Abstract
Signaling by fibroblast growth factor 2 (FGF-2), an autocrine stimulator of glioma growth, is regulated by heparan sulfate proteoglycans (HSPGs) via a ternary complex with FGF-2 and the FGF receptor (FGFR). To characterize glioma growth signaling, we examined whether altered HSPGs contribute to loss of growth control in gliomas. In a screen of five human glioma cell lines, U118 and U251 cell HSPGs activated FGF-2 signaling via FGFR1c. The direct comparison of U251 glioma cells with normal astrocyte HSPGs demonstrated that the glioma HSPGs had a significantly elevated ability to promote FGF-2-dependent mitogenic signaling via FGFR1c. This enhanced activity correlated with a higher level of overall sulfation, specifically the abundance of 2S- and 6S-containing disaccharides. Glioma cell expression of the cell-surface HSPG glypican-1 closely mirrored the FGF-2 coactivator activity. Furthermore, forced expression of glypican-1 in (glypican-1-deficient) U87 glioma cells enhanced their FGF-2 response. Immunohistochemical analysis revealed a highly significant overexpression of glypican-1 in human astrocytoma and oligodendroglioma samples compared with nonneoplastic gliosis. In summary, these observations suggest that altered HSPGs contribute to enhanced signaling of FGF-2 via FGFR1c in gliomas with glypican-1 playing a significant role in this mitogenic pathway.
High-grade gliomas remain among the most deadly cancers. Mechanisms responsible for loss of growth control in this tumor type include growth factor-mediated signaling loops. A considerable amount of evidence implicates the fibroblast growth factor 2 (FGF-2)/FGF receptor 1 (FGFR1) axis in autonomous glioma cell growth and malignant progression.1,2 Glioma cells express FGF-2 at elevated levels and respond to FGF-2 with increased proliferation.3,4 A switch in FGF receptor expression has also been reported during glioma progression: FGFR2 expression is abundant in normal astrocytes and low-grade astrocytomas but absent in malignant astrocytomas. Conversely, FGFR1 is barely detectable in normal white matter but is highly expressed in malignant astrocytomas.5
In addition to FGF-2 and FGF receptors, heparan sulfate proteoglycans (HSPGs) play a critical role in FGF-2 signaling. HSPGs represent a diverse group of molecules that are composed of a core protein and covalently attached heparan sulfate glycosaminoglycan (HSGAG) chains. HSGAGs modulate FGF-2 signaling by participating in a high-affinity ternary complex with FGF-2 and FGFR.6 Mounting evidence indicates that specific structural motifs within HSGAGs regulate FGF signaling.7–9 Thus, qualitative or quantitative changes in HSPGs may have profound consequences on FGF-2-induced cell proliferation.
Alterations of HSPGs have been observed in human cancers compared with normal cells or tissues.10–12 These alterations affect both core protein expression and heparan sulfate chain composition. Evidence also exists that HSPGs are functionally important for cancer growth. For example, suppression of perlecan expression by antisense technology reduces malignant melanoma cell proliferation in response to FGF-2 and decreases migratory and invasive properties.13 Moreover, HSGAGs have been found to play an important role in tumorigenesis, tumor progression, and metastasis.11
Our understanding of expression levels and function of HSPGs in glioma is incomplete. Steck et al14 reported that high-grade glioma-derived cells express significantly increased amounts of hyaluronic acid and heparan sulfate compared with normal glial cells, and the immunofluorescence staining pattern of HSPGs changes from a distinctive punctate staining in normal brain or low-grade astrocytomas to an intense diffuse cell-surface staining. However, it is unclear what effect these HSPG alterations have on FGF-2 signaling.
Here, we report that HSPGs isolated from U251 glioma cells have a greater ability to assemble the FGF-2/FGFR1c signaling complex and promote FGF-2-induced proliferation than HSPGs isolated from normal human astrocytes. We also find that the cell-surface HSPG glypican-1 (Gpc-1) is increased in human U251 glioma cells and in the vast majority of human gliomas in vivo. The fact that overexpression of Gpc-1 in glioma cells, significantly increases their FGF-2 response suggests a specific role for Gpc-1 in glioma cell growth.
Materials and Methods
Heparitinase, chondroitinase ABC, standard heparan sulfate disaccharides, and anti-heparan sulfate (3G10) antibody were purchased from Seikagaku America (Associates of Cape Cod, Falmouth, MA). Yeast-derived human recombinant FGF-2 was provided by Dr. Brad Olwin (University of Colorado, Boulder). Heparin from porcine intestinal mucosa was from Sigma (St. Louis, MO). EZ-Link Sulfo-NHS-Biotin was purchased from Pierce (Rockford, IL). Neutralizing anti-FGF-2 antibody was purchased from R&D Systems (Minneapolis, MN).
Cell Culture
Normal human astrocytes (NHA cells) were purchased from BioWhittaker (East Rutherford, NJ). U87 and U118 human glioma cells were from the American Type Culture Collection (Rockville, MD). U251, U373 human glioma, and rat glioma C6 cells were provided by Dr. Behnam Badie (University of Wisconsin, Madison). U105 human glioma cells were provided by Dr. Peter Steck (University of Texas, M.D. Anderson Cancer Center). Cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. FR1c11 cells, which are BaF3 lymphoid cells transfected with FGFR1c, were provided by Dr. David Ornitz (Washington University, St. Louis, MO). FR1c11 cells were grown in RPMI 1640 containing 10% FBS and 10% WEHI-3 cell conditioned medium as source of interleukin-3 (IL-3). Rat primary astrocytes were kindly provided by Dr. Dandan Sun (University of Wisconsin, Madison).
Glioma Cell Proliferation Assay
Cells were seeded on 24-well tissue culture plates at a density of 10,000 cells per well and allowed to attach overnight in complete DMEM. The cells were starved for 24 hours before the addition of FGF-2 (500 pmol/L). After a 3-day incubation, cells were trypsinized, and the cell number was determined with a Coulter Counter (Beckman, Hayward, CA). The effect of neutralizing anti-FGF-2 antibody (R&D Systems) on U251 cell proliferation was measured by bromodeoxyuridine (BrdU) incorporation assay (BrdU colorimetric; Roche Diagnostics, Indianapolis, IN) according to the manufacturer’s instructions.
FR1c11 Cell Proliferation Assay
The FR1c11 proliferation assay was a modification of a method described by Allen et al.15 FR1c11 cells were added to 96-well flat-bottom tissue culture plates at a density of 2 × 104 cells/well in starvation medium (IL-3-deficient RPMI 1640 containing 10% calf serum) and incubated with or without various concentrations of HSPGs and FGF-2 (10 nmol/L). After a 72-hour incubation, CellTiter 96 Aqueous One Solution reagent (Promega, Madison, WI) was added to quantify the cell number according to the manufacturer’s instructions.
The activity of glioma cell or astrocyte HSPGs to promote FGF-2 signaling in heterotypic contact with FR1c-11 cells was also examined. For these experiments, 2 × 104 glioma cells or normal astrocytes were seeded into each well of a 96-well plate and allowed to attach as confluent monolayer overnight at 37°C in DMEM with 10% FBS. The cell monolayers were fixed in 0.5% glutaraldehyde for 1 hour, followed by three washes in HEPES containing 0.2 mol/L glycine and incubated overnight in starvation medium. Some wells were treated with heparitinase (0.002 U/ml) for 3 hours. FR1c11 cells were added to the monolayers with or without 10 nmol/L FGF-2 in the starvation medium. After a 72-hour incubation, the cell growth assay was performed as described above.
HSPG Extraction
Total glioma cell or normal astrocyte HSPGs were purified as described previously.16,17 In brief, cells were grown in DMEM with 10% FBS until confluent. Then, the cells were placed on ice, washed three times with cold HEPES-buffered saline, extracted with 4 ml of TUT buffer (10 mmol/L Tris, 8 mol/L urea, 0.1% Triton X-100, 1 mmol/L Na2SO4, 1 mmol/L phenylmethylsulfonyl fluoride, and 1 mmol/L N-ethylmaleimide, pH 8.0) for 5 minutes, and scraped into a polypropylene tube. After sonication, diethylaminoethyl (DEAE) beads pre-equilibrated with TUT were added to the tube. Tubes containing DEAE beads and extraction solution were rotated overnight at 4°C. HSPGs were eluted with high-salt HEPES buffer (30 mmol/L HEPES and 1 mol/L NaCl, pH 7.4). Extracted HSPGs were quantified by alcian blue staining as described by Karlsson and Bjornsson.18 Commercially available HSPGs (Sigma) were used as standards.
