibroblasts, smooth muscle cells, and endothelia) from mammary organoids, the pellets were subjected to repeated washes by differential centrifugation (spun at 1000 rpm for 45 s, repeated 10 times). After the last wash, organoids were resuspended in 2 ml of Growth Media and disrupted by pipetting up and down 20 to 25 times with a 1-ml blue tip. After plating in 10-cm plastic dishes, organoids attached and spread as a monolayer of mammary epithelial cells.
Three-Dimensional (3-D) Cultures of Mammary Epithelial Cells
Four to five days after organoid purification, mammary epithelial cell monolayers formed and were trypsinized and resuspended in Assay Media. To obtain a single cell suspension, cells were passed 20 to 25 times through a 1-ml blue tip. Single cells were overlaid onto Matrigel, as previously described.32 Briefly, 5000 cells were diluted into Assay Media supplemented with 2% Matrigel and overlaid in 8-well chamber slides, which were precoated with 40 μl of Matrigel. Chambers were then incubated at 37°C. Acini were cultured either in the absence or presence of EGF (5 ng/ml) or in the absence or presence of estrogen (10 nmol/L). Acini were re-fed with Assay Media supplemented with 2% Matrigel and the appropriate growth factor combination every 4 days. To eliminate the well known estrogenic effect of the pH indicator phenol red, we used phenol red-free media, phenol red-free Matrigel, and charcoal dextran-stripped horse serum. All experiments were performed with primary mammary epithelial cells that were passage 1.
Quantitation of Mammary Acinar Growth
Acinar growth was monitored by imaging day 16 WT and Cav-1 KO acini using an Olympus 1 × 80 microscope with a 10× objective connected with a cooled charge-coupled device camera. Diameters were measured at the middle optical section of each acinus, with the support of Image J software. Greater than 50 acini were scored for each condition and for each genotype.
Western Blot Analysis
Acini lysates were prepared as previously described,32 with minor modifications. Acini were incubated on ice with Lysis Buffer (20 mmol/L Tris-HCl, pH 7.5, 70 mmol/L NaCl, 0.1% SDS, 1% sodium deoxycholate, 1% TritonX-100, 60 mmol/L octyl glucoside, 50 mmol/L NaF, 30 mmol/L sodium pyrophosphate, 10 μmol/L orthovanadate, with a protease inhibitor tablet) for 15 minutes. Lysates were collected in an Eppendorf tube, passed five times through a 26 1/2-gauge needle, and incubated on ice for an additional 15 minutes. Lysates (50 μl) were loaded onto SDS-PAGE gels and transferred to nitrocellulose membranes (Schleicher and Schuell, 0.2 μm). Blots were blocked for 1 hour in TBST (10 mmol/L Tris-HCl, pH 8.0, 150 mmol/L NaCl, 0.2% Tween 20) containing 1% bovine serum albumin and 4% nonfat dry milk (Carnation, Nestlé, Switzerland). Then, membranes were incubated for 2 hours with primary antibodies in a TBST/1% bovine serum albumin solution. Membranes were then washed with TBST and incubated for 45 minutes with the appropriate horseradish peroxidase-conjugated secondary antibodies (diluted 5000-fold in TBST/1% bovine serum albumin; Pierce, Rockford, IL). Signals were detected with an ECL kit (Pierce). Equal loading was assessed by immunoblotting with an epithelial marker, ie, E-cadherin.
Results
LCM and Allele-Specific Real-Time PCR Screening of Human Breast Cancer Tumor Samples
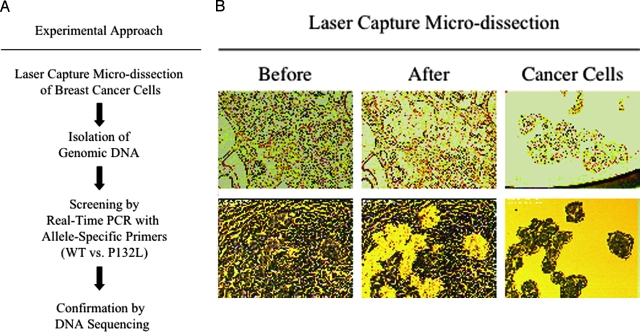
We first established a rapid and sensitive strategy to detect Cav-1 P132L mutations in human breast cancer samples. Interestingly, the Cav-1 P132L mutation has been shown to act in a dominant-negative fashion and to cause complete loss-of-function of the Cav-1 protein. An outline of the experimental approach we used is summarized in Figure 1A. Briefly, to specifically isolate malignant cells from normal cells within mammary gland tissue, LCM was performed on paraffin-embedded mammary tumor sections. Then, genomic DNA was isolated from breast cancer cells and screened by real-time PCR with allele-specific primers. Using this approach, we were able to rapidly and efficiently identify the presence of the Cav-1 P132L mutant allele. Cav-1 P132L mutations were confirmed by direct sequencing (see Materials and Methods).
Figure 1.
Experimental approaches. A: Schematic flow diagram of the strategies used to identify Cav-1 mutations in human breast cancer samples. Briefly, LCM was performed on paraffin-embedded sections derived from archived human breast cancer samples. Then, genomic DNA was purified from isolated breast cancer cells and screened by allele-specific real-time PCR to discriminate between the two different Cav-1 alleles (WT vs. P132L). The presence of Cav-1 mutations was also independently evaluated by direct sequencing. B: Using LCM, human breast cancer cells were isolated from paraffin-embedded breast cancer sections. Two examples are included (upper and lower panels, respectively). Left panels represent sections before LCM. Note that, after LCM, normal cells remains on the slides (middle panels), whereas tumor cells are separated from the rest of the tissue and isolated on a separate polymer film (right panels).
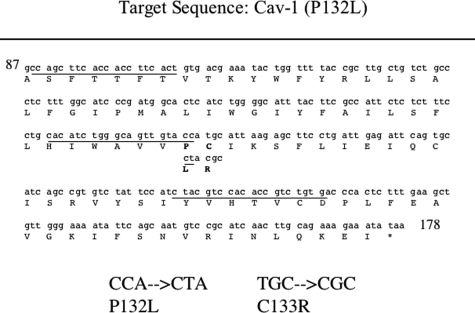
A total of 55 paraffin-embedded breast cancer samples were retrieved from the archive of the Department of Pathology at the Montefiore Medical Center and subjected to LCM to isolate tumor cells from the surrounding normal tissue and stromal cells. Figure 1B shows two examples of breast cancer sections before and after LCM. The left panels represent sections before the procedure. It consists of a mixture of breast cancer and normal cells. The middle panels represent sections depleted of tumor cells, which were isolated and collected on a thin polymer film (right panels). Using this powerful procedure, we were able to extract genomic DNA only from tumor cells, clearly enhancing the ability of detecting cancer-specific gene mutations. Figure 2 shows the sequence within the Cav-1 gene, which was the target of our allele-specific real-time PCR screening. The P132L mutation resides within the Cav-1 central hydrophobic region, a domain that is thought to be critical for both membrane association and protein-protein interactions. Underlined are the sequences of the primers used both for allele-specific real-time PCR, to rapidly discriminate the WT and the P132L alleles, and for conventional PCR, to subject the entire target region to direct sequencing.
Figure 2.
Cav-1 mutations: analysis of human breast cancer samples. The Cav-1 target sequence for PCR amplification from genomic DNA is shown. Specific primer-targeted sequences are underlined (see also Table 1 for details). The positions of the P132L (CCA → CTA) and C133R (TGC → CGC) mutations are shown in bold. For conventional PCR, the forward primer (5′-CCAGCTTCACCACCTTCACT-3′) and reverse primer (5′-CACAGACGGTGTGGACGTAG-3′) were used to amplify a 210-bp DNA fragment corresponding to a 70-amino acid region (amino acids 88–156), which includes the entire transmembrane domain (amino acids 102–134) of the Cav-1 gene.
