Abstract
The Integrated Primate Biomaterials and Information Resource (www.IPBIR.org) provides essential research reagents to the scientific community by establishing, verifying, maintaining, and distributing DNA and RNA derived from primate cell cultures. The IPBIR uses mitochondrial cytochrome c oxidase subunit I sequences to verify the identity of samples for quality control purposes in the accession, cell culture, DNA extraction processes and prior to shipping to end users. As a result, IPBIR is accumulating a database of ‘DNA barcodes’ for many species of primates. However, this quality control process is complicated by taxon specific patterns of ‘universal primer’ failure, as well as the amplification or co-amplification of nuclear pseudogenes of mitochondrial origins. To overcome these difficulties, taxon specific primers have been developed, and reverse transcriptase PCR is utilized to exclude these extraneous sequences from amplification. DNA barcoding of primates has applications to conservation and law enforcement. Depositing barcode sequences in a public database, along with primer sequences, trace files and associated quality scores, makes this species identification technique widely accessible. Reference DNA barcode sequences should be derived from, and linked to, specimens of known provenance in web-accessible collections in order to validate this system of molecular diagnostics.
Keywords: primate, DNA barcoding, mtDNA
1. Introduction
The advent of molecular techniques has opened new possibilities for taxonomic research, which is important given that the vast majority of all extant species are not well characterized morphologically. Taxonomies based on morphological analyses can be problematic due to either convergence in phenotype among unrelated species or the failure to identify ‘cryptic species’ where morphologic divergence has not kept pace with genetic divergence.
In an effort to standardize the approach to species identification using molecular techniques it has been proposed that as many species as possible be characterized for the same genetic markers (Blaxter 2004). The mitochondrial gene, cytochrome c oxidase subunit I (cox1) has been proposed as a candidate locus given its conserved sequence allows for ‘universal’ primers to be used across multiple divergent taxa and its high degree of phylogenetic signal relative to other mtDNA loci that have been used for interspecific analysis (e.g. 12s or 16s ribosomal DNA). This feature is perhaps due to heavy stabilizing selection within species for mitochondrial/nuclear cytochrome protein complexes (Hebert et al. 2003a,b). Thus a sequence of several hundred nucleotides in length acts as a unique identifier for members of a given species, hence the analogy to a computerized barcode label, although the analogy is imperfect given the existence of intraspecific variation so that not all members of a species are expected to be completely identical. Nonetheless the degree of intraspecific variation compared to the degree of interspecies variation would be expected to be low enough such that sequences from polymorphic species would cluster together in a genetic distance based cluster analysis.
Early studies demonstrated the efficacy of using universal primers to amplify and sequence a variety of taxa from divergent phyla (Hebert et al. 2003a), but the use of cox1 sequences as species identifying barcodes has been limited to a study of North American birds (Hebert et al. 2004a) and one complex species of neotropical butterflies (Astraptes fulgerator) from Costa Rica (Hebert et al. 2004b); but see e.g. Janzen et al. (2005), Saunders (2005), Smith et al. (2005) and Armstrong & Ball (2005) for recent examples. To date, the efficacy of molecular barcoding has not been determined within mammalian taxa.
Primates is a mammalian order with worldwide distribution, the members of which are important in conservation, evolutionary and biomedical studies. The taxonomic classification of extant species of the primate order has been agreed upon for several decades with a few interesting exceptions (Le Gros Clark 1954). The placement of tarsiers and the demarcation of the groupings among the hominoidea (including the genus Homo) are two of those exceptions that have been the focus of extensive taxonomic reorganization. Even with the overall structure of primate taxonomy in place there remains much work to be done in understanding the relationships of closely related taxa within many of the major groupings (Ruiz-Garcia & Alvarez 2003; Singer et al. 2003).
In addition, to elucidating relationships among the lower taxonomic levels of primates, there are practical aspects to DNA barcoding. The bush meat trade is threatening many wild populations of primates and other endangered species (Brashares et al. 2004). To effectively prosecute poachers trafficking in meat it would be beneficial to law enforcement and conservation officials to have access to a forensic database of primate cox1 samples so that positive identification of seized contraband could be made. Given that many non-human primates are important in biomedical research (Vandeberg & Williams-Blangero 1996, 1997) it has become increasingly desirable to genetically characterize the various species used in research. Thus far, there are species-specific differences (and even strain differences within species) to pathological aetiology and temperamental differences that may be important in behavioural research (Champoux et al. 1994; Champoux et al. 1997). In addition biomaterial repositories would have a simple and universal means of verifying the species identity of samples submitted to them for inclusion in their collections. Finally, the ability to identify or verify the source of biomaterials from field-collected specimens may be a useful tool to conservation and range scientists as well as a means of identifying the geographical provenience of captive born animals.
