Abstract
Molecular surveys of meiofaunal diversity face some interesting methodological challenges when it comes to interstitial nematodes from soils and sediments. Morphology-based surveys are greatly limited in processing speed, while barcoding approaches for nematodes are hampered by difficulties of matching sequence data with traditional taxonomy. Intermediate technology is needed to bridge the gap between both approaches. An example of such technology is video capture and editing microscopy, which consists of the recording of taxonomically informative multifocal series of microscopy images as digital video clips. The integration of multifocal imaging with sequence analysis of the D2D3 region of large subunit (LSU) rDNA is illustrated here in the context of a combined morphological and barcode sequencing survey of marine nematodes from Baja California and California. The resulting video clips and sequence data are made available online in the database NemATOL (http://nematol.unh.edu/). Analyses of 37 barcoded nematodes suggest that these represent at least 32 species, none of which matches available D2D3 sequences in public databases. The recorded multifocal vouchers allowed us to identify most specimens to genus, and will be used to match specimens with subsequent species identifications and descriptions of preserved specimens. Like molecular barcodes, multifocal voucher archives are part of a wider effort at structuring and changing the process of biodiversity discovery. We argue that data-rich surveys and phylogenetic tools for analysis of barcode sequences are an essential component of the exploration of phyla with a high fraction of undiscovered species. Our methods are also directly applicable to other meiofauna such as for example gastrotrichs and tardigrades.
Keywords: identification, taxonomy, Nematoda, meiofauna, Gulf of California
1. Capturing nematode diversity, identity and morphology
Nematodes are estimated to be the most abundant metazoans on earth (Lambshead 2004). Even though they probably constitute one of the most diverse metazoan phyla in terms of species richness, the overwhelming majority of these species remains unknown (Hugot et al. 2001; Coomans 2002). Contrary to common assumption, many nematode taxa are morphologically highly diverse (De Ley in press), but this diversity remains under-appreciated as it often requires high-resolution light microscopy and electron microscopy to be observed, as well as knowledge of the wide range of forms described in specialized taxonomic literature. Nematode identification with morphological characters is often difficult and laborious because of these same two requirements. To make matters worse, nematode species can be variable in morphology, while the differences between valid species can be obscured by cryptic diagnostic differences (e.g. De Ley et al. 1999). As a result, ecological studies and surveys of nematode diversity are usually restricted to identifications at genus level, since most genera can be identified at first sight, while taxonomic surveys hardly ever approach completeness in identifying all species isolated from all but a few samples.
Aside from the challenges of identification, vouchering of microscopic nematode specimens presents some interesting problems in its own right. A number of well-established protocols exist for preserving and mounting nematodes in permanent slides, and in our experience type specimens may survive over a hundred years of storage. Unfortunately, in practice it turns out that within 5–15 years most vouchers deteriorate substantially, are shattered accidentally or go missing, not least because of the steady erosion of long-term investment in taxonomic collections. Obtaining detailed images of their appearance could mitigate the loss of type specimens. A fundamental limitation in the documentation and vouchering of microscopic invertebrates has always been the critical need for observation with high-resolution microscopy, before any level of accuracy can be attained in interpretation of characters and identification of species. Nematode specimens, as a particular case in point, are not only thin and transparent but are also three-dimensional objects that must be examined de rigueur with multiple magnifications and at multiple focal planes. This multifocal aspect explains why in most cases single photographs are not sufficient to document nematode morphology, and why re-examination of type specimens is often really necessary (though rarely possible) if published drawings and descriptions do not suffice to confirm or reject conspecificity. In order to fully capture the multifocal nature of nematode microscopy, a new approach for recording nematode morphology was therefore proposed by De Ley & Bert (2002). Inspired by the complex software and equipment used to record nematode embryonic development with a technique known as ‘four-dimensional microscopy’ (Thomas et al. 1996; Schnabel et al. 1997), this new approach consists of capturing a multifocal series of images to computer hard disk in the format of a single video clip. This clip can then be edited with any consumer-type video editing program to optimize it for distribution via the Internet or on CD. The resulting technique was dubbed ‘video capture and editing’ (VCE) and examples of some of its many different applications can be seen at http://faculty.ucr.edu/~pdeley/vce.html on the World Wide Web.
The process of capturing morphology of transparent microscopy specimens as multifocal clips is highly adaptable and scaleable. It can be applied at minimal expense in any laboratory equipped with at least one good research microscope, one video camera and one personal computer. In this case, nematode images can be recorded directly as video clips while simply focusing by hand through parts of the specimen. However, with sufficient resources the process can be qualitatively optimized and standardized using an automated computer-controlled microscope and digital camera. A first example of the latter approach was published by Eyualem et al. (2004) to document the findings of an exploratory marine nematode survey in the Gulf of Maine. In this case, nematode morphology was recorded by stepwise capturing of successive focal plans as image stacks, which were subsequently converted into digital video format to produce digital multifocal images (DMI) that are viewable in any multimedia player software. Public access to the resulting clips is available through a versatile online database called NemATOL (http://nematol.unh.edu/).
2. Linking barcoded genotypes with phenotypes
Molecular methods have opened up a wide range of new approaches to nematode surveys, particularly in the context of characterizing short sequence stretches amenable to species identification. Two recent developments are especially significant for barcoding applications. The first of these is the development of a phylogeny-based species concept for nematodes that is both theoretically sound and applicable in diagnosis (Adams 1998, 2001; Nadler 2002). The second is the development of nematode barcoding techniques that attain maximal processivity by foregoing any attempt at morphological vouchering or species identification, focusing instead on assignment to molecular operational taxonomic units (MOTU; Floyd et al. 2002; Blaxter 2004; Blaxter et al. 2005). It has been argued in the context of such surveys that it is impossible to describe biodiversity with traditional approaches (Blaxter 2003), that DNA sequences should become the basis of a new taxonomic reference system (Tautz et al. 2003; Blaxter 2004) and that cytochrome oxidase subunit I (cox1) is an especially appropriate choice for species discrimination in triploblastic Metazoa (Hebert et al. 2003). In the heat of the debate over these exciting new directions and their implied priorities for funding and collaboration, others have strongly cautioned against the dangers of mistaking the huge potential of new approaches for a justification to effectively sideline established methodologies and expertise (Lipscomb et al. 2003; Wheeler 2004).
Despite explicit statements to the contrary by proponents of new DNA-based standards for identification and classification (Hebert et al. 2003; Tautz et al. 2003; Blaxter 2004), there is a clear suspicion among other members of the taxonomic community that such approaches will yet again draw away from the already very meagre resources for morphological studies. This suspicion probably partly derives from comparisons made with current practice in microbial taxonomy, where genotypic criteria for species identification are now seemingly given absolute primacy over older phenotypic standards. In reality, microbiologists approach the thorny issue of prokaryote identification and classification with levels of ambiguity and circumspection (e.g. Torsvik et al. 2002) that are perhaps even greater than those found among eukaryote taxonomists, and it would be erroneous to assume that the former have altogether given up on expressed characters in general and on morphology in particular. Much like prokaryotes, nematodes have the paradoxical reputation of being biologically diverse yet morphologically uniform, to the point where it is tempting to conclude that nematode taxonomy should essentially reset itself and switch to similar principles as those of microbiology. In reality, nematodes harbour endless morphological diversity and the genuine problems with ‘traditional’ nematode identification are instead rooted in quite different obstacles. These obstacles include (but are not limited to) the particular constraints of observing, interpreting and representing nematode morphology through various microscopy techniques. Scanning or transmission electron microscopy often reveals clear diagnostic characters, but is not applicable on the scale required for routine surveys. Light microscopy is much more limited in attainable magnification and resolution, and furthermore imposes specific constraints on the nature of the visual information it generates.
In this paper, we present the next logical development in the use of multifocal recordings: an integrative approach for morphological vouchering of individual nematodes, as well as other meiofauna, in the context of data-rich nematode surveys combining microscopy with molecular barcoding. Although our strategy does not maximize specimen processivity to the extent of barcoding-only approaches (e.g. Floyd et al. 2002), we argue that barcoding combined with morphological vouchering is as much of an essential component of the exploration of biodiversity in nematodes as it is in other organisms. Such a combination not only maintains a direct link with established taxonomy and ecology, but it also provides a set of reference morphologies and sequences that are essential for interpreting the data obtained from barcoding-only surveys. We strongly believe that such an approach is essential for sustained growth of taxonomy as a holistic discipline employing an ever-widening array of tools and benefiting from an ever-increasing range of data sources.