Isolation of Glycosaminoglycans
Glycosaminoglycans (GAGs) were isolated as reported by Lee et al,19 with some minor modifications. Briefly, the cells (NHA and U251), which were 80 to 100% confluent, were extracted with extraction buffer. The proteins and GAGs were precipitated by addition of 3 volumes of 95% ethanol containing 1.3% potassium acetate. The precipitate was digested exhaustively with proteinase K (Invitrogen, Carlsbad, CA) at 65°C for 24 hours. Proteins remaining in the digest were precipitated by 5% trichloroacetic acid (TCA), and the supernatant was once again precipitated by 3 volumes of 95% ethanol containing potassium acetate. The precipitate was dried and desalted on a PD-10 column using pyridine acetate buffer, pH 5.0, as an eluant. The eluted fractions were dried by speedvac and pooled. Quantitation of sulfated GAGs was performed by the dye binding assay of Chandrasekhar et al,20 using dimethylmethelene blue (Aldrich, St. Louis, MO), except that the absorbance was read at 525 nm.
Steady-State Complex Binding Assay
Complex binding was measured to quantitatively analyze the ability of HSPGs or HSGAGs to promote FGF-2 binding to FGFR1c. Extracted HSPGs were biotinylated with EZ-Link Sulfo-NHS-Biotin (Pierce) for 1 hour at room temperature. Biotinylated HSPGs were then bound to DEAE beads to remove free biotin by washing. HSGAGs were freshly biotinylated by biotin-LC-hydrazide (50 mmol/L in dimethylsulfoxide) and 1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide hydrochloride (0.5 mol/L in 100 mmol/L MES, pH 5.5) at room temperature overnight after being exhaustively digested with conventional chondroitinase ABC to remove CS chains. Excess biotinylating agents were removed using centricon columns (molecular weight cutoff 3,000). Biotinylated HSPGs or HSGAGs were immobilized on streptavidin-coated 96-well plates (Pierce), using 150 ng of HSPGs or 50 ng of HSGAGs per well. As the negative control, several wells were treated with heparitinase before the binding assay. FGF-2 at varying concentrations and FR1c-AP (5 nmol/L), a soluble fusion protein containing the extracellular domain of FGFR1c and alkaline phosphatase (AP) as enzyme tag,17,21 were added. The plates were incubated for 1 hour and then washed twice with 400 mmol/L NaCl in TBS to reduce nonspecific binding. AP substrate solution (0.4 mol/L diethanolamine, 1 mmol/L MgCl2, 20 mmol/L l-homoarginine, and 24 mmol/L p-nitrophenylphosphate) was added to each well to detect bound FRIc-AP. Absorbance at 405 nm reflects the amount of ternary complex. A curve fit was performed using nonlinear saturation fitting in the Prism software package (GraphPad Software, San Diego, CA). The program also calculates an apparent Kd for the binding interaction. The equation the software used is Y = Bmax × X/(Kd + X).
Disaccharide Composition Analysis of HSGAG Chains
One to 2 μg of HSGAGs was digested with 3 mIU of heparinase II in the total volume of 10 μl. The digested products were labeled with 2-aminobenzamide (2-AB; Aldrich) according to the procedure of Kinoshita et al.22 The labeled products were analyzed on an amine-bound silica PA03 column (4.6 × 250 mm; YMC-Pack PA; Waters, Milford, MA) using a linear gradient of NaH2PO4 from 16 to 700 mmol/L for a period of 1 hour at a flow rate of 1 ml/min using fluorescent detection (Excitation: 330; Emission: 420).
Gel Electrophoresis and Western Blotting
Extracted HSPGs were digested with heparitinase and chondroitinase ABC (0.002 U/ml) twice for 2 hours each to remove all glycosaminoglycan chains. Samples and prestained molecular mass markers (Bio-Rad, Hercules, CA) were denatured in sample buffer (2% sodium dodecyl sulfate [SDS], 10% glycerol, bromophenyl blue, and 2.5% β-mercaptoethanol) and heated to 100°C for 3 minutes before gel electrophoresis. Samples were then electrophoretically separated on a 3.5 to 15% Tris-borate polyacrylamide gradient gel and transferred to a polyvinylidene difluoride (PVDF) membrane. The blots were probed with anti-Δ heparan sulfate (3G10) antibody (0.03 μg/ml), which reacts with heparan sulfate “stubs” generated by heparitinase treatment. A horseradish peroxidase-conjugated secondary IgG (Sigma) was used for detection. The signal was visualized with SuperSignal West Femto Maximum Sensitivity Substrate (Pierce).
FGF-2/FGFR1c Complex Precipitation
HSPGs promoting FGF-2 binding to FGFR1c were analyzed as described by Mundhenke et al.17 Briefly, FR1c-AP was immobilized on anti-alkaline phosphatase-conjugated agarose beads (Sigma). Total HSPGs (2 × 106 cell equivalent) were incubated with the FR1c-AP-carrying agarose beads in the presence or absence of 10 nmol/L FGF-2 overnight at 4°C. The agarose beads were subsequently washed with 0.5 mol/L NaCl in Tris-buffered saline twice to reduce nonspecific binding and digested with chondroitinase. HSPG core protein was released into solution from the FGF-2/FGFR/heparan sulfate complex by heparitinase digestion. The samples were loaded on a 3.5 to 15% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gradient gel. HSPG core protein was detected with 3G10 antibody. To ascertain heparan sulfate dependence of complex formation, total HSPGs were digested with heparitinase before the binding reaction in control samples. Omission of FGF-2 from the reaction mixture served as an additional negative control.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Quantitative real-time PCR was performed to measure sulfotransferase, HSPG core protein, and FGFR mRNA levels. Total RNA was isolated using RNeasy Protect Mini kit (Qiagen, Valencia, CA). One microgram of total RNA from NHA or human glioma cells was reverse transcribed into cDNA using the ThermoScript RT-PCR System (Invitrogen). Five percent of the RT product was used in the quantitative RT-PCR reaction with SYBR Green PCR master mix on an iCycler instrument (Bio-Rad). β-Actin mRNA level was used as internal control for normalizing different samples.
Cell Transfection
Human Gpc-1 cDNA (kindly provided by Dr. Guido David, University of Leuven, Belgium) was cloned into the pcDNA3 expression vector. U87 glioma cells were stably transfected with either construct or the respective empty vector using TransIT-LT1 transfection reagent (Mirus, Madison, WI). After selection with G418 (400 μg/ml), cell extracts were subjected to HSPG Western blot analysis with 3G10 antibody to determine the levels of HSPG core protein. Proliferation assays were performed as described above.
Immunohistochemical Analysis of Human Tumor Samples
After IRB approval, paraffin blocks from glioma resections and temporal lobectomy specimens from seizure patients were selected from the pathology archives. Cases with insufficient viable tissue were excluded. Tissue arrays containing duplicate 1-mm cores were constructed from these samples using the Arrayer I instrument (Beecher Instruments, Sun Prairie, WI). Immunohistochemical analysis of Gpc-1 was performed on 4-μm sections from the array blocks using monoclonal antibody S1 (kindly provided by G. David23) as described previously.16,17 The array slides were scored independently by two pathologists (S.S. and A.F.) using the method by Allred and colleagues.24 This technique takes both the proportion of positive cells and the staining intensity into account and yields a summary score on a scale from 0 to 8. The agreement between both observers was excellent (r = 0.9183; P < 0.0001). The average of both scores was used for further analysis.
Results
FGF-2 Stimulates Proliferation of Glioma Cells but Not of Normal Astrocytes
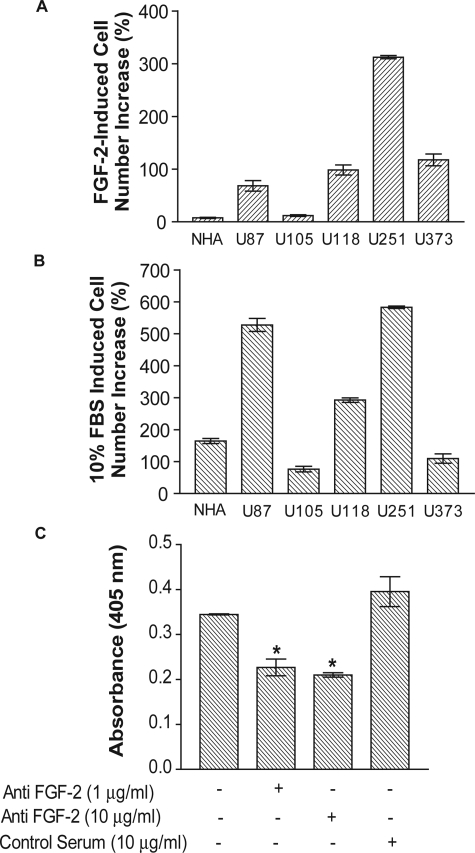
FGF-2 has been identified in human glial tumors and in transformed human glial cell lines.1,2 Previous studies suggest that FGF-2 is involved in an autocrine pathway regulating glioma growth and invasion.4 When we screened a panel of five glioma cell lines, we found that the majority proliferate in response to FGF-2, whereas NHA cells fail to do so (Figure 1A). Among these cell lines, U251 cells had the strongest response to FGF-2. Its cell number increased by 312.5 ± 3.1% when exposed to FGF-2 at a concentration of 500 pmol/L. U373, U118, and U87 cells also showed a robust proliferative response to FGF-2, whereas U105 cells were unresponsive. To exclude the possibility that NHA cells have lost their mitogenic potential in culture, we also measured their proliferation in response to fetal bovine serum (10%). Growth of NHA cells and all glioma cells in the panel was significantly stimulated in response to this mitogen (Figure 1B).