Past efforts to collect, store and develop genetic resources have largely been uncoordinated efforts scattered over different institutions and countries (Savolainen & Reeves 2004). Primates are a target group for scientific and technological development due to their importance in biomedicine and conservation biology, especially given their evolutionary proximity to humans. Within this context, the collection and storage of primate resources covering all branches of their taxonomy is an urgent need to boost primate molecular biology. Such resources include living and preserved collections, tissues, DNA, frozen viable cells and cell lines. The storage of complementary information on the origin, morphology, physiology, ecology, demography or behaviour of the specimens is also crucial to explore the link between gene and function. The coordinated development of these resources will prevent repeated sampling of wild populations, reduce the number of animals used in research, and help to standardize molecular tools and protocols.
The Integrated Primate Biomaterials and Information Resource (www.IPBIR.org) provides essential research reagents to the scientific community by establishing, verifying, maintaining and distributing DNA and RNA derived from primate cell cultures. Proper quality assurance/quality control requires the ability to verify the identity of samples as they move through the accession, culture and extraction processes. At present IPBIR has 97 of the approximately 200 different species (Cheney et al. 1986) of primates representing most of the major taxonomic divisions of the order. Given the taxonomic breadth of the samples in the resource it is important to choose a molecular marker that would work in as many species as possible.
IPBIR uses DNA sequences for routine identification of non-human biomaterials by simple sequence matching. The GenBank public-access database provides a working archive of available sequences, forming a valuable resource for such studies. However, the accuracy and reliability of sequences deposited in GenBank have been questioned, especially when the sequences are not linked to a voucher specimen. The identity of some DNA sequences deposited in public databases is being contested and there is a need to determine if such reports reveal a widespread phenomenon (Bridge et al. 2003).
The publicly available cox1 DNA sequences are distinct for each primate species, but represent a very incomplete data set. Moreover, sequence information can be difficult to interpret for several reasons. First, different levels of variation may occur in the same DNA region in different taxa, making generalized comparisons between taxa problematic. Second, amplification of non-target DNA from contaminants, or numts (nuclear mitochondrial DNA sequences), constitutes a danger. It has been proposed that as many as 500 copies of translocated mtDNA exist in the human nuclear genome (Richly & Leister 2004), ranging in length from 47 to 14 654 base pairs (bp) (Ricchetti et al. 2004). Hence, caution must be taken when amplifying any mitochondrial segment short of the entire mitochondrial genome. Third, sequences derived from unspecified reference materials cannot be validated.
In this study we investigate the feasibility of using cox1 sequence ‘molecular barcode’ data to verify the species designation of 225 individuals representing 56 species of primates (table 1) using both ‘universal’ cox1 primers identified in earlier molecular barcoding studies (Hebert et al. 2003a) as well as primers developed specifically for primate taxa.
Table 1.
Taxonomic distribution of the samples used in this study.
| infraorder | suborder | family | subfamily | genus | Species | number in this study |
|---|---|---|---|---|---|---|
| Haplorhini | Catarrhini | Cercopithecidae | Cercopithecinae | Allenopithecus | nigroviridis | 2 |
| Cercocebus | torquatus | 1 | ||||
| Cercopithecus | ascanius | 2 | ||||
| cephus | 1 | |||||
| lhoesti | 1 | |||||
| mitis | 6 | |||||
| neglectus | 7 | |||||
| petaurista | 1 | |||||
| wolfi | 1 | |||||
| Chlorocebus | aethiops | 27 | ||||
| Erythrocebus | patas | 1 | ||||
| Lophocebus | albigena | 1 | ||||
| Macaca | fascicularis | 1 | ||||
| fuscata | 1 | |||||
| nemestrina | 1 | |||||
| mulatta | 2 | |||||