3. Material and methods
(a) Sample preparation and image capture
In order to maximize taxonomic diversity and optimize our methods, we sought to include as wide a range of nematode species as possible. Specifically, we focused on marine nematodes as an excellent test bed for establishing our approach, since they include some of the most neglected major nematode taxa, they are uniquely challenging in terms of their potentially enormous diversity (Lambshead 1993), their biogeographical record is largely limited to northwestern Europe, and they are currently extremely underrepresented in sequence databases—especially so when compared to terrestrial or parasitic nematode taxa. To test for applicability outside Nematoda, we also included microscopic individuals from four other invertebrate phyla. Specimens were collected mainly in the context of our ongoing surveys in the Gulf of California (Baja California, Mexico) and various coastal locations in California. Additional species and specimens were obtained from various other localities (table 1; see http://nematol.unh.edu for detailed locality data).
Table 1.
Origin of nematodes and other meiofauna from which D2D3 sequences were obtained, along with their best BLAST hits in GenBank.
| specimen ID | provisional taxon ID | sample origin | BLAST % ID; GenBank accession number(s) |
|---|---|---|---|
| nematodes subjected to multifocal imaging | |||
| 1P6K2 | Ascolaimus | Newport beach, CA, USA | 91.6; AF210406 |
| 2P6K2 | Enoplolaimus | Newport beach, CA, USA | 96.2; AF210421 |
| 3P6K2 | Viscosia | Newport beach, CA, USA | 97.4; AF210413 |
| 4P6K2 | Metachromadora | Newport beach, CA, USA | 88.9; AF210410 |
| 2I6K2 | Oncholaimidae | Newport beach, CA, USA | 92.7; AF210404 |
| 4I6K2 | Paracanthonchus | Newport beach, CA, USA | 96.6; AF210421 |
| 5I9K2 | Chromadorida | San Felipe, BC, Mexico | 91.3; AF210416 |
| 5P9K2 | Odontophora | San Felipe, BC, Mexico | 96.3; AF210421 |
| 1I11K2 | Chromadorida | San Felipe, BC, Mexico | 92.2; AF210421 |
| 2I11K2 | Odontophora | San Felipe, BC, Mexico | 96.7; AF210421 |
| 3I11K2 | Enoploides/Enoplolaimus | San Felipe, BC, Mexico | 99.1; AF210409, AF210407 |
| 1P11K2 | Enoploides | San Felipe, BC, Mexico | 93.5; AF210406 |
| 3P11K2 | Xyala | San Felipe, BC, Mexico | 95.1; AF210425 |
| 4P11K2 | Richtersia-like | San Felipe, BC, Mexico | 93.9; AF210410 |
| 2P12K2 | Pomponema-like | San Felipe, BC, Mexico | 96.6; AF210421 |
| 2P9K2 | Enoploides/Enoplolaimus | San Felipe, BC, Mexico | 94.7; AF210409 |
| 9P8K2 | Chromadorida | San Felipe, BC, Mexico | 97.3; U94755, AF226573-89, AY332064 |
| 1P12K2 | Spilophorella-like | San Felipe, BC, Mexico | 86.0; AF210401 |
| 3P12K2 | Monhysterida | San Felipe, BC, Mexico | 89.4; AF210421 |
| 5P12K2 | Richtersia | San Felipe, BC, Mexico | 92.8; AF210410 |
| 7I12K2 | Linhomeus | San Felipe, BC, Mexico | 98.1; U61758, AY652779, AF210421 |
| 8I12K2 | Ceramonematidae | San Felipe, BC, Mexico | 95.5; AY377676 |
| 1I14K2 | Viscosia | San Felipe, BC, Mexico | 92.6; AF210406 |
| 2I14K2 | Metachromadora | San Felipe, BC, Mexico | 93.8; AF210410 |
| 2P15K2 | Latronema | San Felipe, BC, Mexico | 95.7; AF210400 |
| 3P15K2 | Tricoma | San Felipe, BC, Mexico | 86.1; AF210421 |
| 1I12K3 | Rhabdocoma | Sta. Clara, BC, Mexico | 96.9; AF210418 |
| 1P10K3 | Latronema? | Sta. Clara, BC, Mexico | 95.7; AF210400 |
| 2I12K3 | Cyatholaimidae | Sta. Clara, BC, Mexico | 97.2; AF210400 |
| 3I23B4 | Phanoderma | Laguna beach, CA, USA | 90.9; AF210409 |
| 3I24B4 | Pontonema | Laguna beach, CA, USA | 91.3; AF210399 |
| 5I23B4 | Phanoderma | Laguna beach, CA, USA | 89.8; AF210409 |
| 6I23B4 | Pontonema | Laguna beach, CA, USA | 91.5; AF210399 |
| 1I22G4 | Odontopharynx | near Pomponio State beach, CA, USA | 93.7; AB189984 |
| 1M21G4 | Chromadorina? | near Pomponio State beach, CA, USA | 87.5; AY300843 |
| 7I16G4 | Haliplectus | near Pomponio State Beach, CA, USA | 95.9; AY210845, AY364867, AY580056-7, AB087186-7 |
| 8M21G4 | Choanolaimus | near Pomponio State Beach, CA, USA | 98.1; AF210406 |
| 4I22G4 | Helicotylenchus | near Pomponio State Beach, CA, USA | 92.4; AY592987-93 |
| Reference nematodes (no multifocal imaging) | |||
| Hirschmanniella | Hirschmanniella pomponiensis | near Pomponio State Beach, CA, USA | 93.5; U47556 |
| Mesocriconema | Mesocriconema xenoplax | Parlier, Fresno, CA, USA | 99.3; AF133304 |
| Apor BDGW | Aporcelaimellus | Big Sur, CA, USA | 98.5; AY601632 |
| Prion DGW | Prionchulus | Big Sur, CA, USA | 91.6; AY593065 |
| Brevibucca_SB261 | Brevibucca saprophaga | SB 261 petri plate culture | 95.8; AB189984, AY210806 |
| Brevibucca_SB117 | Brevibucca punctata | SB 117 petri plate culture | 96.4; AB189984, AY210806 |
| Basiria A_GPhi | Basiria | UCR campus, CA, USA | 87.6; U47557, AJ545025, AF435797-801 |
| Bunonema | Bunonema | UCR campus, CA, USA | 97.9; AF151919, AY220628 |
| Diphtherophora | Diphtherophora | UCR campus, CA, USA | 94.6; AY377672 |
| Alaimus | Alaimus | Botanical Garden, UCR, CA, USA | 96.9; AB087186 |
| Tylocephalus PP2 | ‘Tylocephalus’ | Sweeney Granite Mountain, CA, USA/petri plate culture | 99.9; AF147068 |
| Other Meiofauna subjected to multifocal imaging | |||
| Tardi oak | Isohypsibius? (terrestrial tardigrade) | Alisal Rd., Solvang, CA, USA | 96.5; AY210826 |
| 1PolyA3 | unidentified polytroch larva (marine polychaete) | Sta. Clara, BC, Mexico | 90.6; AY366515 |
| Macro 1 | Macrodasyida (Marine gastrotrich) | Sta. Clara, BC, Mexico | 95.6; AF062965, AF363322, AY210804 |
| 1FlatA3 | unidentified marine flatworm | Sta. Clara, BC, Mexico | 94.7; AY222200 |
| 1GasIIA3 | Heteroxenotrichula? (marine gastrotrich) | Sta. Clara, BC, Mexico | 96.2; AY338676 |
| Tetra 1 | Thaumastoderma (marine gastrotrich) | Sta. Clara, BC, Mexico | 96.0; AY145396, AJ436879 |
Nematodes were extracted alive from sediment, soil or organic litter samples through decanting and sieving, tray extraction and/or mist chamber extraction. Live specimens were individually mounted and immobilized in various ways under cover slip on ringed fluorescence slides, a Taylor microcompressor Mk II (Taylor 1993), or ordinary glass slides. If the specimen was not immobilized by the pressure of the cover slip alone, temporary paralysis was obtained through a heat shock of 5–10 s at 60–70 °C (depending on the type of sample and size of nematode), or chemical anaesthesia with 7.5% MgCl2 for marine meiofauna. The temporary mounts were placed on a microscope equipped with differential interference contrast optics, and the diagnostically most important body parts were captured at various magnifications as multifocal images. Our respective laboratories avail of three different systems for multifocal imaging. The one used for this study is a more budget-conscious configuration that is manually controlled and writes S-VHS quality video clips directly to hard disk (VCE procedure as described in De Ley & Bert 2002). By comparison, the other two are more sophisticated systems with motorized microscope controls and capture as digital image stacks that are then saved as high definition video clips (cf. Eyualem et al. 2004). Multifocal vouchers of the barcoded specimens included in the analyses below were deposited in NemATOL (http://nematol.unh.edu/) with the specimen ID codes listed in table 1.