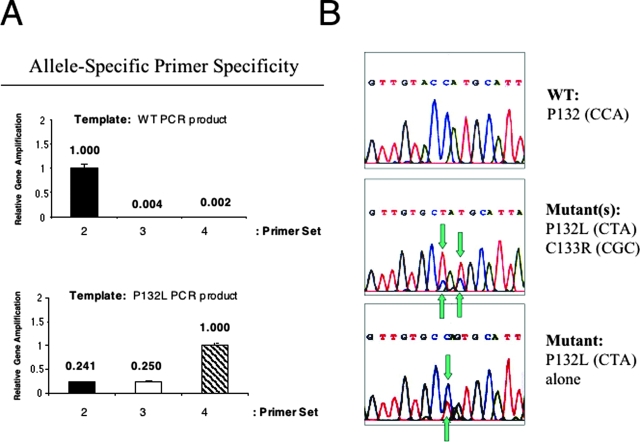
The detailed primer sequences are listed in Table 1. The reverse primer is in common for all of the different primer combinations, whereas the forward primer varies and is specifically tailored to the goal of the amplification. Primer set 1 amplifies the entire region and was used for conventional PCR and direct sequencing. Primer sets 2, 3, and 4 were designed for allele-specific real-time PCR, as they selectively recognize either the WT allele (primer set 2) or the P132L allele (primer sets 3 and 4). Because of a polymorphism at valine 131, the forward primer of set 4 was designed with degeneracy. To test our screening strategy, we first examined the specificity of our allele-specific primer sets 2, 3, and 4 in distinguishing a given allele. To this end, real-time PCR was performed using as templates PCR products of known genetic sequence, ie, Cav-1 WT and Cav-1 P132L PCR products. Figure 3A shows that primer set 2 preferentially recognizes the WT template, whereas primer sets 3 and 4 are unable to identify and amplify the same WT template. On the contrary, primer set 4 displays the highest specificity toward the P132L allele. Because of a polymorphism at position 131, primer set 3 was able to recognize P132L, but not efficiently (25%), and unable to recognize WT132 (0%), and as a consequence it was no longer used throughout the study.
Figure 3.
Demonstration of allele-specific primer specificity and examples of Cav-1 mutations identified (P132L and C133R). A: Evaluating the specificity of allele-specific real-time PCR screening methodology, using PCR products of known genetic sequence as the template, ie a WT PCR template (upper panel) or a P132L PCR template (lower panel). Note that primer set 2 preferentially recognizes the WT template, whereas primer set 4 (with the degenerate forward primer) only recognizes the P132L template. Because of a polymorphism at valine 131, primer set 3 did not efficiently recognize either the WT or P132L template. As such, it was not further used. B: Examples of mutations identified using primer set 1 that amplifies the entire Cav-1 gene target region (amino acid 87–156). Upper, wild-type allele P132 (CCA); middle, double mutant example, P132L (CCA→ CTA) and C133R (TGC→ CGC); lower, single mutant example, P132L (CCA→ CTA). Note that in the middle panel, the P132L allele predominates (P132L > WT). Mutations are indicated by the use of blue-green arrows. [Also, several synonymous nucleotide polymorphisms in the Cav-1 gene were identified in our study, eg, the third nucleotide of P132P (CCA → CCA/G) and S136S (AGC→AGT/C). We discussed the chromatogram sequencing results with the Director of the Sequencing Facility at our institution: although the A and G of the P132 were not completely lined up, its location and surrounding nucleotide sequences exclude the possibility of an insertion, and it thus should be considered as a polymorphism.]
The Cav-1 (P132L) Mutation Is Detected Only in ERα-Positive Breast Cancer Samples
Having established a rapid and selective real-time PCR assay to distinguish between Cav-1 WT and Cav-1 P132L alleles, we proceeded to screen the collection of 55 genomic DNA samples extracted from LCM-isolated breast cancer cells. In all of the positive cases, the presence of the Cav-1 P132L mutation was confirmed by sequencing. Figure 3B shows several sequence examples. The upper panel is representative of a normal WT sequence. In the middle panel, note the concomitant presence of two Cav-1 mutations, ie, P132L and C133R, and the prevalence of the P132L allele over the WT allele. However, we were unable to establish whether the two mutations were allelic, because we did not have access to more samples for RNA extraction. The bottom panel shows an allele harboring only a single mutation at position 132, ie, P132L allele. Interestingly, all of the mutations we detected were “heterozygous,” consistent with the hypothesis that the Cav-1 P132L mutation behaves in a dominant-negative fashion.
The Cav-1 P132L mutation was present in 6 of the 55 patients examined. In the course of direct sequencing, we were able to detect six other novel Cav-1 mutations (missense and premature stop) in the same patient cohort, occurring both in combination with the Cav-1 P132L mutation or in P132L-negative patients (W128Stop, Y118H, S136R, I141T, Y148H, and Y148S). In total, 11 of 55 patients harbored a mutation in the Cav-1 gene. As such, we conclude that the overall incidence of Cav-1 mutations in our limited patient sample is 20%; of course, based on our current findings, future studies with a larger patient population should be performed.
Table 2 shows a summary of patient age, AJCC stage, and time to first relapse of the 55 patients examined. To better delineate a clinical and prognostic picture, ERα status was assessed by immunohistochemistry on mammary tumor sections from all patients. Note that 32 cases were ERα-positive, and 23 cases were ERα-negative. Table 2 also summarizes the number of patients who were positive for Cav-1 mutations. Interestingly, Cav-1 mutations cosegregated only with ERα-positive status, with a total incidence of 34.4% (11 of 32) in ERα-positive breast cancer samples. More specifically, the incidence of the Cav-1 P132L mutation and of other Cav-1 mutations in ERα-positive breast cancer samples was 18.8% (6 of 32) and 15.6% (5 of 32), respectively. Importantly, this is the first demonstration that Cav-1 mutations are associated with ERα-positive breast cancers.
Table 2.
Cav-1 Mutation Incidence in ER (+) and ER (−) Patients
Summary of age, stage, and time to first relapse of the 55 patients examined. Note that 32 cases were ERα-positive (+) and 23 cases were ERα-negative (−). Interestingly, Cav-1 mutations (P132L and others) were identified only in ERα (+) patients.
| Characteristics | ER (+) | ER (−) | Significance | |
|---|---|---|---|---|
| Patients (n) | 32 | 23 | ||
| Age (yrs) | Mean | 58.2 | 54.0 | P ≤ 0.24* |
| Median | 57.7 | 53.0 | ||
| Range | 31.1–83.7 | 38.1–70.8 | ||
| AJCC stage† | 1 | 4 | 4 | P ≤ 0.139* |
| 2A | 14 | 11 | ||
| 2B | 4 | 2 | ||
| 3A | 3 | 2 | ||
| 3B | 3 | 0 | ||
| 4 | 2 | 4 | ||
| Unknown | 1 | 0 | ||
| Time to first relapse (months) | Median | 30.1 | 29.0 | P ≤ 0.4* |
| Range | 3.2–60.1 | 8.8–51.4 | ||
| Cav-1 (P132L) status | # Pos. (%) | 6 (18.8%) | 0 (0%) | P ≤ 0.03‡§ |
| Other Cav-1 mutations | # Pos. (%) | 5 (15.6%) | 0 (0%) | P ≤ 0.05‡§ |
| Total | # Pos. (%) | 11 (34.4%) | 0 (0%) | P ≤ 0.001‡§ |
Student’s t-test.
AJCC, American Joint Committee on Cancer Staging.
Fisher’s exact test.
Indicates statistical significance.
The characteristics of Cav-1 P132L-positive patients are reported in Table 3. Remarkably, in the two patients who presented with distant recurrence (metastasis), the Cav-1 P132L mutated allele was prevalent over the WT allele, and a second C133R mutation was also detected. In addition, in one of these two cases, a third Cav-1 mutation was also detected. Table 4 shows the characteristics of ERα-positive patients harboring Cav-1 mutations other than P132L. Interestingly, the novel Cav-1 mutations (missense and premature stop) were detected independently of the recurrence type. Of the eight mutations identified, only P132L and C133R were previously described in human breast cancers.24 Also, the Cav-1 (I141T) mutation that we identify here is analogous to a I141F mutation previously identified in human squamous cell carcinoma samples.33
Table 3.