| nigra | 1 | |||||
| thibetana | 1 | |||||
| Mandrillus | leucophaeus | 1 | ||||
| sphinx | 3 | |||||
| Miopithecus | talapoin | 1 | ||||
| Papio | anubis | 6 | ||||
| cynocephalus | 54 | |||||
| hamadryas | 2 | |||||
| Theropithecus | gelada | 2 | ||||
| Colobinae | Colobus | guereza | 2 | |||
| polykomos | 1 | |||||
| Nasalis | larvatus | 2 | ||||
| Semnopithecus | entellus | 1 | ||||
| Trachypithecus | francoisi | 1 | ||||
| Hylobatidae | Hylobates | gabriellae | 2 | |||
| agilis | 1 | |||||
| lar | 2 | |||||
| leucogenys | 1 | |||||
| pileatus | 1 | |||||
| syndactylus | 3 | |||||
| Hominidae | Pan | paniscus | 7 | |||
| troglodytes | 25 | |||||
| Gorilla | gorilla | 19 | ||||
| Pongo | pygmaeus | 9 | ||||
| Platyrrhini | Callitrichidae | Callimico | goeldii | 1 | ||
| Leontopithecus | rosalia | 4 | ||||
| Saguinus | fuscicollis | 1 | ||||
| midas | 1 | |||||
| Cebidae | Atelinae | Ateles | geoffroyi | 1 | ||
| Lagothrix | lagotricha | 1 | ||||
| Aotinae | Aotus | nancymaae | 1 | |||
| Pithecinae | Pithecia | pithecia | 1 | |||
| Cebinae | Saimiri | bolivensis | 1 | |||
| oerstedii | 1 | |||||
| Strepsirhini | Daubentoniidae | Daubentonia | madagascariensis | 1 | ||
| Lemuridae | Eulemur | fulvus | 1 | |||
| mongoz | 3 | |||||
| Varecia | variegata | 1 | ||||
| Galagonidae | Galago | moholi | 1 | |||
| Otolemur | garnettii | 1 | ||||
| total | 225 |
2. Material and methods
(a) DNA Extraction
Genomic DNA was extracted from cell pellets obtained from lymphoblastoid cell cultures, fibroblastoid cell cultures or buffy coats obtained from whole blood using standard salting-out techniques (Miller et al. 1988) or from tissue biopsies or plasma/serum using QiaAmp DNA Blood Kits (Qiagen).
(b) PCR Amplification
A region approximately 727-bp long near the 5′ terminus of the cox1 gene was amplified using one of three primer sets (table 2). PCR reactions were done in a total volume of 25 μL and consisted of 2.5 μL of 10× PCR II buffer (Applied Biosystems), 2.5 μL of 25 mM MgCl2, 2.0 μL of 10 mM dNTP mix (2.5 mM each dNTP), 0.2 μL of each primer (25 μM stock) and 0.2 μL of 5 U/μL TaqGold DNA polymerase (Applied Biosystems), 2.0 μL of DNA template (∼50 ng) and dH2O to 25 μL. The thermocycling conditions were as follows: 96.0°C for 10 min to activate the TaqGold and then 35 cycles of 96.0°C for 1 min, 50.0 to 58.0°C for 1 min and 72.0°C for 1 min followed by a final hold of 72.0°C for 10 min. PCR products were visualized on 6% polyacrylamide minigels and the PCR product was purified using QiaQuick 96 PCR Purification Kit (Qiagen). The purified PCR product was eluted into 45 μL buffer AE (Qiagen).
Table 2.
Primer set success by genus.
| taxa | primer set 1 | primer set 2 | primer set 3 |
|---|---|---|---|
| Pan | yes | no | yes |
| Gorilla | yes | no | yes |
| Pongo | no | yes | |
| Hylobates | yes | no | |
| Papio | no | yes | yes |
| Cercopithecus | no | yes | yes |
| Macaca | yes | no | yes |
| Mandrillus | yes | no | yes |
| Chlorocebus | no | no | yes |
| Allenopithecus | yes | yes | yes |
| Leontopithecus | yes | yes | |
| Saimiri | yes | yes | |
| Pithecia | yes | ||
| Varecia | yes | ||
| Daubentonia | yes | no | no |
Primer Set 1 comprised LCOI1490: ggt caa caa atc ata aag ata ttg g and HCOI2198: taa act tca ggg tga cca aaa aat ca.
Primer Set 2 comprised VERTCOIf1: ttc tca acc aac caa caa aga cat tgg and VERTCOIr1: tag act tct ggg tgg cca aag aat ca.
Primer Set 3 comprised OWMCO-If: (A/G)CT (G/C)TT TTC AAC AAA (C/T)CA (C/T)AA AGA C and OWMCO-Ir: GTA (A/G)AC TTC (G/C)GG GTG (A/G)CC (A/G)AA GAA TC.
(c) DNA Sequencing
Cycle sequencing reactions were carried out in 10 μL total volume. A forward and reverse reaction was performed for each sample consisting of 5.4 μL of the purified PCR product, 4 μL of Big Dye Terminator Ready Reaction Mix, v1.1(Applied Biosystems) and 0.6 μL of 2.5 μM primer (same as PCR primer). Cycling conditions were 96.0°C for 1 min and then 25 cycles of 96.0°C for 10 s, 50.0°C for 5 s, 60.0°C for 4 min. Unincorporated fluorescent dye terminators were removed from the cycle sequencing reactions using SigmaSpin 96-well plate (Sigma Aldrich), dried at 37°C for 20 min and resuspended in 10 μL of HiDi formamide (ABI). The cycle sequencing product was detected using an ABI 3730 DNA analyser.