Figure 1.
FGF-2-induced glioma cell proliferation. A: FGF-2-induced glioma cell proliferation. Cells were treated with 500 pmol/L FGF-2 for three days and then trypsinized. Cells were counted with a Coulter Counter. The data points indicate the percent increase in cell number in the presence of FGF-2 compared with a no-treatment control. B: FBS-induced glioma cell proliferation. Cells were treated and counted the same way as shown in A. Data points indicate the percent increase in cell number with 10% FBS compared with the no-treatment control. C: The effect of anti-FGF-2 antibody on the proliferation of U251 cells. Cells were cultured on a 96-well plate with 3,000 cells per well and starved for 24 hours. A polyclonal anti-FGF-2 antibody was added to the starvation medium. FGF-2 was not added during or after the serum starvation. Proliferation was measured with the BrdU incorporation assay according to the manufacturer’s instructions. Data are mean ± SEM. *P < 0.05 versus control.
Next, we examined whether an autocrine growth-promoting loop involving FGF-2 contributes to the high baseline growth of U251 cells. Treatment with goat anti-FGF-2 neutralizing antibody significantly decreased U251 cell baseline proliferation in starvation medium (Figure 1C). U251 cell growth was not inhibited by a goat serum control at the same concentration. This result suggests the existence of an FGF-2-mediated autocrine loop, which is further augmented by the addition of exogenous FGF-2.
One explanation for the differences in FGF-2 signaling between glioma cells and normal astrocytes would be variances in their FGFR expression profiles. A meaningful analysis of FGFR expression must take splice variation into account, because alternative splicing greatly affects ligand-binding specificity.25 Because isoform-specific antibodies are not available, we measured FGFR expression at the mRNA level by quantitative real-time PCR. FGFR1c was the most predominant FGFR expressed among NHA and human glioma cell lines (Table 1). The mRNA level of FGFR1c in U87, U118, and U251 cells was significantly higher than in NHA cells (Table 1). This is consistent with the previous finding that FGFR1 expression is elevated in malignant astrocytomas.5 However, the diversity of FGFR1c levels clearly does not fully explain the differences in FGF-2 response. This prompted us to examine the role of HSPGs as FGF-2 co-stimulators in gliomas and astrocytes.
Table 1.
Quantitation of FGFR mRNA Level in NHA and Human Glioma Cells
| mRNA copy number per 106 copies of β-actin mRNA
|
|||||||
|---|---|---|---|---|---|---|---|
| FGFR1c | FGFR1b | FGFR2c | FGFR2b | FGFR3c | FGFR3b | FGFR4 | |
| NHA | 11,815 ± 202 | 234 ± 10 | 4,347 ± 24 | 12 ± 6 | 125 ± 30 | 66 ± 8 | 250 ± 21 |
| U87 | 35,722 ± 916 | 1,864 ± 291 | 10,512 ± 1234 | 0 | 1 ± 1 | 2 ± 1 | 136 ± 2 |
| U105 | 11,399 ± 268 | 373 ± 6 | 3,211 ± 45 | 2 ± 2 | 12 ± 4 | 24 ± 9 | 164 ± 1 |
| U118 | 26,227 ± 1,682 | 2,886 ± 159 | 7,300 ± 518 | 3 ± 3 | 3 ± 2 | 9 ± 9 | 144 ± 8 |
| U251 | 35,971 ± 985 | 0 | 827 ± 87 | 27 ± 2 | 1,203 ± 164 | 4 ± 3 | 20 ± 2 |
| U373 | 1,848 ± 86 | 129 ± 16 | 2,105 ± 48 | 34 ± 4 | 262 ± 59 | 162 ± 21 | 184 |
Total RNA isolation and quantitative RT-PCR was performed as described in Materials and Methods. Individual standard ranges were used for different genes to ascertain that samples were within the range. All of the copy numbers were normalized to 106 copies of β-actin mRNA. Data are representative of three experiments.
Glioma Cell Heparan Sulfate Proteoglycans Promote FGF-2 Signaling via FGFR1c to a Greater Degree Than Normal Astrocyte HSPGs
Previous studies have shown that HSPG expression and distribution in human glioma are altered compared with normal glial cells.14,26 However, it has been unclear whether such changes affect FGF-2 signaling in gliomas. To test the activity of HSPGs from different cellular sources in a standardized experimental system, we used an assay based on HSPG-deficient BaF3 reporter cells. This cell type has been used extensively to analyze growth factor signaling events.25 Parental BaF3 cells are negative for both HSPGs and FGFR expression and require IL-3 for survival. However, BaF3 cells expressing FGFR bypass the IL-3 requirement if a compatible FGF and suitable heparan sulfate are provided.21 In this study, we chose FR1c11 cells, which express functional FGFR1c—the receptor isoform most prevalent on glioma cells.1,2
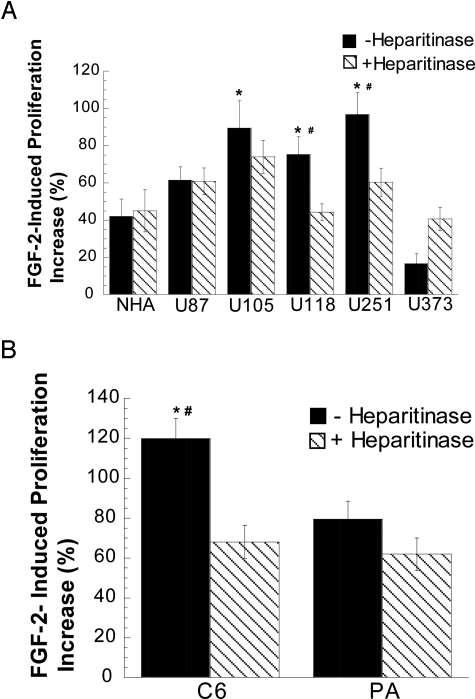
As a first screening test, we examined FR1c11 cell proliferation on fixed monolayers of either glioma cells or normal astrocytes. In this modification of an assay described by Richard et al,27 HSPGs on the adherent glutaraldehyde-fixed cells participate in heterotypic signaling complexes with exogenous FGF-2 and FGFR1c on FR1c11 cells. Heparitinase-sensitive (ie, heparan sulfate-mediated) growth stimulation of FR1c11 cells was observed on monolayers of U251 and U118 glioma cells but not NHA cells or U87, U105, or U373 glioma cells (Figure 2A). This result suggests that HSPGs of U251 and U118 cells have a greater capability to induce FGF-2 signaling than those of NHA and other glioma cells. To test whether this observation is valid across species, we compared rat glioma C6 cells and primary rat cultured astrocytes (PAs). The proliferation of FR1c11 cells on fixed C6 monolayers was significantly higher than on fixed primary rat astrocytes (119 ± 10.01 versus 79.52 ± 8.936%, P < 0.05) in a heparitinase-sensitive manner (Figure 2B). In summary, these results suggest that in a subset of glioma cells, HSPGs quantitatively or qualitatively differ from normal astrocyte HSPGs and promote FGF-2 signaling via the FGFR1c. The observed heterogeneity among the glioma cell lines with respect to signaling pathways confirms prior findings by others.28,29 Our result is also in keeping with the presence of distinct glycosaminoglycan profiles identified in these cell lines by another group.26,30
Figure 2.
FR1c11 cell proliferation on fixed-cell monolayers. Fixed-cell monolayers (A, human cells; B, rat cells) were either treated with heparitinase or without the enzyme as indicated. FR1c11 (20,000 cells/well) cells were added on top of the fixed-cell monolayers in the presence or absence of 10 nmol/L FGF-2. Cell growth was determined after 72 hours using a tetrazolium compound-based (Cell Titer 96 Aqueous) assay. Data points indicate percent increase in absorbance in the presence of 10 nmol/L FGF-2 compared with the no-treatment control. Data indicate mean ± SEM. *P < 0.05 versus NHAs or PAs without heparitinase treatment, #P < 0.05 versus with heparitinase treatment.