Characteristics of Cav-1 (P132L) (+) Patients
Summary of age, stage, and time to first relapse of the 55 patients examined. Note that 32 cases were ERα-positive (+) and 23 cases were ERα-negative (−). Interestingly, Cav-1 mutations (P132L and others) were identified only in ERα (+) patients.
| Patient | Age (yrs)* | AJCC stage | Recurrence type | P132L | C133R | Other mutations |
|---|---|---|---|---|---|---|
| 1 | 34.4 | 1 | Local | WT >P132L | None | W128Stop (TGG → TAG) |
| 2 | 63.2 | 2A | Unknown | WT >P132L | None | |
| 3 | 67.9 | 2A | Distant | P132L >WT | WT >C133R | Y148H (TAT → CAT) |
| 4 | 56.2 | 2A | Regional | WT >P132L | None | |
| 5 | 46.6 | 3A | Local & Distant | WT >P132L | None | |
| 6 | 73.0 | 3B | Distant | P132L >WT | WT >C133R | |
| Mean | 56.9 | |||||
| Median | 59.7 | |||||
| Range | 34–73 |
Age at diagnosis.
TAble 4.
Characteristics of Other ER (+) Patients Harboring Cav-1 Mutations
| Patient | Age (yrs)* | AJCC stage | Recurrence type | Cav-1 mutation(s) |
|---|---|---|---|---|
| 7 | 77.6 | 2A | No relapse | W128Stop (TGG → TAG) |
| 8 | 64.5 | 1 | Local | Y148S (TAT → TCT); S136R (AGC → CGC) |
| 9 | 31.1 | 3A | Regional | Y148S (TAT → TCT); S136R (AGC → CGC) |
| 10 | 80.4 | 3B | Distant | I141T (ATT → ACT) |
| 11 | 61.7 | 2B | Distant | Y118H (TAC → CAC) |
Age at diagnosis.
The occurrence of Cav-1 mutations in ERα-positive breast cancer samples may also be of clinical relevance as a positive predictor of relapse. We noticed that 9 of 11 ERα-positive patients harboring Cav-1 mutations underwent recurrences (local, regional, or distant; ∼81.8%; see Tables 3 and 4). In striking contrast, only 10 of 21 ERα-positive patients lacking detectable Cav-1 mutations showed recurrences (∼47.6%). Thus, the detection of Cav-1 mutations in ERα-positive human breast cancer samples may have certain predictive prognostic value. As such, further prognostic studies with increased numbers of patient samples are clearly warranted.
Multiple Cav-1 mutations were often detected within the same tumor sample (Tables 3 and 4). We believe that the different Cav-1 mutations are occurring singly, within different Cav-1 alleles. To determine whether all of the mutations we identified have dominant-negative activity is beyond the scope of the current study, but it will be interesting to assess their phenotypic behavior in future studies. Importantly, no sequence changes in Cav-1 were observed in cells isolated from adjacent normal tissue. Cav-1 sequence changes were exclusively associated with LCM-isolated breast cancer cells; they were not observed in mammary epithelial cells isolated from adjacent normal breast tissue.
Insights into the pathogenic nature of these breast cancer-related mutations arise from the observation that all these mutations reside within critical Cav-1 domains. Figure 4 shows the complete Cav-1 protein sequence and highlights in red the putative “membrane-spanning” domain (residues 102–134). However, this hydrophobic domain may also function as a protein-protein interaction domain, because it has WW domain-like properties.34 Interestingly, 5 of the 7 mutated residues cluster in a 14-amino acid region near the C-terminal end of the membrane-spanning domain (see boxed residues), clearly suggesting that this domain plays an important in vivo role in Cav-1 functioning. In addition, 3 of 7 mutated amino acids, ie, Y118, W128, and P132, are critical residues of the Cav-1 WW-like domain.34 The WW domain is a small protein-protein interaction domain that is widely distributed among structural, regulatory, and signaling proteins.35,36 Previous in vitro experiments have shown that both Cav-1 and Cav-3 contain a WW-like domain and that residues W128 and P132 in the Cav-1 protein (analogous to W101 and P104 within the Cav-3 sequence) constitute the central core of the WW-like domain.34 In addition, Cav-1 Y118 is believed to be essential for the correct folding of the WW domain.
Figure 4.
Summary of the eight Cav-1 mutations identified in ERα-positive breast cancer samples. The full-length sequence (178 amino acids) of human Cav-1 is shown. The membrane-spanning domain (33 amino acids, residues 102–134) is highlighted in red. Mutations are indicated in bold. Note that 5 of the 7 mutated residues identified cluster within a 14-amino acid region (boxed). This “hot spot” is located near the C-terminal end of the membrane-spanning domain.
Recently, concern has been raised regarding the use of genomic DNA derived from formalin-fixed, paraffin-embedded samples for the detection of mutations, due to the putative DNA damage-induced effects of formalin.29 However, we did not detect any Cav-1 mutations in our ER-negative patient samples. Thus, this serves as an important internal control and directly supports the validity of the methods that we used. Also, although possible, we could not imagine formalin having selective DNA damage effects in tumor cells, but not in non-tumor cells, in the same paraffin tissue blocks.
Up-Regulation of ERα Expression in Cav-1 KO Luminal Mammary Epithelial Cells in Vivo
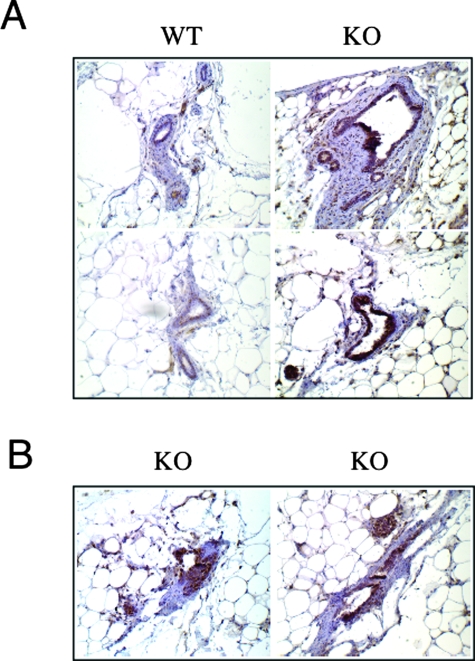
To experimentally mimic Cav-1 functional gene inactivation in the mammary gland, we studied the in vivo behavior of mammary epithelial cells from WT and Cav-1−/− mice. Based on our findings that Cav-1 mutations cosegregate with ERα-positive status in humans, we would predict that ERα levels are up-regulated in Cav-1−/− mouse mammary epithelial cells. To test this hypothesis directly, inguinal mammary glands (4 and 5) were surgically excised from 3-week-old female WT and Cav-1 KO mice. Paraffin-embedded sections were then prepared and immunostained with anti-ERα IgG. Interestingly, Figure 5 (A and B) shows that ERα expression levels were dramatically up-regulated in Cav-1 KO luminal epithelial cells, as predicted. Sections from WT mammary glands are shown for comparison. In addition, note that even at 3 weeks of age, Cav-1-null mice show significant mammary epithelial cell hyperplasia. See also Supplemental Figure 1 at http://ajp.amjpathol.org for lower power images.
Figure 5.
Up-regulation of ERα expression in Cav-1 KO mammary glands in vivo. Inguinal mammary glands (4 and 5) were surgically excised from 3-week-old female WT and Cav-1 KO mice. Formalin-fixed, paraffin-embedded sections were then prepared and immunostained with anti-ERα rabbit IgG (H-184 from Santa Cruz Biotechnology). Bound primary antibodies were detected with a horseradish peroxidase-conjugated secondary antibody (see brown reaction product). Slides were then counterstained with hematoxylin (blue color). Original magnification, 20×. A: Even at 3 weeks of age, Cav-1 KO mice show significant mammary epithelial cell hyperplasia. Note that ERα expression levels are dramatically up-regulated in Cav-1 KO luminal epithelial cells. Sections from WT mammary glands are shown for comparison. B: Two additional examples of ERα immunostaining in Cav-1 KO mammary tissue are shown.