(d) Sequence analysis
The forward and reverse sequence files for each sample were analysed and a consensus sequence for each sample was created using ‘Sequencher’ (GeneCodes). The consensus sequences were in turn aligned using ‘Sequencher’ and exported into a NEXUS file for distance analysis and cluster analysis using PAUP 4.0b10 (Swofford 1998). Mean pairwise differences were computed for all species and genera. In order to validate the cox1 sequences obtained in this study we compared them with cox1 sequences from primate species for which the whole mitochondrial DNA sequence has been established and deposited in GenBank (http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/9347.html) as well as previously archived cox1 sequences (Andrews & Easteal 2000; Wu et al. 2000). In addition we constructed neighbour-joining trees using PAUP and based on Kimura 2p distances to determine whether the sequences cluster as would be expected based on overall taxonomic affinity.
(e) rtPCR
In the case of several old world monkey species, PCR products obtained using the different primers yielded different sequences. The paucity of whole mitochondrial sequences for cercopithecines limited our ability to determine which of the cox1 sequences were derived from mitochondrial cox1 and which were derived from numts. Reverse transcriptase PCR (rtPCR) was performed on Papio anubis DNA obtained from lymphoblasts to ensure amplification of the transcribed mitochondrial cox1 (Collura et al. 1996). Reverse transcription products were amplified in three separate reactions using the three primer sets. The RNA extractions were also amplified with AmpliTaq in place of reverse transcriptase to identify any DNA contamination and a positive control rtPCR reaction was also performed using primers for phosphoglycerate kinase (PGK).
3. Results
All samples in this study amplified with one or more primer sets (table 2). Generally, if samples representing a given species amplified with Primer set 1 (Hebert et al. 2003a) and gave ‘phylogenetically credible’ results they were not assayed with the other two primer sets. In some cases where a species did not amplify with one primer but did with another they were tested with the third primer set in order to increase the probability the sequence was not obtained from numts (Thalmann et al. 2004 and references therein) or other spurious amplicons. In addition to checking clustering for an expected phylogenetic signal to determine whether a sequence was derived from the actual cox1 and not a numt we checked to see that the fragments were identical to, or at least clustered with, the appropriate whole mitochondrial sequences obtained from GenBank. Also, sequences which yielded amino acid transcription interrupted by stop codons were not included in the analysis as they would not likely be derived from the functional mitochondrial cox1 gene. This did occur in a handful of cercopithecine species but the design of primer set 3 did eliminate amplification of spurious cox1 in those species.
The results of the reverse transcription PCR experiment showed that primer sets 2 and 3 amplified cox1 from RNA extracts of P. anubis lymphoblasts (figure 1). Primer set 1 did not amplify a fragment from the RNA preparation indicating that the Primer set 1 fragment amplified from genomic DNA is not from the mitochondrial genome but is possibly from a numt.
Figure 1.
Results of rtPCR performed on RNA extracted from Papio anubis liver. Lanes 1 & 13; Msp I digested pBR322 ladder. Lane 2; PGK positive control. Lane 3; PGK DNA control (no RNA). Lane 4; amplification of genomic DNA using primer set1. Lane 5; rtPCR with primer set 1. Lane 6; ‘mock’ rtPCR using AmpliTaq instead of reverse transcriptase and primer set 1. Lane 7; amplification of genomic DNA using primer set2. Lane 8; rtPCR with primer set 2. Lane 9; ‘mock’ rtPCR using AmpliTaq instead of reverse transcriptase and primer set 2. Lane 10; amplification of genomic DNA using primer set 3. Lane 11; rtPCR with primer set 3. Lane 12; ‘mock’ rtPCR using AmpliTaq instead of reverse transcriptase and primer set 3.