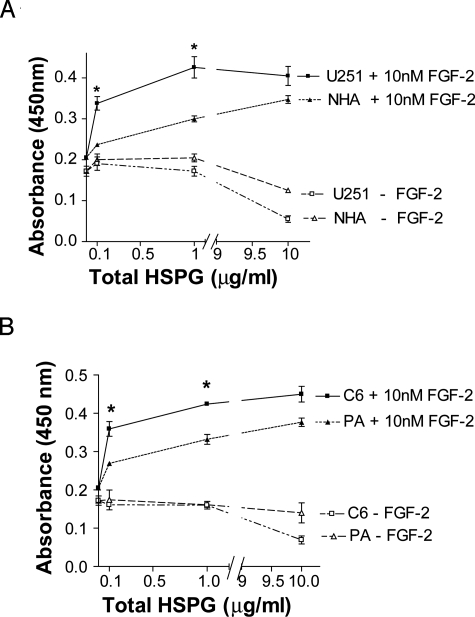
To further study the ability of glioma HSPGs to promote FGF-2 signaling in a more quantitative fashion, we conducted experiments with isolated HSPG preparations. Based on our screening experiments with fixed monolayers, we focused in subsequent studies on a comparison between U251 glioma cells and NHA cells. For an analysis across species, we also compared C6 glioma cells and PA cells. The activity of purified HSPGs was tested directly on FR1c11 reporter cells. Consistent with the result from the monolayer study, extracted U251 and C6 glioma HSPGs displayed a significantly higher activity in promoting FGF-2-induced FR1c11 cell growth than normal astrocyte HSPGs at the concentrations of 0.1 and 1 μg/ml (Figure 3, A and B). At the highest concentration (10 μg/ml), the difference disappeared. This difference in dose response suggests a qualitative difference between HSPGs from normal astrocytes and glioma cells, which can be compensated for by adding higher concentrations of the “activity-deficient” NHA HSPGs.
Figure 3.
FGF-2-induced FR1c11 proliferation with extracted total HSPGs. Extracted total HSPGs (A, human cell HSPGs; B, rat cell HSPGs) were quantified. HSPGs (0, 0.1, 1, and 10 μg/ml) were added to FR1c11 cells in the presence or absence of 10 nmol/L FGF-2. Cell proliferation was determined after 72 hours using a tetrazolium compound-based assay. Data represent mean ± SEM. *P < 0.05 versus NHAs (A) or PAs (B).
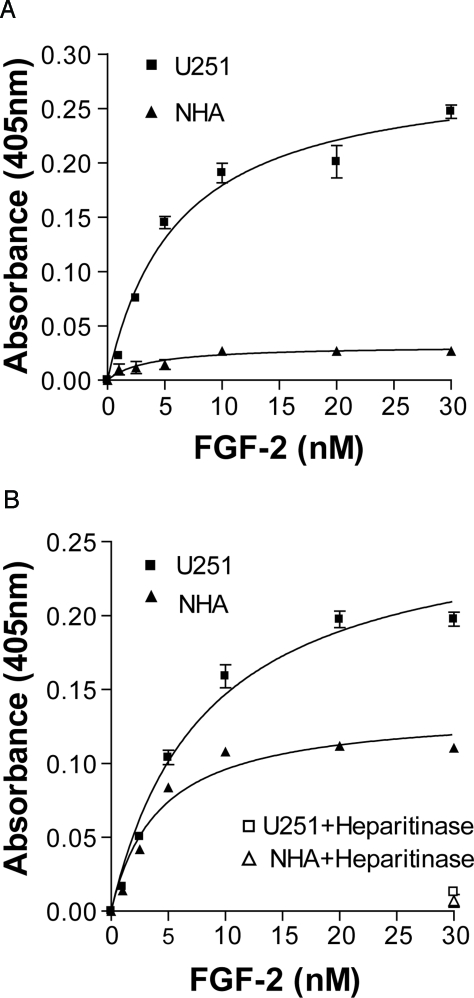
HSPGs regulate the interaction between FGF-2 and FGFR by forming a ternary signaling complex that includes two FGF-2, two FGFR, and heparan sulfate.6,31 The observed elevated activity of U251 and C6 HSPGs in activating the FGF-2-FGFR1c signaling complex may be due to increased affinity and/or binding capacity in the complex. To distinguish between these two possibilities, we examined steady-state binding of FGF-2 to FGFR1c in the presence of HSPGs. In this assay, increasing concentrations of FGF-2 and a fixed amount of FR1c-AP, which is a fusion protein consisting of the extracellular portion of FGFR1c linked to placental AP,21 are added to wells coated with a purified preparation of HSPGs. By measuring the AP activity bound on the plate, we quantitatively tested the ability of HSPGs to promote FGF/FGFR assembly under steady-state conditions. We found that the apparent complex dissociation constant (Kd) on a U251 HSPG-coated plate and a NHA HSPG-coated plate did not differ significantly (6.09 ± 0.973 versus 3.611 ± 1.5 nmol/L; Figure 4A). In contrast, the saturation binding capacity for the FGF-2/HSPG/FGFR1c complex was dramatically higher on U251 HSPG-coated plates (0.288 ± 0.016) compared with NHA HSPG-coated plates (0.032 ± 0.0036, P < 0.05; Figure 4A). This result suggests that the number of heparan sulfate domains capable of mediating a high-affinity interaction between FGF-2 and FGFR1c is increased in glioma cell HSPGs compared with normal astrocyte HSPGs. To confirm this finding and to eliminate potential core protein interference, we performed the steady-state binding assay on HSGAG chains isolated from U251 and NHA cells. Consistent with the HSPG binding results, U251 HSGAG had a significantly higher saturation binding capacity (0.267 ± 0.0097) for the FGF-2/heparan sulfate/FGFR1c complex than NHA heparan sulfate chains (0.136 ± 0.0038, P < 0.05; Figure 4B), although U251 heparan sulfate chains displayed a higher Kd than NHA heparan sulfate chains (8.186 ± 0.79 versus 4.205 ± 0.393 nmol/L). The binding observed was specifically mediated through the heparan sulfate chains because heparitinse digestion completely abolished the binding (Figure 4B).
Figure 4.
FGF-2/FRIc-AP complex assembly on immobilized total HSPGs and HSGAGs. Biotinylated total HSPGs (A) or HSGAGs (B) were quantified and immobilized on streptavidin-coated 96-well plates at 150 ng HSPGs/well or 50 ng HSGAGs/well. Increasing concentrations of FGF-2 and 5 nmol/L FR1-AP were added to the wells as described in Materials and Methods. After coating with HSGAGs, several wells were treated with heparitinase as the negative control for the binding assay. Data represent mean ± SEM. The curves were generated by nonlinear saturation binding curve fit using Prism program (GraphPad Software).
NHA and U251 Cells Express Different HSGAG Sulfotransferases
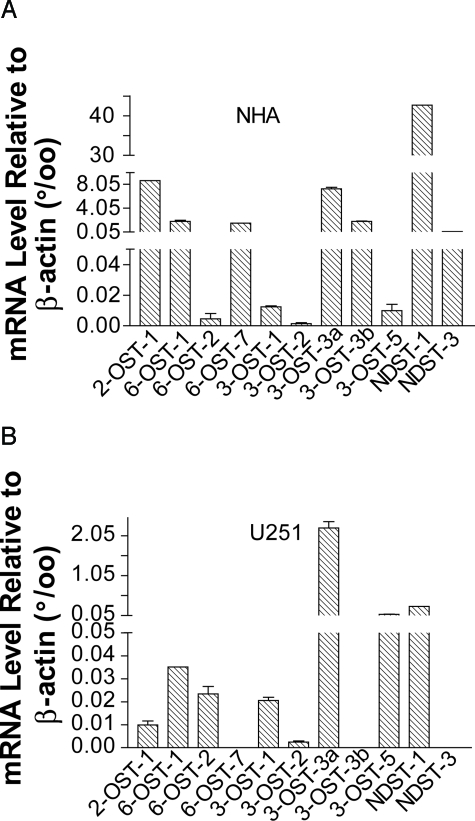
Specific sulfation patterns, and thus protein binding domains, within HSGAG are generated by the concerted action of numerous sulfotransferase enzymes. Many of these enzymes, including isoforms, have only recently been discovered, and antibodies are not available. Recently, a correlation between HSGAG sulfotransferase mRNA levels and HSGAG function during murine brain development has been described.32 We tested by quantitative real-time PCR whether the mRNA expression profile of these enzymes was changed in U251 compared with NHA cells (Figure 5). Both NHA and U251 cells expressed a wide spectrum of HSGAG sulfotransferases, including multiple isoforms of 3-o-sulfotransferase (3-OST) and 6-OST. The expression profiles were distinctly different in NHA versus U251 cells, with NDST-1 being the most abundant enzyme in NHA cells and 3-OST predominating in U251 cells.
Figure 5.
Comparison of sulfotransferase mRNA levels in U251 glioma and NHA cells. Total RNA isolation and quantitative RT-PCR was performed as described in Materials and Methods. Data are present as mRNA level relative to β-actin (A, NHAs; B, U251). Values were calculated as 1000 × 2 to the power of (cycle thresholdβ-actin − cycle thresholdsulfotransferase).