3-D Cultures of Cav-1-Deficient Mammary Epithelial Cells Up-Regulate ERα Expression but Only in the Absence of EGF
Next, we used 3-D cultures of primary mammary epithelial cells derived from WT and Cav-1-null mice. Culturing mammary epithelial cells in a reconstituted basement membrane (Matrigel) recapitulates many features of the mammary epithelium in vivo, including the formation of hollow acini-like spheres, surrounded by a single layer of polarized mammary epithelial cells and the basal deposition of basement membrane components. We have previously shown that Cav-1-deficient mammary epithelial cells retain the ability to form acini-like structures but exhibit several features of abnormal development.37 For example, Cav-1-null acini are larger in size, with immature lumen formation, and undergo EGF-independent growth with hyperactivation of the p42/44 mitogen-activated protein kinase signaling cascade.37
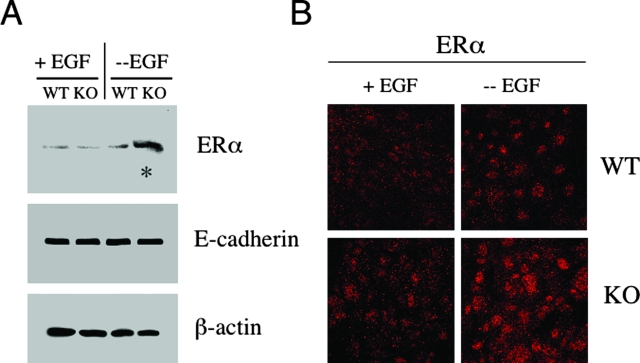
To evaluate the relationship between Cav-1 gene inactivation and increased ERα levels, we first assessed ERα expression levels in WT and Cav-1-deficient 3-D epithelial structures. All experiments were performed with primary mammary epithelial cells at passage 1. Lysates from day 16 WT and Cav-1-null acini were subjected to Western blot analysis using an ERα-specific antibody probe. Interestingly, Figure 6A reveals that Cav-1 null acini up-regulate ERα levels ∼4-fold but only in the absence of EGF. In contrast, ERα expression levels were unchanged in EGF-stimulated Cav-1-null acini, as compared to their WT counterparts.
Figure 6.
Cav-1-deficient mammary epithelial cells up-regulate the ERα under conditions of growth factor deprivation. A: Western blot analysis. WT and Cav-1-deficient mammary epithelial cells were isolated and cultured in Matrigel in the presence or absence of EGF. Under these conditions, mammary epithelial cells are able to form 3-D structures that closely resemble mammary acini. Lysates were then prepared from day 16 acini and subjected to Western blot analysis with anti-ERα specific antibodies (MC-20; Santa Cruz Biotechnology). Note that, under conditions of growth factor deprivation (in the absence of EGF), Cav-1 KO acini display a substantial increase in ERα expression levels (see asterisk). Equal loading was assessed by immunoblotting with an E-cadherin antibody. B: Immunofluorescence. To evaluate ERα localization and expression, WT and Cav-1 KO mammary epithelial cells were grown on glass coverslips for 6 days and cultured in either the absence or presence of EGF overnight. Then, cells were fixed and subjected to immunofluorescence analysis with an antibody directed against ERα. Note that, in the absence of EGF, Cav-1 KO mammary epithelial cells exhibit increased ERα expression levels, as compared to their WT counterparts. We also observed intense ERα nuclear staining in Cav-1-deficient mammary epithelial cells, consistent with ERα receptor activation.
To further evaluate ERα expression and localization, WT and Cav-1 KO mammary epithelial cells were grown on glass coverslips for 6 days and cultured in either the absence or presence of EGF overnight. Cells were then fixed and subjected to immunofluorescence analysis with an antibody directed against ERα. Figure 6B shows that, in the presence of EGF, WT and Cav-1 KO mammary epithelial cells displayed similar low levels of ERα expression. In contrast, when grown in the absence of EGF, Cav-1 KO mammary epithelial cells exhibited increased ERα expression levels, as compared to their WT counterparts. Note the intense ERα nuclear staining in Cav-1-deficient mammary epithelial cells (Figure 6B, lower right panel), consistent with ERα activation.
Estrogen Stimulation Greatly Promotes the Growth of Cav-1-Null Acini in the Absence of EGF
Elevated ERα expression correlates with increased sensitivity to estrogen, which functions as a mitogenic signal in mammary epithelial cells. Next, we assessed whether loss of Cav-1 affects estrogen sensitivity, leading to the enhanced growth of 3-D mammary epithelial structures. To this end, WT and Cav-1-null acini were stimulated with EGF and estrogen, either each alone or in combination, or left untreated. After 16 days of treatment, we scored acinar diameter, as a measure of their growth rate.
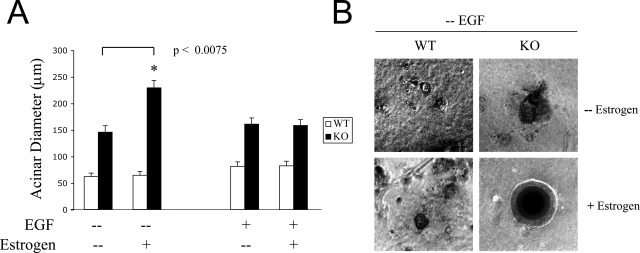
Figure 7A shows a graphic representation of acini growth. Note that Cav-1-deficient acini are larger than WT acini, independent of the type of stimulation. Interestingly, estrogen stimulation does not promote the growth of WT and Cav-1-deficient acini when cultured in the presence of EGF. These data are consistent with Figure 5, showing equal ERα expression levels in EGF-stimulated WT and Cav-1-deficient acini. However, estrogen stimulation greatly promotes the growth of Cav-1-null acini, but not of WT acini, cultured in the absence of EGF. Remarkably, under these conditions (estrogen stimulation in the absence of EGF), Cav-1-null acini grow even larger than when they are estrogen-stimulated in the presence of EGF. These data are consistent with Figure 5, showing that Cav-1-null acini up-regulate ERα expression in the absence of EGF.
Figure 7.
Estrogen stimulation greatly promotes the growth of Cav-1-deficient acini but only during growth factor deprivation. WT and Cav-1 KO mammary epithelial cells were isolated, overlaid onto Matrigel, and allowed to form 3-D acini-like structures. Acini were cultured in the presence or absence of EGF and were either stimulated with estrogen or left untreated. Of course, all estrogen stimulation experiments were performed using phenol red-free media and charcoal-stripped serum (see Materials and Methods). A: Growth curve. Acinar growth was monitored by measuring the diameters of day 16 WT and Cav-1 KO acini. At least 50 epithelial structures were scored for each condition and for each genotype. Note that Cav-1-deficient mammary epithelial cells develop larger acini than WT cells, in the presence or absence of EGF. Remarkably, estrogen treatment greatly stimulates the growth of Cav-1 KO acini in the absence, but not in the presence of EGF, suggesting that a lack of EGF increases the estrogen sensitivity of Cav-1-null acini. Importantly, estrogen treatment did not exert any effects on WT acini. *P < 0.0075. B: Phase contrast images. Micrographs of EGF-deprived WT and Cav-1 KO acini, stimulated with estrogen or left untreated, are shown. Note that, in the absence of EGF, estrogen stimulation does not promote the growth of WT acini. In contrast, EGF-depleted Cav-1-deficient acini become much larger in response to estrogen stimulation.
Figure 7B shows representative phase images of EGF-deprived WT and Cav-1-deficient acini, untreated and after estrogen stimulation. Note that EGF-deprived WT structures are similar in size, in the presence or absence of estrogen treatment. On the contrary, estrogen stimulation greatly promoted the growth of EGF-depleted Cav-1-null acini. These results suggest that growth factor deprivation engages Cav-1-deficient mammary epithelial cells in a pro-proliferative pathway, which is further enhanced by estrogen stimulation.