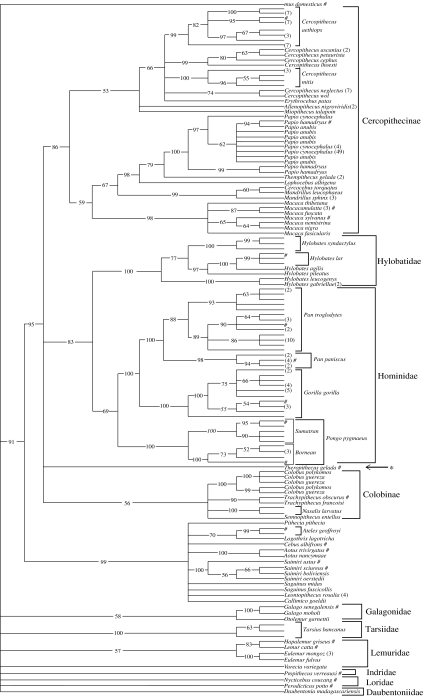
The results of the sequencing analysis for each of the samples were submitted to GenBank (Accession numbers: AY544148–62, AY632376–7, AY671787–98, AY673675, AY972630–808). The sequences were aligned with cox1 sequences obtained from primate whole mitochondrial genomes obtained from GenBank as well as primate cox1 sequences from other studies that have been deposited in GenBank. Sequences less than 400 bp long were excluded from the analysis. A neighbour-joining tree (figure 2) based on Kimura 2p distances (Kimura 1980) was calculated using PAUP, bootstrap values are based on 1000 replicates.
Figure 2.
Bootstrapped neighbour-joining tree calculated from Kimura 2p distances and based on 1000 replicates. The number in parentheses indicates the number of samples that share identical sequences at that position; #, indicates the position of cox1 sequences obtained from GenBank.
The neighbor-joining tree generally agrees with the commonly accepted primate phylogeny with platyrrhine sequences clustering together 99% of the time and catarrhines cluster 95%. The strepsirhines, however, do not form a cohesive cluster; this is not unexpected given the relatively short fragment used in the analysis. There are 17 cox1 sequences from previous studies that were retrieved from GenBank. In ten of the cases the sequences from this study clustered with cox1 sequences obtained from GenBank at the species level (table 3). In four cases the sequences from this study were not represented in GenBank by the same species, however they did cluster together with GenBank derived sequences at the generic level (Saimiri, Macaca sylvanus, Trachypithecus and Galago). In the remaining three cases the GenBank sequences did not cluster with the sequences derived from the same species in this study: i.e. Colobus polykomos and C. guereza did not cluster separately, Papio hamadryas from GenBank clustered with P. anubis in this study and Theropithecus gelada from GenBank did not cluster with T. gelada from this study nor did it cluster with any of the cercopithecine cox1 sequences.
Table 3.
Samples from this study that cluster with cox1 sequences from fully sequenced mtDNAs and cox1 sequences from previous studies obtained from GenBank.
| GenBank acc no. | species | IPBIR sample number | species |
|---|---|---|---|
| gi:4239858 | Ateles geoffroyi | PR00134 | Ateles geoffroyi |
| gi:12484071 | Saimiri sciureus | PR00741 | Saimiri oerstedii |
| gi:12484069 | Saimiri ustus | PR00474 | Saimiri boliviensis |
| gi:4239860 | Colobus polykomos | PR00655 | Colobus guereza |
| gi:60392100 | Colobus guereza | PR00597 | Colobus polykomos |
| PR00980 | Colobus guereza | ||
| gi:60392086 | Trachypithecus obscura | PR01099 | Trachypithecus francoisi |
| gi:49146236 | Macaca mulatta | PR00112 | Macaca mulatta |
| PR00408 | |||
| gi:14010693 | Macaca sylvanus | with all macaques | |
| gi:5835638 | Papio hamadryas | PR00041 | Papio anubis |
| gi:12484067 | Cercopithecus aethiops | BP00219 | Chlorocebus |
| BP00214 | (Cercopithecus) aethiops | ||
| BP00211 | |||
| BP00221 | |||
| BP00220 | |||
| BP00213 | |||
| BP00215 | |||
| gi:5835820 | Hylobates lar | PR00495 | Hylobates lar |
| PR00715 | |||
| gi:5835834 | Pongo pygmaeus abelii | PR00253 | Pongo pygmaeus |
| PR00841 | Sumatran | ||
| PR00054 | |||
| PR01003 | |||
| gi:5835163 | Pongo pygmaeus | PR00276 | Pongo pygmaeus |
| PR01011 | Bornean | ||
| PR00002 | |||
| PR00648 | |||
| PR00488 | |||
| gi:5835121 | Pan troglodytes | PR00744 | Pan troglodytes |
| PR00643 | |||
| PR00660 | |||
| PR00512 | |||
| PR00226 | |||
| PR00953 | |||
| gi:5835135 | Pan paniscus | PR00092 | Pan paniscus |
| PR00446 | |||
| PR00111 | |||
| PR00367 | |||
| PR00366 | |||
| gi:5835149 | Gorilla gorilla | PR00573 | Gorilla gorilla |
| PR00265 | |||
| PR01054 | |||
| PR00807 | |||
| gi:12484065 | Theropithecus gelada | does not cluster with cercopithecines | |
| gi:21449875 | Lemur catta | PR00126 | Lemur catta |
| PR00715 | |||
| gi:4239864 | Galago senegalensis | PR00519 | Galago moholi |
In the cases for which there are no previously reported data to which we can compare the sequences generated in this study we tallied the extent to which our sequences cluster with members of the same species (table 4). There are 10 clusters supported in 100% of the bootstrap replicates in which all members of a given species cluster together. In fact, except for Papio and Colobus as mentioned above, there are no cases where we have multiple specimens of a species in which the sequences do not cluster together.