(b) Molecular analysis of captured specimens
Immediately after image capture, each specimen was recovered alive from its temporary mount, cut into 2–4 pieces (depending on body size) in 20 μl Worm Lysis Buffer (50 mM KCL, 10 mM Tris-Cl pH 8.3, 2.5 mM MgCl2, 0.45% NP40, and 0.45% Tween 20 as described in Williams et al. 1992) transferred to 1–5 microcentrifuge tubes, digested with 2 μl (60 μg ml−1 stock) Proteinase K and stored at −80 °C. In addition to 37 video captured nematodes, the same methods were applied to six animals from four other phyla. Furthermore, 11 individuals from various other nematode genera and species were prepared for freezing and PCR without video capturing, in the course of protocol testing or various soil surveys. These were included as reference taxa in the analysis in order to cover some of the major nematode orders not encountered in our marine samples.
Series of frozen (fragments of) specimens were subsequently thawed and subjected to PCR amplification of the D2 and D3 expansion segments of the LSU rDNA gene. Three of the 11 reference nematodes were first genome-amplified using GenomiPhi™ DNA amplification kit (Amersham-GE Health Care, Sunnyvale, CA, USA) following the manufacturer's protocol, before performing D2D3 amplification. Depending on the size or type of the organism, genome-amplified products are used as undiluted or diluted (10×–1000×) template for D2D3 amplification. PCR amplifications were performed using 2.5 μl genomic DNA as template in 25 μl reaction volume containing 2.5 μl of 10× reaction buffer with MgCl2, dNTP-mix at 0.2 mM each, 1 μM each of primers D2A (>5′-ACAAGTACCGTGAGGGAAAGTTG-3′) and D3B (<5′-TCGGAAGGAACCAGCTACTA-3′) D3b and 1 unit of DyNAzyme EXT DNA polymerase (MJ Research, Waltham, MA 02451, USA) with 40 cycles each involving denaturation at 94 °C for 30 s, annealing at 55 °C for 1 min, and extension at 72 °C for 2 minutes; followed by a 7-minute polymerization at 72 °C. Amplified PCR products were separated in 1% agarose gel (Shelton Scientific, Inc.) stained with 0.003% ethidium bromide (0.02 μg ml−1) with 1 Kbp DNA ladder (Promega Corp., Madison, WI, USA) as size markers. PCR products were purified with exonuclease I and shrimp alkaline phosphatase (USB, Cleveland, OH, USA) following the manufacturer's protocol, or separated on 1.5% SeaPlaque agarose (FMC, Rockland, ME, USA), excised and purified using Qiaquick gel extraction kits (Qiagen, Los Angeles, CA, USA). Sequencing reactions were performed at the UCR Core Instrumentation Facility using Big DyeDeoxy Terminator Cycle Sequencing kits following the manufacturer's protocol prior to fragment analysis using an ABI 3100 automated sequencer (Applied Biosystems Inc., Foster City, CA, USA); and at the Hubbard Center for Genome Studies using the DTCS Quickstart kit and a CEQ-8000 capillary sequencer (Beckman-Coulter, Fullerton, CA, USA).
Sequences were assembled in GeneTool 2.0 (BioTools Inc., Edmonton, AB, Canada) and compared with published data deposited in GenBank by means of a BLAST search (Altschul et al. 1997), and by phylogenetic analysis of all sequences over 500 bp. As our marine nematode survey was still in its early stages and coverage of the taxonomic spectrum was still very fragmentary, we expected substantial problems with alignment of variable regions in D2 and D3 across highly divergent nematode orders, and especially across different phyla. The purpose of our analyses was therefore neither to establish a definitive assignment of barcodes to species, nor to provide a credible topology of deeper phylogenetic relationships. Rather, we set out to explore methods for providing a first assessment of the performance of D2D3, in the context of phylogeny-based barcoding and cross-verification with multifocal recordings of morphology, but also with an eye on applicability and automation in future analyses of much larger datasets with hundreds of barcode sequences. Additional sequences were obtained from GenBank and included to further broaden taxonomic representation and to benchmark the ability of our analyses to match known species identities across different taxa and taxonomic situations. An alignment was created using the server version of Mafft 5.6 (Katoh et al. 2005), an alignment algorithm based on fast Fourier transformation that was developed specifically for large datasets and which is claimed to be more accurate than other automated alignment packages. Mafft was left at the default settings and the iterative refinement chosen was the option ‘E-INS-i’. The result was trimmed at both ends in MacClade 4.0 (Maddison & Maddison 2000) to an alignment width of 1538 characters and then subjected to neighbour-joining and heuristic maximum parsimony analysis (respectively, with 3000 and 1000 bootstraps) in PAUP* 4.0b10 (Phylogenetic analysis using parsimony and other methods; Swofford 1999).
Variable regions were clearly arbitrarily aligned, due to the often great divergence between sequences. Alignment ambiguities are well known to be a major source of error and uncertainty in phylogenetic analyses, and one approach to avoiding such errors is to remove ambiguous positions. In order to examine the usefulness of ambiguity removal algorithms in the context of barcoding applications, we applied the program Cutter (http://goodey.unh.edu) to our manually edited alignment and generated two alignments of reduced length. The first one will henceforth be referred to as Cutter alignment 1 and was created with column threshold setting and wall threshold setting both at 0.10, resulting in trimming down of the original alignment to 570 positions. The second alignment, hereafter referred to as alignment 2, was created with a column threshold setting of 0.05 and a wall threshold setting of 0.65. This alignment retained 662 positions. Mean differences (corrected for missing data) were calculated between all sequence pairs for each alignment, using PAUP*. In order to estimate possible matches of our barcoded specimens with published D3 sequences of nematode species sequenced by Litvaitis et al. (2000), we also created an alignment of just the D3 expansion segment. This alignment only included our barcoded specimens and the sequences deposited in GenBank by Litvaitis et al. (2000). We again analysed this with neighbour-joining and heuristic maximum parsimony. All unpublished sequences used in the above analyses were deposited in GenBank with accession numbers DQ077749–DQ077803.
4. Results
(a) Quality and practicality of multifocal images
At 720 by 480 pixels, the video clips obtained with our VCE system were clearly lower in resolution than the level of detail that could be observed by eye directly through the microscope. Nevertheless, the clips generally captured all the detail that matters to identification. Significantly, the ability to scroll through successive focal planes greatly facilitates interpretation of the shape, location and orientation of structures, especially when compared to single still images. For example, sensory organs like amphids and setae are some of the most important diagnostic structures that are difficult to adequately capture in a single photograph, particularly if a specimen is not oriented at a perfect angle to fit the structure in question in a single optical section (see figures 1 and 2 for examples). By providing continuity across adjacent focal planes, we find that multifocal clips typically make it much easier to recognize such structures—in a way that cannot be adequately reproduced or demonstrated on a printed page.