Up-Regulation of Cyclin D1 in EGF-Deprived Cav-1-Deficient Acini after Estrogen Stimulation
Several studies have demonstrated increased cyclin D1 levels during mammary tumorigenesis.38 For example, cyclin D1 is amplified in >20% of mammary carcinomas, and the cyclin D1 protein is overexpressed in ∼50% of human breast cancer samples. Importantly, in human breast cancers, cyclin D1 overexpression strongly correlates with positive ERα expression.39,40 In vitro and in vivo studies have shown that cyclin D1 levels are linked to the steroid-induced proliferation of mammary epithelial cells, suggesting that cyclin D1 acts as a downstream mediator of estrogen action. For example, when ERα-positive cells are deprived of estrogen, the levels of cyclin D1 are reduced. Conversely, estrogen treatment increases cyclin D1 mRNA and protein.41
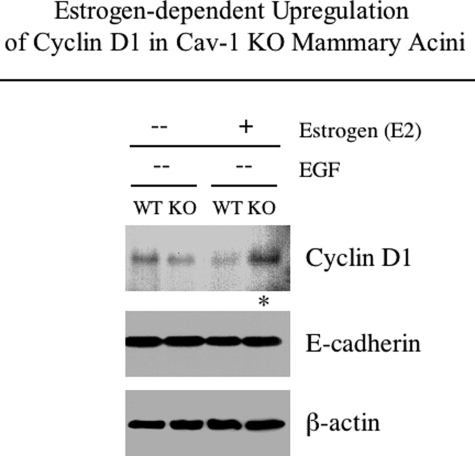
We next attempted to gain insight into the mechanisms downstream of the estrogen-induced growth of EGF-depleted Cav-1-null acini. Lysates from unstimulated or estrogen-stimulated WT and Cav-1 KO acini cultured in the absence of EGF were subjected to Western blot analysis with antibodies directed against cyclin D1. Interestingly, Figure 8 reveals that estrogen stimulation augments cyclin D1 levels in Cav-1-deficient, but not WT, acini cultured in the absence of EGF. No significant differences in cyclin D1 levels were observed in unstimulated WT and Cav-1 KO acini.
Figure 8.
Estrogen-stimulated Cav-1 KO acini up-regulate cyclin D1 levels in the absence of EGF. EGF-deprived WT and Cav-1 KO acini were either estrogen-stimulated starting from day 0 or left untreated. After 16 days, acinar lysates were prepared and subjected to Western blot analysis with anti-cyclin D1 antibodies. Note that estrogen stimulation increases cyclin D1 expression levels in Cav-1-deficient 3-D epithelial structures cultured in the absence of EGF (see asterisk). Equal loading was assessed by immunoblotting with an E-cadherin antibody.
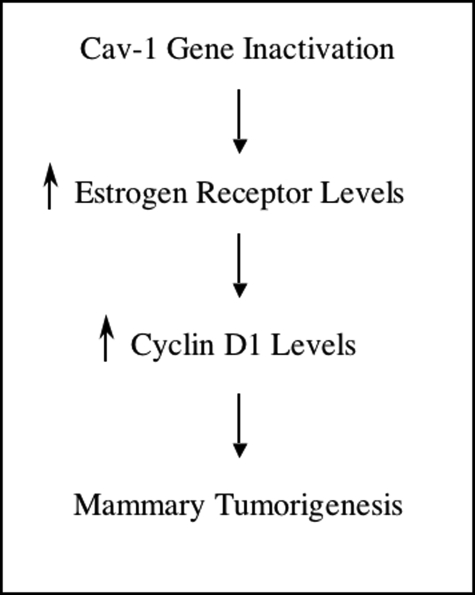
These results reveal a novel pathway leading toward the development of mammary tumors, whereby Cav-1 deficiency drives the up-regulation of both ERα and cyclin D1 levels in mammary epithelial acini (Figure 9). Because both ERα and cyclin D1 play pivotal roles in the development and progression of human breast cancers, understanding their dynamic regulation by Cav-1 gene inactivation may be clinically significant.
Figure 9.
Summary: Cav-1 functional inactivation in mammary tumorigenesis. Cav-1 inactivation (either by loss-of-function mutations or by genetic ablation) in mammary epithelial cells induces increases in ER levels, correlating with enhanced estrogen sensitivity. Up-regulation of ERα promotes the growth of Cav-1-deficient acini and increases cyclin D1 levels. Thus, our results suggest a novel pathway leading toward mammary tumorigenesis.
Up-Regulation of Cyclin D1 in ERα-Positive Breast Cancer Patient Samples
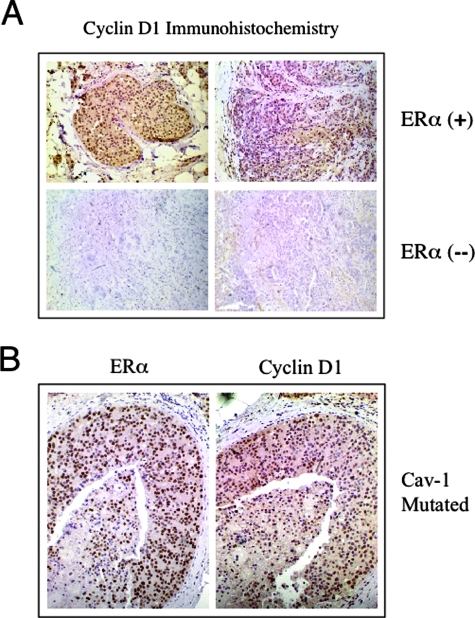
It is previously well documented that ERα-positive breast cancer samples show up-regulation of cyclin D1 gene expression; conversely, ERα-negative breast cancers show little or no cyclin D1 expression (reviewed in Ref. 38). Thus, we next examined cyclin D1 expression in our clinical samples containing Cav-1 mutations by immunostaining. Briefly, formalin-fixed, paraffin-embedded sections were prepared from ERα-positive and ERα-negative breast cancer samples and immunostained with anti-cyclin D1 IgG.
Importantly, Figure 10 shows that the ERα-positive samples containing Cav-1 mutations are also cyclin D1-positive. In contrast, little or no cyclin D1 immunostaining was observed in ERα-negative samples, as predicted. Representative images are shown in panel A. Note that the ERα-positive patient samples shown harbor Cav-1 mutations. Consecutive sections from a patient harboring a Cav-1 P132L mutation were immunostained with anti-ERα IgG and anti-cyclin D1 IgG and are shown in panel B. See also Supplemental Figure 2 at http://ajp.amjpathol.org for negative control images.
Figure 10.
Up-regulation of cyclin D1 in ERα-positive breast cancer patient samples. Formalin-fixed, paraffin-embedded sections, prepared from ERα-positive and ERα-negative breast cancer samples, were immunostained with anti-cyclin D1 rabbit IgG (Ab-3 from NeoMarkers). Bound primary antibodies were detected with an horseradish peroxidase-conjugated secondary antibody (see brown reaction product). Slides were then counterstained with hematoxylin (blue color). Note that, as expected, cyclin D1 immunostaining is highly positive in ERα-positive breast cancer samples (see positive nuclear staining); in contrast, little or no cyclin D1 immunostaining was observed in ERα-negative samples. Representative images are shown in A. Note that the ERα-positive patient samples shown harbor Cav-1 mutations. Virtually identical results were obtained with other ERα-positive patients (not shown). In B, consecutive sections from a patient harboring a Cav-1 P132L mutation were separately immunostained with anti-ERα rabbit IgG (H-184 from Santa Cruz Biotechnology) and anti-cyclin D1 rabbit IgG (Ab-3 from NeoMarkers).