Table 4.
Samples from this study that cluster consistently in a neighbour-joining bootstrap analysis based on Kimura 2p distances.
| bootstrap value | IPBIR sample number | species |
|---|---|---|
| 100 | PR00786 | Leontopithecus rosalia |
| PR00960 | ||
| PR00961 | ||
| PR00963 | ||
| 100 | PR00232 | Mandrillus leucophaeus |
| PR00295 | ||
| 100 | PR00398 | Mandrillus sphinx |
| PR01048 | ||
| PR00718 | ||
| 100 | PR00566 | Cercopithecus ascanius |
| PR00634 | ||
| 100 | PR00710 | Cercopithecus mitis |
| PR00993 | ||
| PR00995 | ||
| PR00987 | ||
| PR00991 | ||
| PR00997 | ||
| 100 | PR01121 | Cercopithecus neglectus |
| PR00981 | ||
| PR00983 | ||
| PR00985 | ||
| 100 | PR00100 | Allenopithecus nigrovirids |
| PR00198 | ||
| 100 | PR00969 | Hylobates syndactylus |
| PR00598 | ||
| PR00721 | ||
| 100 | PR00381 | Hylobates gabriellae |
| PR00652 | ||
| 100 | PR00370 | Eulemur mongoz |
| PR00338 | ||
| PR00288 |
4. Discussion
Using a segment of the 5′ region of cox1 we are able to identify the appropriate species from which a biomaterial submitted to the Integrated Primate Biomaterials and Information Resource was derived. This ability to generate a ‘molecular barcode’ is useful in our case for quality control and the management of the IPBIR. It allows us to verify the identity of samples, as reported by the submitter, as they move through each stage of the accession, cell culture, DNA extraction and aliquoting processes. Since the samples comprising the IPBIR collection are from identified specimens of known species, the cox1 sequences derived from the IPBIR resource have the potential to serve as a forensic database for the identification of primate biomaterials such as those seized in the bush meat trade.
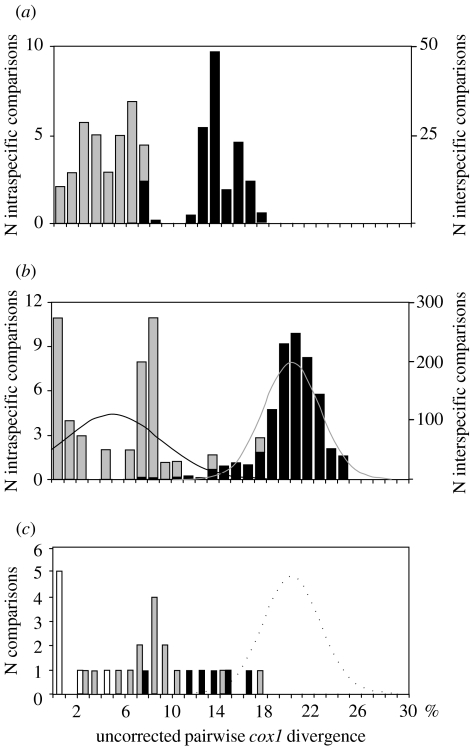
The success of DNA barcoding depends on the amount of intraspecific variation relative to the amount of interspecific variation present among species across their range. The amount of intraspecific variation, measured as mean pairwise difference, in the present study varies (figure 3) from none (Eulemur mongoz, Leontopithecus rosalia, Cercopithecus neglectus) to 0.038 for Pongo pygmaeus (mean for all species =0.011, s.e. =0.004). The amount of intraspecific variation at cox1 will depend on the degree to which the samples reflect the geographic diversity of widely dispersed species (e.g. Pan troglodytes and Chlorocebus aethiops), the amount of gene flow among subpopulations and also whether the species is perhaps an amalgam of multiple species. For example it has been argued that the Sumatran (P. pygmaeus abelii) and Bornean orangutans (P. p. pygmaeus) actually are distinct species (Xu & Arnason 1996; Warren et al. 2001); in fact the orangutans in this study cluster robustly by subspecies (100% of bootstrap replicates) and the degree of genetic divergence is comparable to that that exists between Pan paniscus and P. troglodytes, which lends support for the case that the genus Pongo includes two separate species.