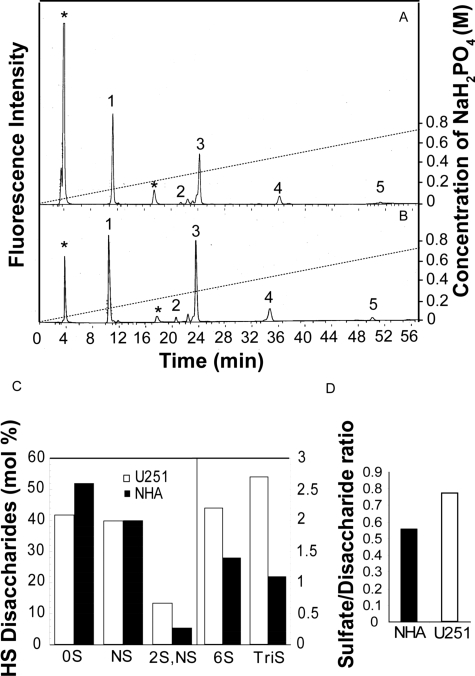
U251 Cell HSGAGs Have a Higher Level of Overall and 6-O Sulfation Than NHA Cells
Sulfotransferase mRNA levels do not allow a direct prediction of HSGAG structure or function. Therefore, we proceeded with a disaccharide analysis of NHA and U251 HSGAGs. High performance liquid chromatography (HPLC) analysis after digestion with a heparinase mixture and 2-AB labeling revealed that HSGAGs from NHA and U251 cells were differentially sulfated (Figure 6). The disaccharide peaks obtained were identified based on the retention times of the au-thentic standard disaccharides and by co-chromatog-raphy. The HSGAGs from U251 cells had a higher degree of sulfated disaccharides especially with respect to ΔHexA-GlcNAc(6-OSO3) (2.2 versus 1.4 mol %) and ΔHexA(2-OSO3)-GlcNSO3 (13.4 versus 5.5 mol %) compared with NHA cells (Figure 6C). Additionally, HSGAG chains from NHA cells had higher proportions of unsulfated disaccharides consequently resulting in a lower sulfate-to-disaccharide ratio compared with U251 cells (0.56 versus 0.77; Figure 6D). There was no recognizable correlation between sulfotransferase mRNA levels and the HSGAG sulfation patterns.
Figure 6.
Disaccharide composition analysis of heparan sulfate chains. GAGs from NHA (A) and U251 (B) cells were digested with heparinase and labeled with 2-AB and analyzed by anion-exchange HPLC on a PA-03 column using a gradient from 16 to 700 mmol/L NaH2PO4 over a period of 1 hour and monitored by fluorescence detection as detailed in Materials and Methods. Based on the elution positions of the standard heparan sulfate disaccharides and comigration, the peaks were identified as follows: 1, 2AB-ΔHexA-GlcNAc; 2, 2AB-ΔHexA-GlcNAc(6-OSO3); 3, 2AB-ΔHexA-GlcNSO3; 4, 2AB-ΔHexA(2-OSO3)-GlcNSO3; and 5, 2AB-ΔHexA(2-OSO3)-GlcNSO3(6-OSO3). The peaks marked by an asterisk are derived from 2-AB reagent. C: Heparan sulfate (HS) disaccharides (mole %) in U251 and NHA cells. D: The sulfate-to-disaccharide ratio of heparan sulfate. The experiment was performed twice with very similar results. Data from one representative experiment are shown.
Gpc-1 Expression Is Elevated in U251 Glioma Cells Compared with NHA Cells
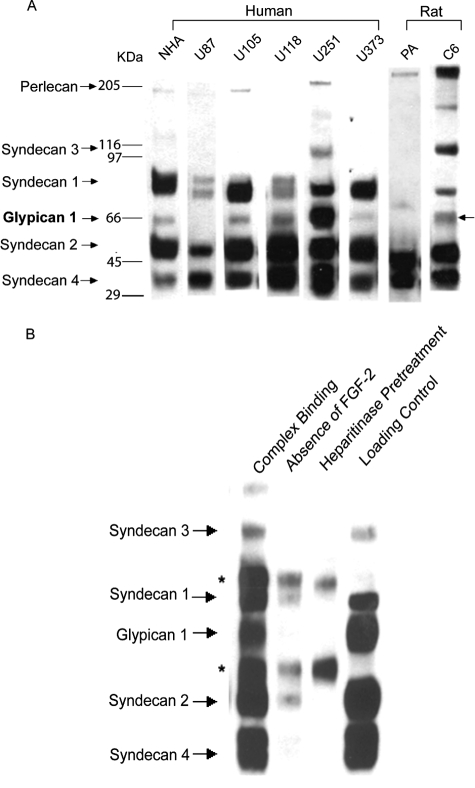
HSPGs can be divided into the general categories of cell-associated forms, which include the syndecans and glypicans, and secreted extracellular matrix forms, such as perlecan. Previous studies indicate that a change in expression of specific HSPG core proteins may result in enhanced mitogenic response of tumor cells to FGF-2. For example, Gpc-1 is overexpressed in human pancreatic cancer and regulates FGF-2 action in the cells.10 We have recently described a specific role for Gpc-1 in promoting FGF-2 signaling in glioma endothelial cells.16 To examine whether expression of specific HSPGs is associated with activity in FGF-2 signaling, we determined HSPG core protein profiles in glioma cells and normal astrocytes. Purified HSPGs were analyzed by gradient SDS-PAGE gel electrophoresis. The blots were probed with an antibody (3G10) that recognizes the heparan sulfate stubs remaining after heparitinase digestion and thus reacts with all HSPGs independent of the identity of the core protein.33 We found that both normal astrocytes and glioma cells expressed a broad spectrum of HSPG core proteins (Figure 7A). Syndecan-2 and -4 were abundant in normal astrocytes (NHAs and PAs) and all of the glioma cell lines tested. Interestingly, the Gpc-1 protein expression level was higher in U251 and in U118 cells compared with the other glioma cells and NHAs, mirroring their HSPG activity in FGF-2 signaling. Also, C6 glioma had an increased Gpc-1 expression compared with PA. The migration pattern of HSPGs had previously been established with core protein-specific antibodies.16 The conditioned medium contained predominantly high molecular weight (presumably extracellular matrix) HSPGs and some Gpc-1 (not shown).
Figure 7.
HSPG core protein expression and FGF-2/FGFR complex precipitation. A: Core protein expression in normal astrocytes and glioma cells. HSPGs (200,000 cell equivalents) were digested with heparitinase and chondroitinase to degrade GAG chains and then analyzed by SDS-PAGE using a 3.5 to 15% gradient gel and transferred to a PVDF membrane as described in Materials and Methods. Cell equivalents (2 × 105) of each type of cell were loaded onto the gel. The blot was probed with 3G10 antibody. B: Fractionation of HSPGs from U251 cells according to their ability to promote the FGF-2/FGFR complex. HSPGs isolated from U251 cells (2 × 106 cell equivalents) were incubated with FGFR1c-alkaline phosphatase (FRIc-AP) fusion protein immobilized on agarose beads in the presence of 10 nmol/L FGF-2. After washing and digestion with heparitinase and chondroitinase, HSPGs complexed to the beads were analyzed by SDS-PAGE. The membrane was probed with 3G10 antibody. A total HSPG loading control was included. Negative controls included digestion of the total HSPGs with heparitinase before complex formation and omission of FGF-2 from the binding reaction. The asterisks indicate the bands generated by interaction of the secondary detection antibody with heavy and light chain of the anti-AP antibody.
We next performed quantitative RT-PCR to examine HSPG core expression at the mRNA level (Table 2). mRNA and protein levels of NHA and glioma cells were in general agreement with the exception of syndecan-3, which showed a poor correlation. Gpc-1 was the most dominant HSPG in U251 cells with mRNA copy numbers exceeding those of NHA cells by a factor of approximately 30.
Table 2.
Quantitation of HSPG mRNA Level in NHA and Human Glioma Cells
| mRNA copy number per 106 copies of β-actin mRNA
|
||||||
|---|---|---|---|---|---|---|
| Glypican-1 | Perlecan | Syndecan-1 | Syndecan-2 | Syndecan-3 | Syndecan-4 | |
| NHA | 9,728 ± 2,197 | 10,306 ± 282 | 17,056 ± 797 | 9,611 ± 361 | 138,889 ± 5,300 | 10,944 ± 681 |
| U87 | 4,882 ± 788 | 2,286 ± 166 | 5,963 ± 425 | 7,501 ± 7 | 2,654 ± 311 | 21,765 ± 1,219 |
| U105 | 6,683 ± 1,704 | 4,598 ± 277 | 7,716 ± 210 | 8,733 ± 371 | 34,711 ± 2,649 | 8,460 ± 260 |
| U118 | 7,667 ± 2,430 | 3,056 ± 662 | 1,745 ± 87 | 16,275 ± 748 | 521 ± 58 | 12,843 ± 276 |
| U251 | 318,596 ± 28,176 | 36 ± 11 | 5 ± 2 | 19,663 ± 730 | 14 ± 3 | 1,926 ± 131 |
| U373 | 17,596 ± 2,872 | 521 ± 42 | 6,542 ± 410 | 12,724 ± 210 | 124,038 ± 4,441 | 4,054 ± 120 |
Total RNA isolation and quantitative RT-PCR was performed as described in Materials and Methods. Individual standard ranges were used for different genes to ascertain that samples were within the range. All of the copy numbers were normalized to 106 copies of β-actin mRNA. Data indicate mean ± SEM.