Discussion
In 2001, a Japanese study reported that up to 16% of breast cancer tumor samples harbor a sporadic mutation, namely P132L, within the human CAV-1 gene.24 However, since this initial report, several other groups have failed to confirm the existence of the Cav-1 P132L mutation (or any other Cav-1 mutations) in human breast cancers.42–44 This may reflect the patient populations examined. Alternatively, the methods used may not have been sensitive enough to detect mutations. Thus, the existence of Cav-1 mutations in human breast cancers remains controversial. Here, to increase the sensitivity of detection, we used 1) LCM to enrich for transformed mammary tumor cells and 2) a real-time allele-specific PCR approach to detect the P132L mutation. Interestingly, we show for the first time that Cav-1 mutations are selectively associated with only ERα-positive breast cancers. Importantly, this finding could explain why other groups failed to detect Cav-1 mutations, because they may have screened only ERα-negative breast tumor samples.
Our ex vivo reconstitution experiments using 3-D cultures of primary mammary epithelial cells provide a rational basis to mechanistically explain why Cav-1 mutations exclusively cosegregate with ER-positive breast cancers. Our data strongly suggest the existence of a correlation between Cav-1 loss-of-function and the dysregulation of mammary epithelial proliferation. We show that a Cav-1−/− deficiency is one of the mechanisms responsible for increased ERα levels, sequentially leading to enhanced mammary acinar growth and increased cyclin D1 levels in 3-D mammary epithelial structures that have acquired EGF-independent growth capabilities. It is noteworthy that all these steps require growth factor deprivation. We have previously shown that Cav-1−/− acini undergo EGF-independent growth, with hyperactivation of the Ras-p42/44 mitogen-activated protein kinase cascade.37 Growth factor deprivation is likely to engage Cav-1-deficient mammary epithelial cells in a pro-proliferative survival pathway that is necessary to induce increased estrogen sensitivity, thereby promoting mammary acinar growth.
In an attempt to elucidate the downstream signaling events, we show here that the enhanced growth of estrogen-stimulated, growth factor-depleted Cav-1-null mammary acini correlates with the up-regulation of cyclin D1 expression. Previous studies in fibroblasts have shown that Cav-1 transcriptionally represses cyclin D1 gene expression, whereas antisense-mediated Cav-1 down-regulation increases cyclin D1 levels.45 In addition, genetic ablation of Cav-1 was shown to increase cyclin D1 expression levels in a variety of in vivo experimental models, including oncogene-induced mammary tumors13 and carcinogen-induced epidermal hyperplasia.46 Of course, we cannot exclude that, in our system, Cav-1 deficiency leads to cyclin D1 up-regulation by directly releasing the inhibition of cyclin D1 transcription. However, because cyclin D1 levels are virtually identical in WT and Cav-1 KO mammary acini under steady-state nonstimulated conditions, and cyclin D1 up-regulation occurs only after estrogen stimulation, we believe that cyclin D1 overexpression is mediated by increased ERα transcriptional activity in Cav-1-null acini.
The mechanisms that regulate ERα expression during mammary epithelial proliferation and transformation remain largely unknown. In the normal mammary gland, ERα is expressed mainly in nonproliferating luminal epithelial cells that reside adjacently to the ERα-negative dividing cells.27 However, ERα becomes overexpressed at the very early stages of mammary tumorigenesis, with most proliferating lesions displaying a clear increase in ERα expression.28 As such, mammary transformation is associated with an increasing inability to down-regulate ERα expression or to restrain the proliferation of ERα-positive cells. In this report, we provide novel evidence that Cav-1 loss-of-function may be one of the mechanisms responsible for increased ERα activation and dysregulated proliferation during early mammary tumorigenesis. First, we show that, in breast cancer patients, loss-of-function mutations in the Cav-1 gene are exclusively found in ERα-positive tumors, suggesting that Cav-1 may normally control ERα expression levels in the mammary gland. In addition, we show that Cav-1 gene inactivation increases ERα expression in vivo and in vitro and promotes the acinar growth of 3-D cultures of primary mouse mammary epithelial cells. As such, Cav-1 may normally function as a “switch” to regulate the critical balance between ERα expression and proliferation in mammary epithelial cells.
Our results are consistent with a recent study showing that Cav-1 haplo-insufficiency induces the constitutive activation of ERα expression in an immortalized “normal” human mammary epithelial cell line, namely MCF-10A cells. In addition, estrogen stimulation was shown to promote the anchorage-independent growth of Cav-1 haplo-insufficient MCF-10A cells in vitro and to stimulate their ability to form tumors in nude mice.47 Consistent with these results, we show here that Cav-1 gene inactivation leads to increased ERα expression levels, both in primary human breast cancers and in mouse primary mammary epithelial cells, and promotes the growth of estrogen-stimulated 3-D epithelial cultures.
How loss of Cav-1 expression induces ERα overexpression remains an open question. An emerging theory in cancer research proposes that adult stem cells may play a role in the development of human cancers, including breast cancers.48,49 Adult stem cells represent a cellular subpopulation with the dual ability to self-renew and to differentiate into specific lineages depending on the tissue type.50 Stem cells are normally involved in morphogenesis, tissue repair, and remodeling. In particular, breast stem cells play a vital role in the development, differentiation, and function of the mammary gland, which undergoes important cycles of transformation during adult life. Because proliferation potential decreases with differentiation, stem cells and progenitor cells are likely candidates for accumulating genetic alterations associated with tumorigenesis. Mutations in stem cells may affect the delicate balance between self-renewal and differentiation.
In contrast to studies suggesting that ERα-positive cells are quiescent, several studies have suggested that a small subset of ERα-positive cells constitute a slowly proliferating subpopulation, with characteristics of mammary stem cells.51 These ERα-positive breast stem cells have the ability to self-renew through asymmetric cell division and to generate adjacent proliferating cells, which represent a transient amplifying population, with a loss of ERα expression. After a few cell divisions, transient amplifying cells exit from the cell cycle and differentiate into either myoepithelial or luminal epithelial cells.51
Dysregulation of the normal self-renewal process may cause the increased symmetrical division of stem cells, with increased proportions of proliferating ERα-positive cells, in both precancerous and cancerous breast lesions. As such, ERα-positive breast tumor cells may arise from stem cells that have lost the ability to divide asymmetrically, instead undergoing symmetric divisions to produce two identical ERα-positive daughter cells.
The fact that Cav-1 expression increases during cellular differentiation, with its highest expression in terminally differentiated cells, suggests the possibility that Cav-1 expression plays a key role during differentiation. For example, Cav-1 expression is up-regulated during the differentiation of primary cultures of human alveolar epithelial cells.52 Moreover, Cav-1 was found to be selectively up-regulated during the in vitro differentiation of embryonic stem cells to vessel-like endothelial structures.53 In the mammary gland, Cav-1 loss-of-function may interfere with the differentiation of a subset of breast stem cells and lead to an increased population of ERα-positive progenitor cells. This conclusion remains speculative but merits further investigation. However, in direct support of this notion, Cav-1-deficient mice have been shown to possess an increased mammary stem cell population.54 Furthermore, Cav-1−/− mammary epithelial cells show increased “plasticity”, with the ability to undergo a spontaneous epithelial mesenchymal transition or to undergo lactogenic differentiation, in the absence of a lactogenic stimulus.37,55
Future studies will be necessary to address the detailed phenotypic behavior of the various Cav-1 mutations that we describe in this report. We believe that these mutations will ultimately be shown to act in a dominant-negative fashion, thereby mimicking a Cav-1-null mammary acinar phenotype. For this purpose, these Cav-1 mutations will need to be stably expressed in an estrogen-responsive normal human mammary epithelial cell line. To this end, we have expressed Cav-1 (WT and P132L) in hTERT-HME1 cells. However, unfortunately, the parental hTERT-HME1 cells do not form regular 3-D acinar structures with a hollow lumen. Instead, they form large aggregates of variable size without a lumen (data not shown). This may be due to the fact that they are already immortalized/partially transformed by the expression of telomerase.