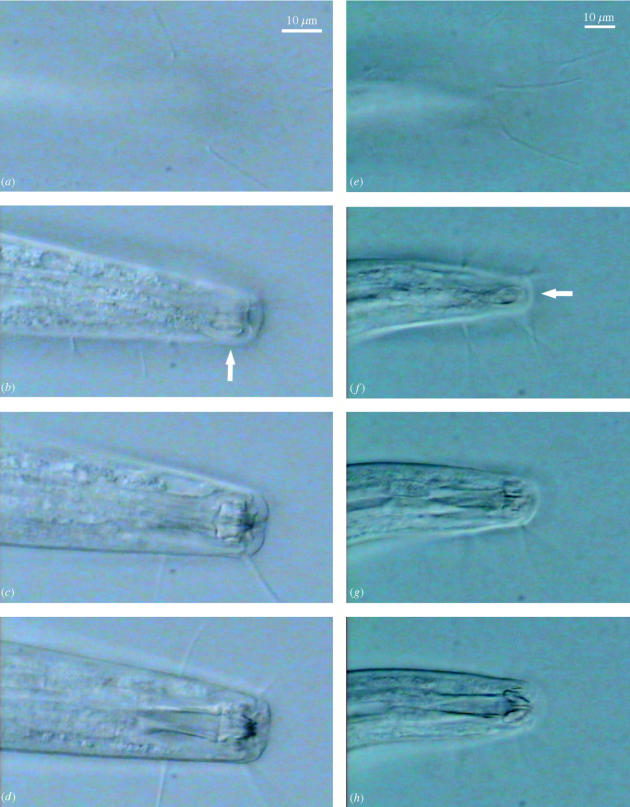
Figure 1.
Five frames (images) extracted from a multifocal video clip of the lip region of barcoded nematode specimen 2P12K2, identified as the genus Pomponema. Note the various setae revealed in different focal planes, as well as the different aspect of the faint but tightly coiled amphid in successive frames (a–c) and the presence of a subventral tooth (e). The placement and shape of the amphids and most of the setae would be impossible to record in a single photograph in this specimen. The position of the tooth would be impossible to ascertain without determining the angle of view, which information must be gleaned from other focal planes (e.g. from the relative position of the amphid, which is located laterally).
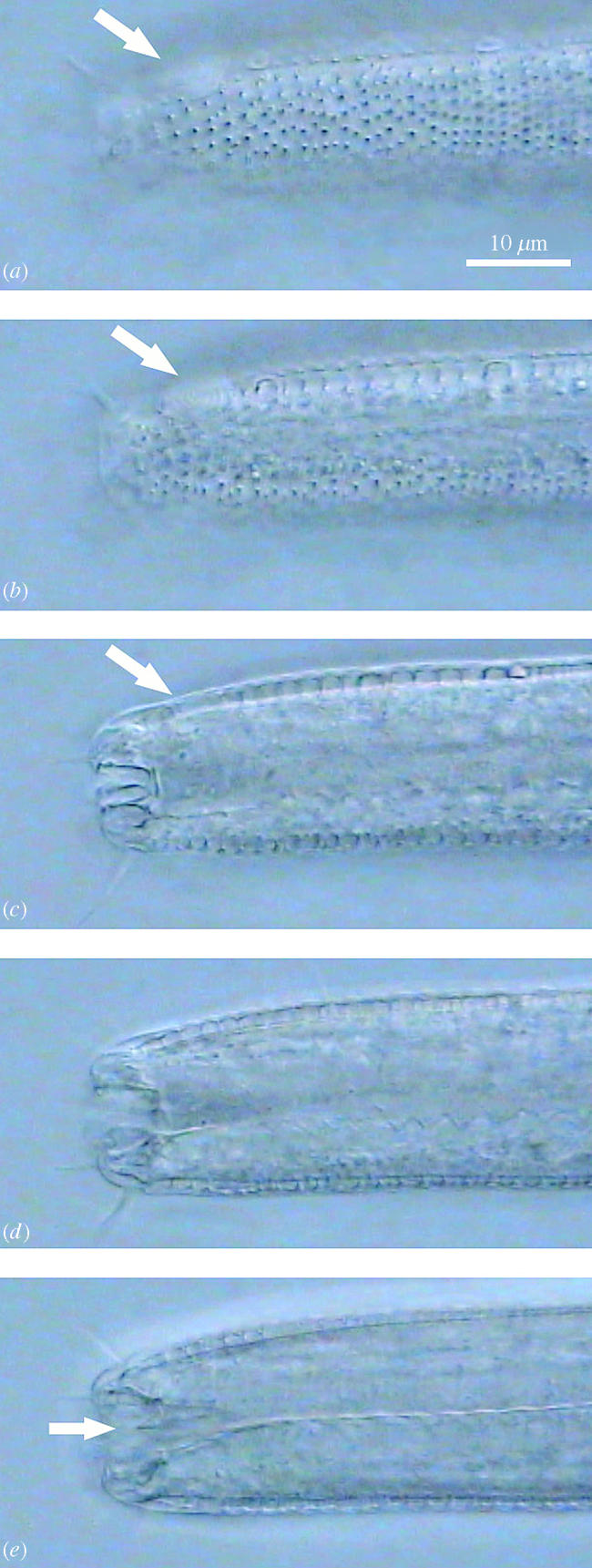
Figure 2.
Four successive frames from a multifocal video clip of the lip region of barcoded specimens 2I11K2 and 5P9K2, both identified as belonging in the genus Odontophora. Note the extremely long setae pressing against the coverslip (a, e) as well as the horseshoe-shaped amphid (b, f) and the radially arranged teeth (c, d, g, h). The placement of the setae, shape of the amphid and arrangement of the teeth is not recordable with a single (or even a few) still photograph(s). Both these specimens cluster together with Pomponema 2P12K2 (shown in figure 1) in phylogenetic analyses (see figure 3) but they are clearly not conspecific, illustrating the ability of multifocal images to pinpoint potential errors in barcode-based species delineations.
In terms of storage, the file size of one uncompressed video clip at S-VHS resolution is usually around 4 to 20 megabytes, an order of magnitude roughly comparable to that of a single uncompressed still image captured at resolutions around 2 megapixels. This is well within the handling capacity of current desktop and laptop computers. If necessary it is relatively easy to optimize file sizes with further manipulations such as batch compression. Our multifocal imaging procedures as described above (with manual focusing and analogue video camera) typically added about 15–30 min to the processing time of each specimen prior to DNA extraction, thus allowing one person to capture the morphology and prepare frozen lysates of between 10 and 20 nematodes during an average working day.
(b) First results with barcoding of nematodes from the Gulf of California
Multifocal capturing followed directly by PCR and direct sequencing yielded good D2D3 sequences for over 80% of all specimens. Genomic amplification was successful and the resulting DNA amplicons easily allowed PCR and direct sequencing of the D2D3 locus. By comparison, PCR and/or sequencing of other loci were less consistently successful and clearly need more optimizing (data not shown). Of the 43 barcode sequences obtained (including six from other phyla), only one matched GenBank data with a BLAST percent identity score just above 99% (table 1). Trees produced from all alignments suggested six pairs or groups of putative matches within our set of nematode barcodes (marked by horizontal blue bars in figure 3). A number of other interesting sequence pairs also occurred among the sequences downloaded from GenBank or determined without prior video capturing (marked by grey bars in figure 3). We will discuss these in light of strength and weaknesses of our approach as confronted with the complications of actual identification. Four sets of sequences were scored with 0% mean divergence in all three alignments. Two of these consisted of pairs of identical sequences known to correspond to either one species (Acrobeloides bodenheimeri AF147064-5) or one single individual (2I6K2 Oncholaimidae was sequenced twice for quality control purposes). A third identical pair included Acrobeloides thornei and a cultured nematode originally presumed to belong in the genus Tylocephalus. Apparently A. thornei was accidentally carried over into the latter culture (a mistake that could easily have been avoided had we first video imaged the nematode in question). All three identical pairs show that at least the Mafft alignment correctly matched up identical sequences.