To study whether Gpc-1 directly participates in the FGF-2 signaling complex, we analyzed the composition of HSPG core proteins found in FGF-2/FGFR1c complexes formed in the presence of U251 cell HSPGs in vitro. Gpc-1 and other U251 glioma cell HSPGs co-immunoprecipitated with FGFR1c in the presence of FGF-2 (Figure 7B). Complex formation was FGF-2 and heparan sulfate dependent, because it was completely abolished by omitting FGF-2 from the reaction or degrading heparan sulfate with heparitinase before binding. This result indicates that all glioma cell HSPG core proteins are decorated with heparan sulfate chains similarly capable of promoting stable binding of FGF-2 to FGFR1c, albeit subtle differences between HSPG types cannot be excluded with this qualitative assay.
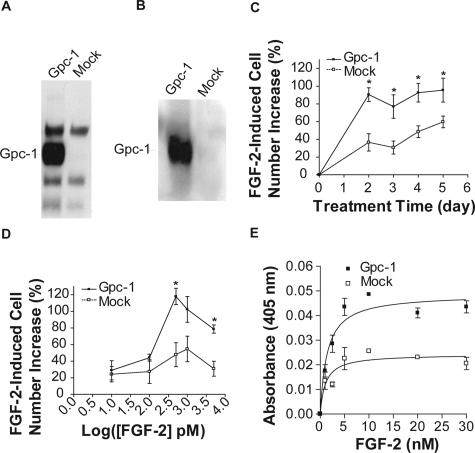
Gpc-1 Overexpression Increases the FGF-2 Response of U87 Glioma Cells
To further confirm an important role of Gpc-1 in FGF-2 signaling, we examined whether expression of this HSPG core protein in a glioma cell line deficient in Gpc-1 would increase the FGF-2 response. Stable transfection of U87 cells with human Gpc-1 cDNA leads to elevated Gpc-1 protein levels (Figure 8, A and B). Compared with the mock-transfected cells, Gpc-1-transfected cells showed an enhanced response to FGF-2. After 2 days of treatment with 500 pmol/L FGF-2, Gpc-1-transfected cells already reached their maximum response, whereas control cell proliferation peaked after 5 days of treatment with FGF-2 (Figure 8C). Gpc-1 overexpressing cells also showed a greater response to FGF-2 than control cells in a dose-response experiment (Figure 8D). In the FGF-2/HSGAG/FGFR steady-state complex binding analysis, the apparent complex dissociation constant (Kd) was similar for HSPGs from Gpc-1-transfected cells and mock-transfected cells (Figure 8E; 1.418 ± 0.386 versus 1.07 ± 0.488 nmol/L). However, the maximal binding capacity of HSPG from Gpc-1-transfected cells was significantly increased compared with HSPGs from control cells (0.049 ± 0.0027 versus 0.024 ± 0.002; P < 0.05).
Figure 8.
The effect of Gpc-1 overexpression on U87 FGF-2 response. A: The Gpc-1 protein level was significantly increased in U87 cells stably transfected with Gpc-1 cDNA. Total HSPGs (500,000 cell equivalents) were extracted from both mock-transfected and Gpc-1-transfected cells. After digestion with heparitinase and chondroitinase, HSPG extracts were analyzed by SDS-PAGE using a 3.5 to 15% gradient gel and transferred to a PVDF membrane as described in Materials and Methods. The blot was probed with 3G10 antibody. B: The blot shown in A was re-probed with anti-human Gpc-1 antibody. C: Time course of FGF-2 response of Gpc-1-transfected U87 and mock-transfected U87 cells. After 24 hours of starvation, cells were treated with FGF-2 (500 pmol/L). Cells were trypsinized and counted with a Coulter Counter at the days indicated. Data points indicate cell number increase of FGF-2-treated cells compared with the no-treatment control. *P < 0.05. D: The FGF-2 dose response of Gpc-1-transfected U87 and mock-transfected U87 cells. Cells were treated with 0, 10, 100, 500, 1000, and 5000 pmol/L FGF-2 for 3 days and then counted. *P < 0.05 versus mock-transfected cells. E: FGF-2/FR1-AP complex assembly on immobilized HSPGs from Gpc-1-transfected or mock-transfected U87 cells. Binding was performed according to the description of Figure 4. Data represent mean ± SEM. The curves were generated by nonlinear saturation binding curve fit using Prism program (GraphPad Software).
Gliomas Overexpress Gpc-1
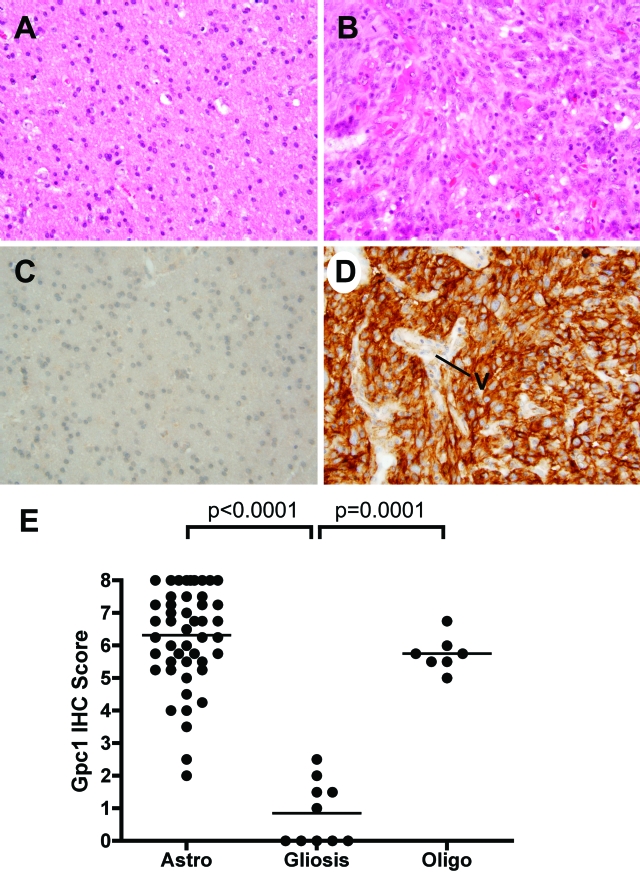
The correlation between Gpc-1 overexpression and HSPG activity in the cell lines prompted us to examine Gpc-1 levels in human glioma samples. A tissue array was assembled, containing duplicate samples from 49 astrocytomas (4 grade I, 2 grade II, 5 grade III, and 39 grade IV tumors), 7 oligodendrogliomas, and 10 temporal lobe gliosis cases as controls. Gpc-1 was either undetectable or weakly expressed by astrocytes in the non-neoplastic samples (mean score, 0.85 ± 0.97; Figure 9, A, C, and E). In contrast, astrocytomas and oligodendrogliomas expressed Gpc-1 at elevated levels (mean scores, 6.3 ± 1.5 and 5.75 ± 0.54, respectively; Figure 9, B, D, and E). Eleven astrocytomas (22%) showed strong expression diffusely throughout the tumor (Figure 9D). The difference in immunohistochemistry scores between the astrocytomas and the non-neoplastic controls was highly significant (P < 0.0001; Figure 9E). Only two astrocytomas overlapped with the expression range seen in non-neoplastic gliosis. There was no significant correlation between Gpc-1 expression and tumor grade, albeit the number of lower grade tumors was low. Gpc-1 expression in tumor vessel endothelial cells was seen in 20 astrocytomas (41%), which is in keeping with our previous observations.16 In summary, Gpc-1 overexpression is an almost universal feature of gliomas.
Figure 9.
Expression levels of Gpc-1 in human tumor samples. Tissue array slides containing samples from both gliomas and non-neoplastic gliosis cases were stained with hematoxylin-eosin (H&E) and immunolabeled with an antibody to Gpc-1 (mouse monoclonal antibody S1, 5 μg/ml). Original magnification for all photomicrographs is ×400. A: Temporal lobe gliosis; H&E stain. B: Grade IV astrocytoma, H&E stain. C: Temporal lobe gliosis (same case as A) immunolabeled for Gpc-1. D: Grade IV atrocytoma (same case as B) immunolabeled for Gpc-1. The tumor cells show diffuse and strong staining; tumor vessel (V) endothelial cells stain weakly in this sample. E: Gpc-1 IHC scores of 49 astrocytomas, 10 gliosis cases, and 7 oligodendrogliomas. The scores were significantly different by nonparametric Mann Whitney test as indicated. The horizontal lines indicate the means.