In summary, in this study we provide novel evidence showing that Cav-1 mutations are found in ∼35% of estrogen receptor-positive breast cancer samples. Using 3-D cultures of primary mammary epithelial cells from WT and Cav-1−/− mice, we show that loss of Cav-1 increases ERα levels, promotes mammary acinar growth, and induces the up-regulation of cyclin D1 expression levels. Thus, a novel Cav-1-related pathway may be involved as an initiating event in human breast cancers.
Supplementary Material
Footnotes
Address reprint requests to Dr. Michael P. Lisanti, Department of Cancer Biology, Kimmel Cancer Center, Bluemle Life Sciences Building, Room 933, 233 S. 10th Street, Philadelphia, PA 19107. E-mail: michael.lisanti@jefferson.edu.
Supported by grants from the National Institutes of Health and the American Heart Association, and a Hirschl/Weil-Caulier Career Scientist award (all to M.P.L.).
T.L. and F.S. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol. 2005;288:C494–C506. doi: 10.1152/ajpcell.00458.2004. [DOI] [PubMed] [Google Scholar]
- Bouras T, Lisanti MP, Pestell RG. Caveolin-1 in breast cancer. Cancer Biol Ther. 2004;3:82–92. doi: 10.4161/cbt.3.10.1147. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Baltimore D, Lisanti MP. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA. 1995;92:1381–1385. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Lee RJ, Karnezis A, Bearss DJ, Webster M, Siegel P, Muller WJ, Windle JJ, Pestell RG, Lisanti MP. Reciprocal regulation of Neu tyrosine kinase activity and caveolin-1 protein expression in vitro and in vivo. Implications for mammary tumorigenesis. J Biol Chem. 1998;273:20448–20455. doi: 10.1074/jbc.273.32.20448. [DOI] [PubMed] [Google Scholar]
- Lee SW, Reimer CL, Oh P, Campbell DB, Schnitzer JE. Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene. 1998;16:1391–1397. doi: 10.1038/sj.onc.1201661. [DOI] [PubMed] [Google Scholar]
- Park DS, Razani B, Lasorella A, Schreiber-Agus N, Pestell RG, Iavarone A, Lisanti MP. Evidence that Myc isoforms transcriptionally repress caveolin-1 gene expression via an INR-dependent mechanism. Biochemistry. 2001;40:3354–3362. doi: 10.1021/bi002787b. [DOI] [PubMed] [Google Scholar]
- Fiucci G, Ravid D, Reich R, Liscovitch M. Caveolin-1 inhibits anchorage-independent growth, anoikis and invasiveness in MCF-7 human breast cancer cells. Oncogene. 2002;21:2365–2375. doi: 10.1038/sj.onc.1205300. [DOI] [PubMed] [Google Scholar]
- Zhang W, Razani B, Altschuler Y, Bouzahzah B, Mostov KE, Pestell RG, Lisanti MP. Caveolin-1 inhibits epidermal growth factor-stimulated lamellipod extension and cell migration in metastatic mammary adenocarcinoma cells (MTLn3). Transformation suppressor effects of adenovirus-mediated gene delivery of caveolin-1. J Biol Chem. 2000;275:20717–20725. doi: 10.1074/jbc.M909895199. [DOI] [PubMed] [Google Scholar]
- Sloan EK, Stanley KL, Anderson RL. Caveolin-1 inhibits breast cancer growth and metastasis. Oncogene. 2004;23:7893–7897. doi: 10.1038/sj.onc.1208062. [DOI] [PubMed] [Google Scholar]
- Lee H, Park DS, Razani B, Russell RG, Pestell RG, Lisanti MP. Caveolin-1 mutations (P132L and null) and the pathogenesis of breast cancer: Caveolin-1 (P132L) behaves in a dominant-negative manner and caveolin-1 (-/-) null mice show mammary epithelial cell hyperplasia. Am J Pathol. 2002;161:1357–1369. doi: 10.1016/S0002-9440(10)64412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Lee H, Cheung MW, Cohen AW, Razani B, Iyengar P, Scherer PE, Pestell RG, Lisanti MP. Combined loss of INK4a and caveolin-1 synergistically enhances cell proliferation and oncogene-induced tumorigenesis: Role of INK4a/CAV-1 in mammary epithelial cell hyperplasia. J Biol Chem. 2004;279:24745–24756. doi: 10.1074/jbc.M402064200. [DOI] [PubMed] [Google Scholar]
- Williams TM, Cheung MW, Park DS, Razani B, Cohen AW, Muller WJ, Di Vizio D, Chopra NG, Pestell RG, Lisanti MP. Loss of caveolin-1 gene expression accelerates the development of dysplastic mammary lesions in tumor-prone transgenic mice. Mol Biol Cell. 2003;14:1027–1042. doi: 10.1091/mbc.E02-08-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Medina F, Badano I, Hazan RB, Hutchinson J, Muller WJ, Chopra NG, Scherer PE, Pestell RG, Lisanti MP. Caveolin-1 gene disruption promotes mammary tumorigenesis and dramatically enhances lung metastasis in vivo. Role of Cav-1 in cell invasiveness and matrix metalloproteinase (MMP-2/9) secretion. J Biol Chem. 2004;279:51630–51646. doi: 10.1074/jbc.M409214200. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zhang XL, Lisanti MP. Genes encoding human caveolin-1 and -2 are co-localized to the D7S522 locus (7q31.1), a known fragile site (FRA7G) that is frequently deleted in human cancers. FEBS Lett. 1998;436:403–410. doi: 10.1016/s0014-5793(98)01134-x. [DOI] [PubMed] [Google Scholar]
- Zenklusen JC, Bieche I, Lidereau R, Conti CJ. (C-A)n microsatellite repeat D7S522 is the most commonly deleted region in human primary breast cancer. Proc Natl Acad Sci USA. 1994;91:12155–12158. doi: 10.1073/pnas.91.25.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen JC, Thompson JC, Troncoso P, Kagan J, Conti CJ. Loss of heterozygosity in human primary prostate carcinomas: a possible tumor suppressor gene at 7q31.1. Cancer Res. 1994;54:6370–6373. [PubMed] [Google Scholar]
- Zenklusen JC, Thompson JC, Klein-Szanto AJ, Conti CJ. Frequent loss of heterozygosity in human primary squamous cell and colon carcinomas at 7q31.1: evidence for a broad range tumor suppressor gene. Cancer Res. 1995;55:1347–1350. [PubMed] [Google Scholar]
- Zenklusen JC, Weitzel JN, Ball HG, Conti CJ. Allelic loss at 7q31.1 in human primary ovarian carcinomas suggests the existence of a tumor suppressor gene. Oncogene. 1995;11:359–363. [PubMed] [Google Scholar]
- Achille A, Biasi MO, Zamboni G, Bogina G, Magalini AR, Pederzoli P, Perucho M, Scarpa A. Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res. 1996;56:3808–3813. [PubMed] [Google Scholar]
- Koike M, Takeuchi S, Yokota J, Park S, Hatta Y, Miller CW, Tsuruoka N, Koeffler HP. Frequent loss of heterozygosity in the region of the D7S523 locus in advanced ovarian cancer. Genes Chromosomes Cancer. 1997;19:1–5. doi: 10.1002/(sici)1098-2264(199705)19:1<1::aid-gcc1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Shridhar V, Sun QC, Miller OJ, Kalemkerian GP, Petros J, Smith DI. Loss of heterozygosity on the long arm of human chromosome 7 in sporadic renal cell carcinomas. Oncogene. 1997;15:2727–2733. doi: 10.1038/sj.onc.1201448. [DOI] [PubMed] [Google Scholar]
- Huang H, Qian C, Jenkins RB, Smith DI. Fish mapping of YAC clones at human chromosomal band 7q31.2: identification of YACS spanning FRA7G within the common region of LOH in breast and prostate cancer. Genes Chromosomes Cancer. 1998;21:152–159. doi: 10.1002/(sici)1098-2264(199802)21:2<152::aid-gcc11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Jenkins RB, Qian J, Lee HK, Huang H, Hirasawa K, Bostwick DG, Proffitt J, Wilber K, Lieber MM, Liu W, Smith DI. A molecular cytogenetic analysis of 7q31 in prostate cancer. Cancer Res. 1998;58:759–766. [PubMed] [Google Scholar]