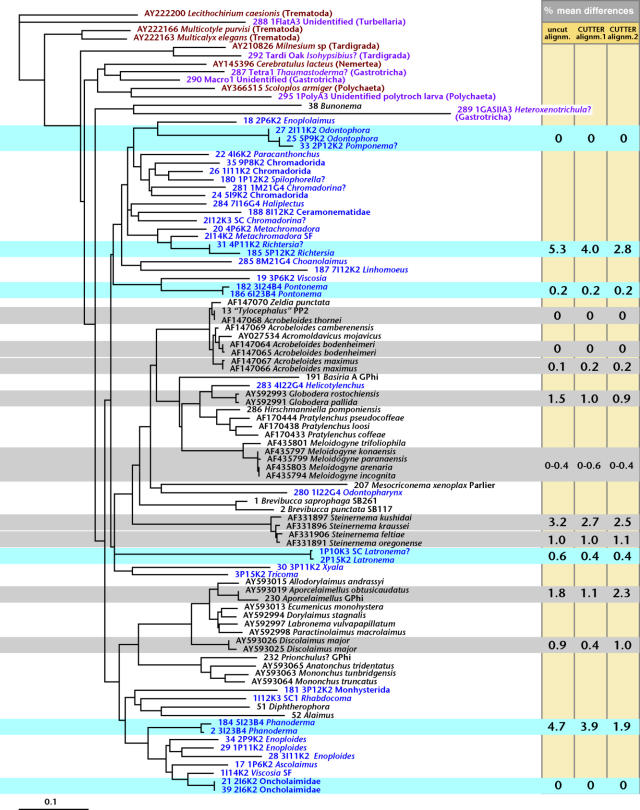
Figure 3.
Neighbour-joining phylogram based on alignment 2 of the D2D3 expansion segment of barcoded and reference organisms (aligned with Mafft 5.6 using setting E-INS-i followed by ambiguity trimming with Cutter using column setting=5, wall setting=65). Taxon names are coloured as follows: red for downloaded sequences of non-nematode taxa, purple for newly video vouchered and sequenced non-nematode taxa, blue for newly video vouchered and sequenced nematode taxa, black for nematode taxa of which sequences were downloaded or newly determined without video capturing. Deeper relationships are not necessarily meaningfully resolved as the D2D3 locus is too short and too divergent for that purpose. Interesting benchmark sequences as discussed in the text are highlighted with horizontal bars (blue for video vouchered nematodes, grey for others). Percentage mean difference values for the relevant sequence pairs are listed on the right for the original Mafft alignment without any removal of ambiguities, for alignment 1 (Mafft followed by Cutter using column setting=10, wall setting=10) and for alignment 2.
The fourth set with 0% mean differences included not two but three sequences: two of these came from individuals identified as Odontophora (figure 2) while the third nematode was identified as Pomponema (figure 1). These two genera are morphologically quite different from one another, traditionally being classified in the orders Monhysterida and Chromadorida respectively. The obtained multifocal images strongly suggest that in this case the chosen locus and/or species-matching algorithm did not perform optimally. The sequences themselves were not identical but the only differences consisted of a few unresolved nucleotides.
Among the video captured nematodes, two sequence pairs appear to constitute bona fide matches: Pontonema specimens 3I24B4 and 6I23B4 derived from the same sample and had a mean divergence of 0.2% in all three alignments, while Latronema 1P10K3 and 2P15K2 came from different sites but had a mean divergence of 0.6% or less. Two other sequence pairs were more problematical, each clustering together as sister taxa with maximal bootstrap values in the trees (data not shown) but nevertheless differing by higher mean percentages. Richtersia 5P12K2 was an adult male, while 4P11K2 was a juvenile and could not be morphologically matched with certainty to the same species or even genus; their sequences differed by 2.8–5.3% and it seems possible that these could actually represent two different species. Phanoderma 3I23B4 and 5I23B4 matched more clearly to the same species morphologically, for example in sharing presence of eyespots, but nevertheless had a mean difference in D2D3 sequence of 1.0–4.7% in our three alignments.
Among the nematode sequences that did not derive from video captured specimens, a number of other groupings are noteworthy with respect to species identity versus mean sequence differences. The included species in the entomopathogenic genus Steinernema and the plant parasitic genera Globodera and Meloidogyne are morphologically confounding among their respective congeners, but nevertheless considered valid species based on life history characters among others. The respective mean difference values between D2D3 sequences are small to very small (especially for Meloidogyne), especially when compared to sequences from putative conspecific individuals such as our two Latronema individuals, or two sequences of Discolaimus major as downloaded from GenBank.
Clearly, D2D3 mean differences do not correlate consistently with conspecificity across the phylum, and external data (such as morphological vouchers) are needed to corroborate putative matches. Overall, we cautiously estimate that our 37 video captured and barcoded nematode specimens represent 32–34 different species, none of which matches any previously sequenced species. Trees derived from the D3-only alignment suggested not a single match with any of the sequences of Litvaitis et al. (2000), and placed AF210426 Tricoma among Ceramonematidae, well away from our 3P15K2 Tricoma (data not shown), the former sequence presumably deriving from a misidentified specimen.
(c) Phylogenetic signal or noise
Phylogenetic reliability of the obtained trees was clearly not very high. The untrimmed Mafft alignment as well as Cutter alignment 1 yielded trees in which non-nematode sequences were inserted in at least three separate places in between nematode taxa. Only alignment 2 yielded a relatively coherent grouping of the other phyla (figure 3). Various other dubious placements occurred, e.g. with respect to separation of Enoplolaimus 2P6K2 from other Enoplidae, placement of Oncholaimidae (including Viscosia and Pontonema) in two separate clades, as was also the case for 3P12K2 (unidentified member of Monhysterida) versus 3P11K2 Xyala. Heuristic maximum parsimony analysis yielded respectively 61, 57 and 84 best trees for the untrimmed alignment, Cutter alignment 1 and Cutter alignment 2. In view of all this evidence for poor resolution of deeper nodes, which is undoubtedly connected with the fragmentary taxonomic representation in our data, we did not at this stage attempt species assignment by autapomorphy analysis as proposed by Adams (1998).
5. Discussion
(a) Are multifocal images useful for barcoding studies?
Nematodes and many other microscopic Metazoa are optically unusual as three-dimensional objects in that they are both minute and largely transparent. As a result, nearly all internal features are visible without requiring dissections or special staining techniques—but these features are also transparent in their own right and must be observed at the highest magnifications in optical conditions where focal depth is reduced to a single optical section. In order to understand any particular anatomical structure, one must therefore both detect subtle differences in transparency as well as examine multiple two-dimensional focal planes, before one can mentally visualize the three-dimensional shape of the feature in question. This visualization problem also applies to any images that are created to record and represent nematode morphology: unlike for example a photograph of an insect wing, no single photograph of a nematode body part can accurately embed and reflect its three-dimensional contents and context. On the other hand, line drawings can represent structures at multiple levels of focus, but require substantial personal skill and are incapable of accurately representing subtle contrasts or shades of transparency. Video and digital camera technology have opened up new possibilities for morphological vouchering through analogue or DMI. Many cameras used in scientific applications are optimized for applications that require the highest resolution and/or the highest sensitivity in low light conditions. The question therefore arises whether such cameras are also suitable for capturing the subtle nuances of transparency that reveal the outline and internal organization of nematode structures. Based on our material, we feel this is definitely the case—even for relatively inexpensive video cameras of limited resolution.
PCR can be optimized for small fragments and/or for formalin-fixed material (Thomas et al. 1997; Dorris et al. 2002), but high-throughput DNA extraction of large numbers of taxonomically diverse individual animals usually requires the destruction of entire specimens, thereby obliterating all morphological characters. Molecular barcoding without morphological vouchering results in the loss of all other biological data, creating at best a first record that will not be available for further study until future rediscovery of specimens in nature. Morphological vouchering through multifocal imaging cuts through this Gordian knot, and is actually easier to implement with specimens that are small enough to require complete destruction during DNA extraction. Furthermore, it reverses the traditional problem of time-consuming species identifications or descriptions in taxonomically challenging phyla such as nematodes. Identifications can now be deferred until molecular barcodes are obtained, and identification effort can be prioritized based on both the outcome of barcoding analysis and any apparent morphological novelty, or particular significance to other studies. Also, vouchering through multifocal imaging provides a direct link to existing keys, classifications and hypotheses based on previously documented morphologies and ecologies. The virtual vouchers are not likely to suffice in and of themselves for descriptions of new species, but they do provide an effective way of linking molecular data from destroyed specimens with unsequenced (and generally unsequenceable) type material in permanent slides, through comparison of their respective morphologies.
(b) Do barcoding studies need named species?