Discussion
Previous studies by other investigators have shown that FGF-2 signaling in glioma cells is altered compared with normal astrocytes, because gliomas contain elevated levels of FGF-2, switch their FGFR repertoire to optimally respond to FGF-2, and are stimulated in growth by an FGF-2-mediated autocrine loop.3–5 We now show that HSPG coreceptors are also altered in glioma cells, contributing to increased FGF-2 responsiveness. In contrast, normal astrocyte HSPG is ill suited to support an FGF-2-mediated mitogenic signal. Alterations in glioma cell HSPGs appear to be quantitative in nature at the level of core protein expression and qualitative at the level of heparan sulfate structure and function.
HSPGs conducive to FGF-2 signaling were found in two of five human glioma cell lines and in C6 rat glioma cells. FGFR1c, the principal FGF-2 signaling receptor, was co-expressed at high levels, setting the stage for an autocrine and/or paracrine mitogenic pathway. These results illustrate that a functional FGF-2 signaling pathway exists in a considerable proportion of, but not all, glioma cell lines. The heterogeneity displayed by human glioma cell lines with respect to FGF-2 signaling could be due to variances in the spectrum of HSPGs and FGFRs. During glioma cell progression, the requirement for HSPG coreceptors may be bypassed by the activation of alternative signaling pathways. The HSPG-independent EGF signaling axis is turned on in U87 and U373 cells, which may dominate over FGF-2 signaling events.28,29 Mitogenic loops involving PDGF and TGF have also been reported.34–36 Glioma growth regulation can also be perturbed downstream of growth factor receptors. The tumor suppressor phosphatase and tensin homolog (PTEN), located on chromosome 10, is frequently lost in gliomas, leading to constitutive phosphorylation of AKT and autonomous growth.36
In this study, we describe a striking overexpression of Gpc-1 in the majority of human gliomas compared with non-neoplastic astrocytes. In glioma cell lines, a correlation between Gpc-1 expression and FGF-2 co-stimulator activity was identified. Glypicans have been found to play a crucial role in FGF-2 signaling,16,37,38 Wnt signaling,39 and mammalian development.40 The observations on glioma cells reported here fit into the emerging picture of a special role for Gpc-1 in malignancy. Gpc-1 is overexpressed in pancreatic cancers, and enzymatic removal of glycosylphosphotidylinositol-anchored cell-surface molecules or silencing of Gpc-1 expression with antisense RNA inhibits pancreatic carcinoma cell growth.10 Similarly, in breast carcinomas, Gpc-1 was required for the activity of “heparin binding” growth factors heregulin and hepatocyte growth factor/scatter factor.41 We have recently shown that Gpc-1 overexpression in glioma vessel endothelial cells contributes to FGF-2 responsiveness and tumor angiogenesis.16
Gpc-1 is clearly equipped with heparan sulfate chains capable of promoting FGF-2 binding to FGFR1c, but other cell-surface HSPGs may also be involved (Figure 7B). In intact cells, Gpc-1 may have a special role in FGF-2 signaling by virtue of its GPI anchor, which serves as a constitutive cell membrane lipid raft localization signal. FGFR substrate 2, an obligatory FGFR adaptor molecule, is also exclusively found in lipid rafts, and thus the two molecules form a “signaling scaffold” in these membrane microdomains. However, we find that Gpc-1-rich glioma cell HSPGs promote FGF-2 signaling even when they are removed from the cell surface as purified preparations. This observation could be explained by a recent finding by Chen and Lander42 that the Gpc-1 globular domain steers GAG synthesis toward a higher content of heparan sulfate versus chondroitin sulfate. Interestingly, this effect was not limited to GAG on the Gpc-1 core protein but applied to the other core proteins in a “trans” effect.
Our in vitro assays with FR1c-11 cells demonstrate that U251 glioma HSGAGs are qualitatively superior to NHA HSGAGs in promoting FGF-2 signaling via FGFR1c. This elevated activity can likely be attributed to the higher degree of sulfation found in the glioma cell HSGAGs and, specifically, the elevated levels of 6-O sulfate substitution on N-acetylglucosamine. Several lines of evidence point toward a crucial role for this sulfate group in FGF-2 signaling, although other investigators identified a privileged role for 2-O-sulfate on uronate.43,44 Selective removal of 6-O sulfate from heparin fragments or HSGAG oligosaccharides renders these molecules inactive in FGF-2 signaling.45,46 Overexpression of HSulf1 and/or HSulf2, endosulfatases with substrate specificity toward 6-O sulfate, diminishes ternary FGF-2/HSGAG/FGFR1c complex binding on the cell surface and suppresses FGF-2-induced signaling and mitogenesis.47,48 There is mounting evidence that relatively subtle differences in HSGAG sulfation can have dramatic effects on FGF signaling.44,49
In summary, the results of our study indicate that HSPGs from a subset of human glioma cells and rat glioma cells have a greater capability of promoting FGF-2 signaling than HSPGs from normal human or primary rat astrocytes. The enhanced HSPG activity correlated with high expression levels of Gpc-1 and structural HSGAG alterations. These observations indicate the importance of HSPG coreceptors in autocrine growth stimulation by FGF-2. The heterogeneity of mitogenic signaling pathways in gliomas suggests that it may be futile to search for a universally active glioma treatment. Improved therapy success for this devastating disease may be achieved by the characterization of disregulated mitogenic pathways in individual tumors followed by targeted and individualized treatment.
Acknowledgments
We thank Karianna Decker for excellent technical assistance in complex binding experiment. This material is based on work supported by the Office of Research and Development, Department of Veterans Affairs.
Footnotes
Address reprint requests to Andreas Friedl, University of Wisconsin-Madison, Department of Pathology and Laboratory Medicine, Clinical Sciences Center K4/850, 600 Highland Ave., Madison, WI 53792-8550. E-mail: afriedl@wisc.edu.
Supported in part by National Institutes of Health grant R01 NS048921-01 and American Cancer Society grant RSG-01-068-01-CSM.
References
- Takahashi JA, Mori H, Fukumoto M, Igarashi K, Jaye M, Oda Y, Kikuchi H, Hatanaka M. Gene expression of fibroblast growth factors in human gliomas and meningiomas: demonstration of cellular source of basic fibroblast growth factor mRNA and peptide in tumor tissues. Proc Natl Acad Sci USA. 1990;87:5710–5714. doi: 10.1073/pnas.87.15.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagzag D, Miller DC, Sato Y, Rifkin DB, Burstein DE. Immunohistochemical localization of basic fibroblast growth factor in astrocytomas. Cancer Res. 1990;50:7393–7398. [PubMed] [Google Scholar]
- Saxena A, Ali IU. Increased expression of genes from growth factor signaling pathways in glioblastoma cell lines. Oncogene. 1992;7:243–247. [PubMed] [Google Scholar]
- Gross JL, Morrison RS, Eidsvoog K, Herblin WF, Kornblith PL, Dexter DL. Basic fibroblast growth factor: a potential autocrine regulator of human glioma cell growth. J Neurosci Res. 1990;27:689–696. doi: 10.1002/jnr.490270429. [DOI] [PubMed] [Google Scholar]
- Yamaguchi F, Saya H, Bruner JM, Morrison RS. Differential expression of two fibroblast growth factor-receptor genes is associated with malignant progression in human astrocytomas. Proc Natl Acad Sci USA. 1994;91:484–488. doi: 10.1073/pnas.91.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- Raman R, Venkataraman G, Ernst S, Sasisekharan V, Sasisekharan R. Structural specificity of heparin binding in the fibroblast growth factor family of proteins. Proc Natl Acad Sci USA. 2003;100:2357–2362. doi: 10.1073/pnas.0437842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull J, Powell A, Guimond S. Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 2001;11:75–82. doi: 10.1016/s0962-8924(00)01897-3. [DOI] [PubMed] [Google Scholar]
- Raman R, Sasisekharan V, Sasisekharan R. Structural insights into biological roles of protein-glycosaminoglycan interactions. Chem Biol. 2005;12:267–277. doi: 10.1016/j.chembiol.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Kleeff J, Ishiwata T, Kumbasar A, Friess H, Buchler MW, Lander AD, Korc M. The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J Clin Invest. 1998;102:1662–1673. doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- Filmus J. Glypicans in growth control and cancer. Glycobiology. 2001;11:19R–23R. doi: 10.1093/glycob/11.3.19r. [DOI] [PubMed] [Google Scholar]