Figure 3.
Mean pairwise differences (Kimura 2p) at various taxonomic levels.
It is interesting to note that there are three cases where the cox1 sequences derived from the complete mitochondrial genome sequences did not cluster with the same species from this study. In the first case GenBank sequence gi:4049475 is listed as being obtained from P. hamadryas but the two P. hamadryas from this study PR00440 and PR00559 cluster together outside of the P. cynocephalus / P. anubis group whereas gi:4049475 clusters with P. anubis samples. Since it is known that P. anubis and P. hamadryas do interbreed (Szmulewicz et al. 1999) it is possible that the GenBank sample represents a hybrid individual. In any event this study supports Newman et al.'s (2004) finding that P. cynocephalus and P. anubis are not monophyletic clades but rather cluster together.
The second case involves C. polykomos (gi:4239860) and C. guereza (gi:60392100). These two cox1 sequences were submitted independently to GenBank; they differ at only 8 positions out of the 1545 bp that constitute the whole cox1 sequence, in fact in the first 700 bp which constitute the region used for molecular barcoding they differ at only a single position. Clearly these two individuals fall within the range of variation of a single species. One of the samples from this study (PR00655, C. guereza) clusters 100% of the time with these two GenBank sequences. Two other samples (PR00597, C. polykomos and PR00980, C. guereza) also cluster together 100% of the time. The two Colobus samples cluster together as well 100% of the time. Thus we will need to determine whether the species named associated with these samples are indeed correct or whether cox1 will not differentiate these species as in the P. anubis/P. cynocephalus case.
In the third case a specimen from GenBank (gi:12484065, listed as T. gelada) does not cluster with the T. gelada from this study nor does it even cluster with other African cercopithecids, perhaps representing a numt, contamination or a misidentification of the original sample. This sample points out the importance of having barcode sequences linked to a morphologically vouchered specimen so that such anomalies can be resolved by returning to the original sample (Ruedas et al. 2000).
The generation of incorrect sequences appears to happen for several reasons, not the least of which being the misidentification of the original material. Other problems involve primer specificity and the amplification or co-amplification of numts. Still other problems for the repository involve contamination of the cell culture in the laboratory. Non-target DNA from contaminants or numts can easily result in the extraction or amplification of incorrect or chimeric DNA sequences.
For molecular barcoding to have forensic value, reference barcode sequences should be derived from, and linked to, voucher specimens in web-accessible collections. NCBI maintains a number of databases (including GenBank, PubMed, Taxonomy and others) that are linked together in the Entrez indexing and retrieval engine. The LinkOut program allows outside groups to maintain sets of hotlinks from objects in Entrez back to specific locations on their web sites. The Integrated Primate Biomaterials and Information Resource has indexed holdings in the taxonomy domain of Entrez and indexed barcode sequences derived from the repository specimens in the sequence domain of Entrez (GenBank). This is a simple and practical approach to the problem of linking biological specimens with the biological data and research that are derived from them. The individual barcodes validate the species identification of specimens submitted to the repository and collectively form a publicly available reference database for primate molecular diagnostics.
Although GenBank encourages the separate submission of identical sequences obtained from multiple specimens, current practice in phylogenetics research often involves reporting only the variable haplotypes. For barcoding to assume a quantitative approach to species diagnosis, barcode sequences have been submitted for all specimens in the study. Moreover, data quality is of crucial importance to barcoding if it is to develop into a forensic tool. The Consortium for the Barcode of Life (CBOL; www.barcoding.si.edu) Database Working Group is calling for the deposition of barcode sequences in GenBank together with the primers that were used to generate them, their trace files and associated quality scores.
With the characterization of the IPBIR collection, we have expanded the number of primate barcodes from about a dozen sequences from unvalidated source material in GenBank (derived from primate whole mtDNA sequences), to include 56 species of the approximately 200 species in the order primates. This work sheds light on the reliability of the existing data and represents a significant increase in the potential for DNA barcoding to be employed as a tool for molecular diagnostics of primates.
Prior to this study, a major practical concern for DNA barcoding was the relatively few sequences deposited for which a specimen was available for re-examination. The deposition of materials in IPBIR, a public-access collection, provides a mechanism to allow verification of potentially problematic data and the re-examination of source material as advocated by Ruedas et al. (2000). The DNA barcode data generated by IPBIR represents sequences not previously found in GenBank. Thus, we hope the quality control efforts performed on the samples provided to the IPBIR will contribute significantly to the expansion of data available for the study of primate genetic diversity.