In a number of cases, the advantages listed above will clearly outweigh the benefits of maximal processing speed that could be attained with a voucher-less MOTU approach. Although maximal processing speed is excellent for applying mathematical biodiversity analyses, the sacrificing of morphology disallows comparisons with species-based alpha diversity surveys. Combined acquisition of molecular and morphological data greatly enhances the strengths of each while bypassing many of their respective weaknesses. For example, juveniles can be detected through microscopy but are rarely accurately identifiable, since most morphological keys are based only on adults (cf. our two Richtersia specimens). Conversely, barcoding works with both adults and juveniles, but does not reveal developmental stage unless morphological data are collected as well.
Floyd et al. (2002) have noted that biology of sequenced but voucher-less MOTUs can be inferred from proximity to known species. However, this process can easily lead to a significant fraction of erroneous conclusions in poorly sampled phyla, while phylogenetic analyses are themselves volatile and typically generate multiple topologies with at least some unresolved areas. Furthermore, in order to be able to make any inferences on biology, MOTU approaches must necessarily rely on a reasonable representative set of barcodes (the ‘stepping-stone sequences’ of Blaxter 2004) from species identified with independent criteria. Multifocal imaging combined with barcoding of specimens provides a structured approach to establishing such a set of reference points. This selected set must necessarily grow at a slower pace than MOTU studies that maximize on high throughput of specimens. Nevertheless, a combined strategy such as the one outlined in this paper will dramatically expand the coverage and accuracy of biological inferences by phylogenetic proxy, even in habitats where the large majority of species are unknown and unlikely ever to be formally named and described.
Similar to the spectacular diversity currently being discovered in prokaryotes (see e.g. Torsvik et al. 2002) and protists (Countway et al. 2005), MOTU surveys of nematodes can play a very useful role in generating first-line data on ‘operational’ biodiversity in different sites or habitats, thereby providing an objective basis for e.g. selecting samples that need to be prioritized for identification and description of species. However, the fundamental problem of nematode surveys is the identification of species, and this problem clearly will not be tackled by sidestepping the theoretical and diagnostic challenges of species concepts in nematodes. Instead, a genuine solution to the nematode corner of the taxonomic impediment will have to squarely face the challenges of diagnosing species, as opposed to diagnosing operational units. In a recent review of the operational aspects of species diagnosis, Sites & Marshall (2004) noted that no method would invariably succeed in delimiting species boundaries properly. They explicitly caution against reliance on any single data set or method. Similarly, in a barcoding study of cave-dwelling eyeless spiders, Paquin & Hedin (2004) demonstrated the need for external information to allow for the discovery of incongruence between different data types, in order to make sense of otherwise one-dimensional molecular taxonomies.
(c) Do barcoding studies need phylogenies?
Phylogenetic analysis has become essential to the delineation of species in nematodes. The practical application of species concepts used to be a major stumbling block to nematode taxonomy, and it was not unusual for leading experts to declare the problem to be basically insurmountable (e.g. Heyns 1983; see review in Coomans 2002). This conceptual paralysis has changed substantially in recent years, with the proposal of a tree- and autapomorphy-based species concept (Adams 1998) that is rapidly gaining ground in nematode taxonomy (see e.g. De Ley et al. 1999; Stanton et al. 2001, Coomans 2002; Nadler 2002; Troell et al. 2003). This has in many ways opened the doors for more objective approaches to nematode surveys. Even though occasional failures to properly diagnose species are to be expected (Sites & Marshall 2004), the current state of the art in nematode taxonomy explicitly requires species allocations to incorporate phylogenetic criteria. However, as shown by our data the application of autapomorphy analysis in barcoding surveys may not be all that easy, at least until a more comprehensive set of sequences is available for constructing more robust alignments.
The use of short DNA sequences as identification tags of species is a unifying concept with multiple uses. Existing or projected applications include: development of portable identification technology for walkabout surveys in the field; conservation inventory management of listed species (which are by definition known species); comparative biodiversity analyses of alpha and beta diversity; supporting taxonomic discovery by generating a first baseline for quantifying and selecting novel taxa for description. It is important to note that these different applications do not all present the exact same requirements in terms of optimal choice of barcoding locus. For example, the former two would benefit from a single uniform standard barcoding locus across all organisms, which may well require compromises in terms of attainable resolution and representation for some particular groups. In contrast, the latter two applications typically involve more methods development along the way, to determine which loci (if any) are truly optimal for species discrimination among the particular organisms studied.
Barcoding of taxa with a high ratio of unknown to known species is unlikely to be worth the effort if no phylogenetic relationships can be established before a formal morphological study is undertaken. Phylogenies effectively provide road maps through a potentially vast space of undiscovered taxa (De Ley 2000). In the absence of any phylogenetic context, the information conveyed by the barcode alone is purely archival and will only be relevant to future studies if the exact same taxon happens to be encountered again. With a phylogenetic context and a virtual voucher, morphology and DNA sequences can be reciprocally verified, reducing errors and maximizing the chances of discovery of biologically interesting patterns of diversity. Even more importantly, a barcode that carries meaningful phylogenetic signal will ensure relevance to other surveys in the present and in future, because related taxa can be recognized even if they are not identical, while morphological vouchers can establish links with past studies through comparison with type specimens.
(d) Choices of barcoding loci for nematodes and other meiofauna
Our experience with nematodes alone already suggests that no single barcoding locus will perform optimally across all forms of life, and it is likely that different loci should be used for barcoding different types of organisms. For example, using the same primer pair for macroscopic as for microscopic organisms is likely to be counter-effective because it could well lead to mixed PCR products due to the ubiquity of symbionts. Nuclear genes are likely to be more diagnostically useful than mitochondrial loci because they are not subject to lineage sorting. Compared to rDNA, protein genes offer an additional layer of information with respect to different evolutionary rates according to codon positions, and translation into amino acids, but they also present potential problems due to occurrence and variability of introns. Furthermore, the choices for barcoding of nematodes and other very poorly covered taxa are not only constrained by methodological considerations, but also by the availability of research interest and funding. Nematodes are not likely to show up on any lists of endangered species. They are therefore also unlikely to receive sufficiently focused and sustained support to allow an expansive database to be compiled for the specific use in barcoding of any single locus. Thus, the most appropriate loci from the point of view of funding priorities will also be those that can serve multiple tasks, e.g. by being useful in phylogenetics as well as in diagnostics. In the longer term, however, the situation will undoubtedly change quite drastically as sequencing technology improves. The barcoding gene may rapidly become superseded by the advent of ‘genomic barcodes’ and ‘Genomic Operational Taxonomic Units’ constructed by instantaneous sequencing of numerous species-diagnostic markers sampled across entire genomes. For the time being, the following loci appear to be most relevant to barcoding efforts in nematodes and other microscopic invertebrates:
small subunit (SSU) rDNA has received the greatest attention as a barcoding locus in recent literature (Floyd et al. 2002; Blaxter 2004; Powers 2004). SSU targets usually have a high success rate with PCR, but may require primer optimization for different nematode taxa, and for use in other phyla. The gene is not known to be subject to significant amounts of polymorphism; in nematodes it has a wide range of diagnostic resolution but it tends to work better for separating species in some groups than in others. The locus has a high phylogenetic information content and often works well for resolving relationships, especially at family and order levels (e.g. Blaxter et al. 1998; Félix et al. 2000; Rusin et al. 2003). The public record of partial and complete SSU sequences is the single most abundant component of all known nematode sequences, and it is likely to remain the best sampled gene in nematodes, due to its widespread application in both phylogenetic studies and molecular surveys.