- Hayashi K, Matsuda S, Machida K, Yamamoto T, Fukuda Y, Nimura Y, Hayakawa T, Hamaguchi M. Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res. 2001;61:2361–2364. [PubMed] [Google Scholar]
- Koehler KF, Helguero LA, Haldosen LA, Warner M, Gustafsson JA. Reflections on the discovery and significance of estrogen receptor beta. Endocr Rev. 2005;26:465–478. doi: 10.1210/er.2004-0027. [DOI] [PubMed] [Google Scholar]
- Clarke RB, Howell A, Potten CS, Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 1997;57:4987–4991. [PubMed] [Google Scholar]
- Shoker BS, Jarvis C, Sibson DR, Walker C, Sloane JP. Oestrogen receptor expression in the normal and pre-cancerous breast. J Pathol. 1999;188:237–244. doi: 10.1002/(SICI)1096-9896(199907)188:3<237::AID-PATH343>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Shoker BS, Jarvis C, Clarke RB, Anderson E, Hewlett J, Davies MP, Sibson DR, Sloane JP. Estrogen receptor-positive proliferating cells in the normal and precancerous breast. Am J Pathol. 1999;155:1811–1815. doi: 10.1016/S0002-9440(10)65498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievre A, Landi B, Cote JF, Veyrie N, Zucman-Rossi J, Berger A, Laurent-Puig P. Absence of mutation in the putative tumor-suppressor gene KLF6 in colorectal cancers. Oncogene. 2005;24:7253–7256. doi: 10.1038/sj.onc.1208867. [DOI] [PubMed] [Google Scholar]
- Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, Di Vizio D, Hou H, Jr, Kneitz B, Lagaud G, Christ GJ, Edelmann W, Lisanti MP. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem. 2001;276:38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- Simian M, Hirai Y, Navre M, Werb Z, Lochter A, Bissell MJ. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Han SE, Park KH, Lee G, Huh YJ, Min BM. Mutation and aberrant expression of Caveolin-1 in human oral squamous cell carcinomas and oral cancer cell lines. Int J Oncol. 2004;24:435–440. [PubMed] [Google Scholar]
- Sotgia F, Lee JK, Das K, Bedford M, Petrucci TC, Macioce P, Sargiacomo M, Bricarelli FD, Minetti C, Sudol M, Lisanti MP. Caveolin-3 directly interacts with the C-terminal tail of beta-dystroglycan. Identification of a central WW-like domain within caveolin family members. J Biol Chem. 2000;275:38048–38058. doi: 10.1074/jbc.M005321200. [DOI] [PubMed] [Google Scholar]
- Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module–the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- Sotgia F, Williams TM, Schubert W, Medina F, Minetti C, Pestell RG, Lisanti MP. Caveolin-1 deficiency (-/-) conveys premalignant alterations in mammary epithelia, with abnormal lumen formation, growth factor independence, and cell invasiveness. Am J Pathol. 2006;168:292–309. doi: 10.2353/ajpath.2006.050429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- van Diest PJ, Michalides RJ, Jannink L, van der Valk P, Peterse HL, de Jong JS, Meijer CJ, Baak JP. Cyclin D1 expression in invasive breast cancer. Correlations and prognostic value. Am J Pathol. 1997;150:705–711. [PMC free article] [PubMed] [Google Scholar]
- Joe AK, Memeo L, McKoy J, Mansukhani M, Liu H, Avila-Bront A, Romero J, Li H, Troxel A, Hibshoosh H. Cyclin D1 overexpression is associated with estrogen receptor expression in Caucasian but not African-American breast cancer. Anticancer Res. 2005;25:273–281. [PubMed] [Google Scholar]
- Prall OWJ, Rogan EM, Sutherland RL. Estrogen regulation of cell cycle progression in breast cancer cells. J Steroid Biochem Mol Biol. 1998;65:169–174. doi: 10.1016/s0960-0760(98)00021-1. [DOI] [PubMed] [Google Scholar]
- Chen ST, Lin SY, Yeh KT, Kuo SJ, Chan WL, Chu YP, Chang JG. Mutational, epigenetic and expressional analyses of caveolin-1 gene in breast cancers. Int J Mol Med. 2004;14:577–582. [PubMed] [Google Scholar]
- Zenklusen JC, Conti CJ, Green ED. Mutational and functional analyses reveal that ST7 is a highly conserved tumor-suppressor gene on human chromosome 7q31. Nat Genet. 2001;27:392–398. doi: 10.1038/86891. [DOI] [PubMed] [Google Scholar]
- Hurlstone AF, Reid G, Reeves JR, Fraser J, Strathdee G, Rahilly M, Parkinson EK, Black DM. Analysis of the CAVEOLIN-1 gene at human chromosome 7q31.1 in primary tumours and tumour-derived cell lines. Oncogene. 1999;18:1881–1890. doi: 10.1038/sj.onc.1202491. [DOI] [PubMed] [Google Scholar]
- Hulit J, Bash T, Fu M, Galbiati F, Albanese C, Sage DR, Schlegel A, Zhurinsky J, Shtutman M, Ben-Ze’ev A, Lisanti MP, Pestell RG. The cyclin D1 gene is transcriptionally repressed by caveolin-1. J Biol Chem. 2000;275:21203–21209. doi: 10.1074/jbc.M000321200. [DOI] [PubMed] [Google Scholar]
- Capozza F, Williams TM, Schubert W, McClain S, Bouzahzah B, Sotgia F, Lisanti MP. Absence of caveolin-1 sensitizes mouse skin to carcinogen-induced epidermal hyperplasia and tumor formation. Am J Pathol. 2003;162:2029–2039. doi: 10.1016/S0002-9440(10)64335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shen P, Coleman M, Zou W, Loggie BW, Smith LM, Wang Z. Caveolin-1 down-regulation activates estrogen receptor alpha expression and leads to 17beta-estradiol-stimulated mammary tumorigenesis. Anticancer Res. 2005;25:369–375. [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Dontu G, El-Ashry D, Wicha MS. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab. 2004;15:193–197. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Gudjonsson T, Villadsen R, Bissell MJ, Ronnov-Jessen L. Epithelial progenitor cell lines as models of normal breast morphogenesis and neoplasia. Cell Prolif. 2003;36(Suppl 1):33–44. doi: 10.1046/j.1365-2184.36.s.1.4.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RB, Spence K, Anderson E, Howell A, Okano H, Potten CS. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol. 2005;277:443–456. doi: 10.1016/j.ydbio.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Hollins AJ, Laue M, Schaefer UF, Roemer K, Gumbleton M, Lehr CM. Differentiation of human alveolar epithelial cells in primary culture: morphological characterization and synthesis of caveolin-1 and surfactant protein-C. Cell Tissue Res. 2003;311:31–45. doi: 10.1007/s00441-002-0653-5. [DOI] [PubMed] [Google Scholar]
- Ng YS, Ramsauer M, Loureiro RM, D’Amore PA. Identification of genes involved in VEGF-mediated vascular morphogenesis using embryonic stem cell-derived cystic embryoid bodies. Lab Invest. 2004;84:1209–1218. doi: 10.1038/labinvest.3700150. [DOI] [PubMed] [Google Scholar]
- Sotgia F, Williams TM, Cohen AW, Minetti C, Pestell RG, Lisanti MP. Caveolin-1-deficient mice have an increased mammary stem cell population with upregulation of Wnt/beta-catenin signaling. Cell Cycle. 2005;4:1808–1816. doi: 10.4161/cc.4.12.2198. [DOI] [PubMed] [Google Scholar]
- Sotgia F, Schubert W, Pestell RG, Lisanti MP. Genetic ablation of caveolin-1 in mammary epithelial cells increases milk production and hyper-activates STAT5a signaling. Cancer Biol Ther. 2006;5:292–297. doi: 10.4161/cbt.5.3.2390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.