- Adatia R, Albini A, Carlone S, Giunciuglio D, Benelli R, Santi L, Noonan DM. Suppression of invasive behavior of melanoma cells by stable expression of anti-sense perlecan cDNA. Ann Oncol. 1997;8:1257–1261. doi: 10.1023/a:1008243115385. [DOI] [PubMed] [Google Scholar]
- Steck PA, Moser RP, Bruner JM, Liang L, Freidman AN, Hwang TL, Yung WK. Altered expression and distribution of heparan sulfate proteoglycans in human gliomas. Cancer Res. 1989;49:2096–2103. [PubMed] [Google Scholar]
- Allen BL, Filla MS, Rapraeger AC. Role of heparan sulfate as a tissue-specific regulator of FGF-4 and FGF receptor recognition. J Cell Biol. 2001;155:845–858. doi: 10.1083/jcb.200106075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao D, Meyer K, Mundhenke C, Drew SA, Friedl A. Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 signaling in brain endothelial cells: specific role for glypican-1 in glioma angiogenesis. J Biol Chem. 2003;278:16045–16053. doi: 10.1074/jbc.M211259200. [DOI] [PubMed] [Google Scholar]
- Mundhenke C, Meyer K, Drew S, Friedl A. Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 receptor binding in breast carcinomas. Am J Pathol. 2002;160:185–194. doi: 10.1016/S0002-9440(10)64362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Bjornsson S. Quantitation of proteoglycans in biological fluids using Alcian blue. Methods Mol Biol. 2001;171:159–173. doi: 10.1385/1-59259-209-0:159. [DOI] [PubMed] [Google Scholar]
- Lee PH, Trowbridge JM, Taylor KR, Morhenn VB, Gallo RL. Dermatan sulfate proteoglycan and glycosaminoglycan synthesis is induced in fibroblasts by transfer to a three-dimensional extracellular environment. J Biol Chem. 2004;279:48640–48646. doi: 10.1074/jbc.M407241200. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar S, Esterman MA, Hoffman HA. Microdetermination of proteoglycans and glycosaminoglycans in the presence of guanidine hydrochloride. Anal Biochem. 1987;161:103–108. doi: 10.1016/0003-2697(87)90658-0. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Yayon A, Flanagan JG, Svahn CM, Levi E, Leder P. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol Cell Biol. 1992;12:240–247. doi: 10.1128/mcb.12.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Sugahara K. Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high-performance liquid chromatography: application to disaccharide composition analysis and exosequencing of oligosaccharides. Anal Biochem. 1999;269:367–378. doi: 10.1006/abio.1999.4027. [DOI] [PubMed] [Google Scholar]
- David G, Van der SB, Marynen P, Cassiman JJ, Van Den BH. Molecular cloning of amphiglycan, a novel integral membrane heparan sulfate proteoglycan expressed by epithelial and fibroblastic cells. J Cell Biol. 1992;118:961–969. doi: 10.1083/jcb.118.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Glimelius B, Norling B, Westermark B, Wasteson A. Composition and distribution of glycosaminoglycans in cultures of human normal and malignant glial cells. Biochem J. 1978;172:443–456. doi: 10.1042/bj1720443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard C, Liuzzo JP, Moscatelli D. Fibroblast growth factor-2 can mediate cell attachment by linking receptors and heparan sulfate proteoglycans on neighboring cells. J Biol Chem. 1995;270:24188–24196. doi: 10.1074/jbc.270.41.24188. [DOI] [PubMed] [Google Scholar]
- Kasza A, Kowanetz M, Poslednik K, Witek B, Kordula T, Koj A. Epidermal growth factor and pro-inflammatory cytokines regulate the expression of components of plasminogen activation system in U373-MG astrocytoma cells. Cytokine. 2001;16:187–190. doi: 10.1006/cyto.2001.0957. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang Y, Cao L, Chen L, Lee V, Zheng PS, Kiani C, Adams ME, Ang LC, Paiwand F, Yang BB. Identification of the motif in versican G3 domain that plays a dominant-negative effect on astrocytoma cell proliferation through inhibiting versican secretion and binding. J Biol Chem. 2001;276:14178–14186. doi: 10.1074/jbc.M100618200. [DOI] [PubMed] [Google Scholar]
- Glimelius G, Norling B, Westermark B, Wasteson A. Turnover of cell surface associated glycosaminoglycans in cultures of human normal and malignant glial cells. Exp Cell Res. 1978;117:179–189. doi: 10.1016/0014-4827(78)90440-8. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- Turnbull J, Drummond K, Huang Z, Kinnunen T, Ford-Perriss M, Murphy M, Guimond S. Heparan sulphate sulphotransferase expression in mice and Caenorhabditis elegans. Biochem Soc Trans. 2003;31:343–348. doi: 10.1042/bst0310343. [DOI] [PubMed] [Google Scholar]
- David G, Bai XM, Van der SB, Cassiman JJ, Van Den BH. Developmental changes in heparan sulfate expression: in situ detection with mAbs. J Cell Biol. 1992;119:961–975. doi: 10.1083/jcb.119.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto M, Takahashi JA, Murai N, Ueba T, Kono K, Nakatsu S. Induction of apoptosis in glioma cells: an approach to control tumor growth by blocking basic fibroblast growth factor autocrine loop. Anticancer Res. 2000;20:4059–4065. [PubMed] [Google Scholar]
- Tang P, Steck PA, Yung WK. The autocrine loop of TGF-alpha/EGFR and brain tumors. J Neurooncol. 1997;35:303–314. doi: 10.1023/a:1005824802617. [DOI] [PubMed] [Google Scholar]
- Kapoor GS, O’Rourke DM. Mitogenic signaling cascades in glial tumors. Neurosurgery. 2003;52:1425–1434. doi: 10.1227/01.neu.0000065135.28143.39. [DOI] [PubMed] [Google Scholar]
- Steinfeld R, Van Den BH, David G. Stimulation of fibroblast growth factor receptor-1 occupancy and signaling by cell surface-associated syndecans and glypican. J Cell Biol. 1996;133:405–416. doi: 10.1083/jcb.133.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HH, Shi W, Filmus J. OCI-5/rat glypican-3 binds to fibroblast growth factor-2 but not to insulin-like growth factor-2. J Biol Chem. 1997;272:7574–7577. doi: 10.1074/jbc.272.12.7574. [DOI] [PubMed] [Google Scholar]
- Lin X, Perrimon N. Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature. 1999;400:281–284. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- Song HH, Filmus J. The role of glypicans in mammalian development. Biochim Biophys Acta. 2002;1573:241–246. doi: 10.1016/s0304-4165(02)00390-2. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Maruyama H, Guo F, Kleeff J, Itakura J, Matsumoto Y, Lander AD, Korc M. Glypican-1 is overexpressed in human breast cancer and modulates the mitogenic effects of multiple heparin-binding growth factors in breast cancer cells. Cancer Res. 2001;61:5562–5569. [PubMed] [Google Scholar]
- Chen RL, Lander AD. Mechanisms underlying preferential assembly of heparan sulfate on glypican-1. J Biol Chem. 2001;276:7507–7517. doi: 10.1074/jbc.M008283200. [DOI] [PubMed] [Google Scholar]
- Ishihara M, Kariya Y, Kikuchi H, Minamisawa T, Yoshida K. Importance of 2-O-sulfate groups of uronate residues in heparin for activation of FGF-1 and FGF-2. J Biochem (Tokyo) 1997;121:345–349. doi: 10.1093/oxfordjournals.jbchem.a021593. [DOI] [PubMed] [Google Scholar]
- Berry D, Shriver Z, Natke B, Kwan CP, Venkataraman G, Sasisekharan R. Heparan sulphate glycosaminoglycans derived from endothelial cells and smooth muscle cells differentially modulate fibroblast growth factor-2 biological activity through fibroblast growth factor receptor-1. Biochem J. 2003;373:241–249. doi: 10.1042/BJ20021760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye DA, Vives RR, Turnbull JE, Hyde P, Gallagher JT. Heparan sulfate oligosaccharides require 6-O-sulfation for promotion of basic fibroblast growth factor mitogenic activity. J Biol Chem. 1998;273:22936–22942. doi: 10.1074/jbc.273.36.22936. [DOI] [PubMed] [Google Scholar]
- Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salmivirta M, Claesson-Welsh L. Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J Biol Chem. 2000;275:24653–24660. doi: 10.1074/jbc.M908930199. [DOI] [PubMed] [Google Scholar]
- Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, Smith DI, Kaufmann SH, Roberts LR, Shridhar V. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, Shriver Z, Venkataraman G, Sasisekharan R, Sanderson RD. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280:40066–40073. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- Pan C, Nelson MS, Reyes M, Koodie L, Brazil JJ, Stephenson EJ, Zhao RC, Peters C, Selleck SB, Stringer SE, Gupta P. Functional abnormalities of heparan sulfate in mucopolysaccharidosis-I are associated with defective biologic activity of FGF-2 on human multipotent progenitor cells. Blood. 2005;106:1956–1964. doi: 10.1182/blood-2005-02-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]