The LSU rDNA gene has been in use for almost ten years as a source of diagnostic sequences in nematodes, particularly the region that spans the D2 and D3 expansion segments (Thomas et al. 1997). In nematodes it covers about 600–1000 bp, fairly close to the 5′ end of the gene. As with the other expansion segments of LSU, sequence divergence of D2 and D3 between related species is often high. By contrast, the conserved regions alternating with D2 and D3 are highly constant, even across phyla, and provide very robust primer sites. In our experience, the D2D3 primer pair has the highest success rate when applying PCR amplification to a phylum-wide selection of nematodes, and based on our limited testing it also works well in other phyla of microscopic metazoans. The locus is not known to be subject to significant amounts of intraspecific polymorphism, and provides very good separation of cryptic species in some groups (De Ley et al. 1999). Previous studies have included phylogenetic applications of the D2 or D3 alone (cf. Litvaitis et al. 2000). The combination of both expansion segments generally provides more robust signal, at a level corresponding roughly to species, genus and family in local taxonomic context, although it is probably overall too variable for meaningful analysis at deeper levels in classification and phylogeny. Our data presented here also indicate that mean differences in D2D3 do not always correspond reliably with accepted species boundaries. Published sequences of partial or complete LSU are more limited in number than SSU, but this part of the overall nematode dataset is likely to expand considerably due to increasing interest in LSU sequence data for phylogenetic studies.
The internal transcribed spacer region (ITS) initially received great attention as a potentially diagnostic tool in nematodes (Powers et al. 1997). It consists of two variable regions (the spacers) alternating with the much more conserved SSU, 5.8S and LSU rDNA genes, in an arrangement rather similar to the alternation of conserved and variable sequence stretches of the D2/D3 region. However, in our experience no single primer pair seems capable of allowing robust PCR success across all major nematode groups. To make matters worse, several recent studies have revealed occurrence of ITS haplotypes, sometimes with such levels of polymorphism that species boundaries are confounded and direct sequencing becomes impossible (Hugall et al. 1999). As a result, in some nematode groups the ITS region may prove to be a much more interesting marker for population genetics than for species diagnostics. Nematode ITS data in public databases are less numerous than SSU sequences but more so than LSU data. The sequence pool for this region will undoubtedly continue to grow, due to its frequent application in phylogenetic studies of selected taxa (e.g. Chilton et al. 2001; Subbotin et al. 2001), in studies of potential hybridizations (Hugall et al. 1999) or in analyses of intraspecific diversity (Kaplan et al. 2000; Elbadri et al. 2002; Ye et al. 2004).
No phylum-wide primers are available for the mitochondrial cytochrome oxidase I gene in nematodes, in our experience PCR success rates are well below 50% across various taxa within the phylum. Reasons for these problems may relate to the emerging evidence that nematode mitochondrial genomes are highly diverse, displaying unusual properties such as recombination (Lunt & Hyman 1997), insertional editing (Vanfleteren & Vierstraete 1999) and multipartitioning (Armstrong et al. 2000). Thus, designing nematode primers for mitochondrial protein genes may present many complications if these are to be useful for barcoding across the phylum. At present, little is known about the applicability of nematode coxI in phylogenetic analyses and very little is available in public databases in terms of sequences.
(e) Conclusions and wider perspectives
Multifocal imaging opens up possibilities for vouchering that can bring molecular surveys of nematodes closer to the protocols of barcoding studies of larger organisms. However, the flavour of barcoding surveys of nematodes is likely to remain quite different from those focusing on e.g. insects, because it is also determined by the high preponderance of undescribed species, by greater debate about the relevance of morphology, and by differences in theoretical developments with respect to species concepts. We are still in the early stages of exploring the biodiversity of meiofaunal, interstitial and other microscopic Metazoa. Although we can make certain predictions about the most appropriate search strategy for charting this vast taxonomic wilderness, it is unlikely that such predictions will be very accurate on the basis of known species, which might easily constitute less than 1% of all existing microscopic Metazoa. As with geographic exploration of uncharted continents and planets, it therefore makes sense to try multiple strategies of exploration and to maximize data exchange between these different approaches. We consider a data-rich, somewhat less processive approach to be an essential component of this overall process, documenting morphological and molecular diversity in formats that are easily archived and distributed electronically. We are therefore also concentrating our efforts on setting up the necessary infrastructure to facilitate data exchange and to foster new collaborations and linkages. Communication and integration between different surveys will be mediated by websites such as NemATOL, and these will in turn integrate through higher-level web resources. The probable outcome will be an emergent network of Internet resources dedicated to the discovery and documentation of biodiversity.
Acknowledgments
This research was supported by a UCMEXUS/CONACYT joint project award and by the National Science Foundation through award no. DEB-0315829. We are indebted to Matt Bemus for development of Cutter. We also wish to thank Nicola Debenham, John Lambshead and Ian King for help with provisional genus identifications, and are grateful to Ian King and the anonymous reviewers for many constructive suggestions during the writing of this paper.
Footnotes
One contribution of 18 to a Theme Issue ‘DNA barcoding of life’.
References
- Adams B.J. Species concepts and the evolutionary paradigm in modern nematology. J. Nematol. 1998;30:1–21. [PMC free article] [PubMed] [Google Scholar]
- Adams B.J. The species delimitation uncertainty principle. J. Nematol. 2001;33:153–160. [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F, Madden T.L, Schäffer A.A, Zhang J, Zhang Z, Miller W, Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong M.R, Blok V.C, Phillips M.S. A multipartite mitochondrial genome in the potato cyst nematode Globodera pallida. Genetics. 2000;154:181–192. doi: 10.1093/genetics/154.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. Counting angels with DNA. Nature. 2003;421:122–124. doi: 10.1038/421122a. 10.1038/421122a [DOI] [PubMed] [Google Scholar]
- Blaxter M.L. The promise of a DNA taxonomy. Phil. Trans. R. Soc. B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. 10.1098/rstb.2003.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M.L, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. 10.1038/32160 [DOI] [PubMed] [Google Scholar]
- Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, Abebe E. Defining operational taxonomic units using DNA barcode data. Phil. Trans. R. Soc. B. 2005;360:1935–1943. doi: 10.1098/rstb.2005.1725. 10.1098/rstb.2005.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton N.B, Newton L.A, Beveridge I, Gasser R.B. Evolutionary relationships of trichostrongyloid nematodes (Strongylida) inferred from ribosomal DNA sequence data. Mol. Phylogenet. Evol. 2001;19:367–386. doi: 10.1006/mpev.2001.0938. 10.1006/mpev.2001.0938 [DOI] [PubMed] [Google Scholar]
- Coomans A. Present status and future of nematode systematics. Nematology. 2002;4:573–582. 10.1163/15685410260438836 [Google Scholar]
- Countway P.D, Gast R.J, Savai P, Caron D.A. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J. Eukarot. Microbiol. 2005;52:95–106. doi: 10.1111/j.1550-7408.2005.05202006.x. 10.1111/j.1550-7408.2005.05202006.x [DOI] [PubMed] [Google Scholar]
- De Ley P. Lost in worm space: phylogeny and morphology as road maps to nematode diversity. Nematology. 2000;2:9–16. 10.1163/156854100508854 [Google Scholar]
- De Ley, P. In press. A quick tour of nematode diversity and the backbone of nematode phylogeny. In WormBook (ed. The C.elegans Research Community). WormBook. http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- De Ley P, Bert W. Video capture and editing as a tool for the storage, distribution and illustration of morphological characters of nematodes. J. Nematol. 2002;34:296–302. [PMC free article] [PubMed] [Google Scholar]
- De Ley P, Félix M.-A, Frisse L.M, Nadler S.A, Sternberg P.W, Thomas W.K. Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae) Nematology. 1999;1:519–612. [Google Scholar]
- Dorris M, Viney M.E, Blaxter M.L. Molecular phylogenetic analysis of the genus Strongyloides and related nematodes. Int. J. Parasitol. 2002;32:1507–1517. doi: 10.1016/s0020-7519(02)00156-x. 10.1016/S0020-7519(02)00156-X [DOI] [PubMed] [Google Scholar]
- Elbadri G.A.A, De Ley P, Waeyenberge L, Vierstraete A, Moens M, Vanfleteren J. Intraspecific variation in Radopholus similis isolates assessed with RFLP and DNA sequencing of the internal transcribed spacer region of the ribosomal RNA cistron. Int. J. Parasitol. 2002;32:199–205. doi: 10.1016/s0020-7519(01)00319-8. 10.1016/S0020-7519(01)00319-8 [DOI] [PubMed] [Google Scholar]
- Eyualem A, Grizzle R.E, Hope D, Thomas W.K. Nematode diversity in the Gulf of Maine, USA, and a Web-accessible, relational database. J. Mar. Biol. Assoc. UK. 2004;84:1159–1167. 10.1017/S0025315404010604h [Google Scholar]
- Félix M.-A, De Ley P, Sommer R.J, Frisse L.M, Nadler S.A, Thomas W.K, Vanfleteren J.R, Sternberg P.W. Evolution of vulva development in the Cephalobina (Nematoda) Dev. Biol. 2000;221:68–86. doi: 10.1006/dbio.2000.9665. 10.1006/dbio.2000.9665 [DOI] [PubMed] [Google Scholar]
- Floyd R, Eyualem A, Papert A, Blaxter M.L. Molecular barcodes for soil nematode identification. Mol. Ecol. 2002;11:839–850. doi: 10.1046/j.1365-294x.2002.01485.x. 10.1046/j.1365-294X.2002.01485.x [DOI] [PubMed] [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyns J. Problems of species delimitation in the genus Xiphinema, with special reference to monosexual species. In: Stone A.R, Platt H.M, Khalil L.F, editors. Concepts in nematode systematics. Academic Press; London: 1983. pp. 163–174. [Google Scholar]
- Hugall A, Stanton J, Moritz C. Reticulate evolution and the origins of ribosomal internal transcribed spacer diversity in apomictic Meloidogyne. Mol. Biol. Evol. 1999;16:157–164. doi: 10.1093/oxfordjournals.molbev.a026098. [DOI] [PubMed] [Google Scholar]
- Hugot J.P, Baujard P, Morand S. Biodiversity in helminths and nematodes as a field of study: an overview. Nematology. 2001;3:199–208. 10.1163/156854101750413270 [Google Scholar]
- Kaplan D.T, Thomas W.K, Frisse L.M, Sarah J.-L, Stanton J.M, Speijer P.R, Marin D.H, Opperman C.H. Phylogenetic analysis of geographically diverse Radopholus similis via rDNA sequence reveals a monomorphic motif. J. Nematol. 2000;32:134–142. [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambshead J. Recent developments in marine benthic biodiversity research. Oceanis. 1993;19:5–24. [Google Scholar]
- Lambshead P.J.D. Marine nematode biodiversity. In: Chen Z.X, Chen S.Y, Dickson D.W, editors. Nematode morphology, physiology and ecology. vol. 1. Tsinghua University Press; Tsinghua: 2004. pp. 438–492. [Google Scholar]
- Lipscomb D, Platnick N, Wheeler Q. The intellectual content of taxonomy: a comment on DNA taxonomy. Trends Ecol. Evol. 2003;18:65–66. 10.1016/S0169-5347(02)00060-5 [Google Scholar]
- Litvaitis M.K, Bates J.W, Hope W.D, Moens T. Inferring a classification of the Adenophorea (Nematoda). from nucleotide sequences of the D3 expansion segment (26/28S rDNA) Can. J. Zool. 2000;78:911–922. 10.1139/cjz-78-6-911 [Google Scholar]
- Lunt D.H, Hyman B.C. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. 10.1038/387247a0 [DOI] [PubMed] [Google Scholar]
- Maddison D.R, Maddison W.P. Sinauer Associates Inc; Sunderland, MA: 2000. MacClade v.4.0. [Google Scholar]
- Nadler S.A. Species delimitation and nematode biodiversity: phylogenies rule. Nematology. 2002;4:615–625. 10.1163/15685410260438908 [Google Scholar]
- Paquin P, Hedin M. The power and perils of ‘molecular taxonomy’: a case study of eyeless and endangered Cicurina (Araneae: Dictynidae) from Texas caves. Mol. Ecol. 2004;13:3239–3255. doi: 10.1111/j.1365-294X.2004.02296.x. 10.1111/j.1365-294X.2004.02296.x [DOI] [PubMed] [Google Scholar]
- Powers T. Nematode molecular diagnostics: from bands to barcodes. Annu. Rev. Phytopathol. 2004;42:367–383. doi: 10.1146/annurev.phyto.42.040803.140348. 10.1146/annurev.phyto.42.040803.140348 [DOI] [PubMed] [Google Scholar]
- Powers T.O, Todd T.C, Burnell A.M, Murray P.C.B, Fleming C.C, Szalanski A.L, Adams B.A, Harris T.S. The internal transcribed spacer region as a taxonomic marker for nematodes. J. Nematol. 1997;29:441–450. [PMC free article] [PubMed] [Google Scholar]
- Rusin L.Y, Aleshin V.V, Tchesunov A.V, Atrashkevich G.I. The 18S ribosomal RNA gene of Soboliphyme baturini Petrow, 1930 (Nematoda: Dioctophymida) and its implications for phylogenetic relationships within Dorylaimia. Nematology. 2003;5:615–628. 10.1163/156854103322683337 [Google Scholar]
- Schnabel R, Hutter H, Moerman D, Schnabel H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev. Biol. 1997;184:234–265. doi: 10.1006/dbio.1997.8509. 10.1006/dbio.1997.8509 [DOI] [PubMed] [Google Scholar]
- Sites J.W, Marshall J.C. Operational criteria for delimiting species. Annu. Rev. Ecol. Evol. Syst. 2004;35:199–227. 10.1146/annurev.ecolsys.35.112202.130128 [Google Scholar]
- Stanton J, Mundo-Ocampo M, Baldwin J.G, Kaplan D.T. Radopholus musicola n. sp., a new pathogenic species from Australia (Nematoda: Pratylenchidae) Nematology. 2001;3:689–698. 10.1163/156854101753536064 [Google Scholar]
- Subbotin S.A, Vierstraete A, De Ley P, Rowe J, Waeyenberge L, Moens M, Vanfleteren J.R. Phylogenetic relationships within the cyst-forming nematodes (Nematoda Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Mol. Phylogenet. Evol. 2001;21:1–16. doi: 10.1006/mpev.2001.0998. 10.1006/mpev.2001.0998 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates Inc; Sunderland, MA: 1999. Phylogenetic analysis using parsimony (*and other methods). Version 4. [Google Scholar]
- Tautz D, Arctander P, Minelli A, Thomas R.H, Vogler A.P. A plea for DNA taxonomy. Trends Ecol. Evol. 2003;18:71–74. 10.1016/S0169-5347(02)00041-1 [Google Scholar]
- Taylor H.L. The Taylor microcompressor mark II. Microscope. 1993;41:19–20. [Google Scholar]
- Thomas C, De Vries P, Hardin J, White J. Four-dimensional imaging: computer visualization of 3D movements in living specimens. Science. 1996;273:603–607. doi: 10.1126/science.273.5275.603. [DOI] [PubMed] [Google Scholar]
- Thomas W.K, Vida J.T, Frisse L.M, Mundo M, Baldwin J.G. DNA sequences from formalin-fixed nematodes, integrating molecular and morphological approaches to taxonomy. J. Nematol. 1997;29:250–254. [PMC free article] [PubMed] [Google Scholar]
- Torsvik V, Øvreøås L, Thingstad T.F. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. 10.1126/science.1071698 [DOI] [PubMed] [Google Scholar]
- Troell K, Mattsson J.G, Alderborn A, Hoglund J. PyrosequencingTM analysis identifies discrete populations of Haemonchus contortus from small ruminants. Int. J. Parasitol. 2003;33:765–771. doi: 10.1016/s0020-7519(03)00068-7. 10.1016/S0020-7519(03)00068-7 [DOI] [PubMed] [Google Scholar]
- Vanfleteren J.R, Vierstraete A.R. Insertional RNA editing in metazoan mitochondria: The cytochrome b gene in the nematode Teratocephalus lirellus. RNA. 1999;5:622–624. doi: 10.1017/s135583829999009x. 10.1017/S135583829999009X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler Q. Taxonomic triage and the poverty of phylogeny. Phil. Trans. R. Soc. B. 2004;359:571–583. doi: 10.1098/rstb.2003.1452. 10.1098/rstb.2003.1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.D, Schrank B, Huynh C, Shownkeen R, Waterston R.H. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Szalanski A.L, Robbins R.T. Phylogenetic relationships and genetic variation in Longidorus and Xiphinema Species (Nematoda: Longidoridae) using ITS1 sequences of nuclear ribosomal DNA1. J. Nematol. 2004;36:14–19. [PMC free article] [PubMed] [Google Scholar]