Acknowledgments
We thank the Fannie E. Rippel Foundation for their generous support for the primate DNA barcoding initiative. We thank Patrick K. Bender, Donald Coppock, Brian Fisher and Vincent Savolainen for constructive reviews of this manuscript. This material is based upon work supported by the National Science Foundation under Grant Nos BCS0094928 and BCS0094993.
Footnotes
One contribution of 18 to a Theme Issue ‘DNA barcoding of life’.
References
- Andrews T.D, Easteal S. Evolutionary rate acceleration of cytochrome c oxidase subunit in simian primates. J. Mol. Evol. 2000;50:562–569. doi: 10.1007/s002390010059. [DOI] [PubMed] [Google Scholar]
- Armstrong K.F, Ball S.L. DNA barcodes for biosecurity: invasive species identification. Phil. Trans. R. Soc. B. 2005;360:1813–1823. doi: 10.1098/rstb.2005.1713. 10.1098/rstb.2005.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M.L. The promise of a DNA taxonomy. Phil. Trans. R. Soc. B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. 10.1098/rstb.2003.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brashares J.S, Arcese P, Sam M.K, Coppolillo P.B, Sinclair A.R.E, Balmford A. Science. 2004;306:1180–1183. doi: 10.1126/science.1102425. 10.1126/science.1102425 [DOI] [PubMed] [Google Scholar]
- Bridge P.D, Roberts P.J, Spooner B.M, Pancha G. On the unreliability of published DNA sequences. New Phytol. 2003;160:43–48. doi: 10.1046/j.1469-8137.2003.00861.x. 10.1046/j.1469-8137.2003.00861.x [DOI] [PubMed] [Google Scholar]
- Champoux M, Suomi S.J, Schneider M.L. Temperament differences between captive Indian and Chinese–Indian hybrid rhesus macaque neonates. Lab. Anim. Sci. 1994;44:351–357. [PubMed] [Google Scholar]
- Champoux M, Higley J.D, Suomi S.J. Behavioral and physiological characteristics of Indian and Chinese–Indian hybrid rhesus macaque infants. Dev. Psychobiol. 1997;31:49–63. 10.1002/(SICI)1098-2302(199707)31:1%3C49::AID-DEV5%3E3.0.CO;2-U [PubMed] [Google Scholar]
- Cheney D.L, Seyfarth R, Smuts B, Wrangham R. In: The study of primate societies. Primate societies. Smuts B, Cheney D.L, Seyfarth R, Wrangham R, Struhsaker T, editors. University of Chicago Press; Chicago, IL: 1986. pp. 1–8. [Google Scholar]
- Collura R.V, Auerbach M.R, Stewart C.B. A quick, direct method that can differentiate expressed mitochondrial genes from their nuclear pseudogenes. Curr. Biol. 1996;6:1337–1339. doi: 10.1016/s0960-9822(02)70720-3. 10.1016/S0960-9822(02)70720-3 [DOI] [PubMed] [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003a;270:313–321. doi: 10.1098/rspb.2002.2218. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Ratnasingham S, deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B. 2003;270(Suppl. 1):S96–S99. doi: 10.1098/rsbl.2003.0025. 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Stoeckle M.Y, Zemlak T.S, Francis C.M. Identification of birds through DNA barcodes. PloS Biol. 2004a;2:1657–1663. doi: 10.1371/journal.pbio.0020312. 10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Penton E.H, Burns J.M, Janzen D.H, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004b;101:14812–14817. doi: 10.1073/pnas.0406166101. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen D.H, Hajibabaei M, Burns J.M, Hallwachs W, Remigio E, Hebert P.D.N. Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1835–1845. doi: 10.1098/rstb.2005.1715. 10.1098/rstb.2005.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- LeGros Clark W.E. British Museum; London: 1954. History of the primates: an introduction to the study of fossil man. [Google Scholar]
- Miller S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nuc. Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T.K, Jolly C.J, Rogers J. Mitochondrial phylogeny and systematics of baboons (Papio) Am. J. Phys. Anthropol. 2004;124:17–27. doi: 10.1002/ajpa.10340. 10.1002/ajpa.10340 [DOI] [PubMed] [Google Scholar]
- Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PloS Biol. 2004;2:E273. doi: 10.1371/journal.pbio.0020273. 10.1371/journal.pbio.0020273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol. Biol Evol. 2004;21:1081–1084. doi: 10.1093/molbev/msh110. 10.1093/molbev/msh110 [DOI] [PubMed] [Google Scholar]
- Ruedas L.A, Salazar-Bravo J, Dragoo J.W, Yates T.L. The importance of being earnest: what, if anything, constitutes a specimen examined? Mol. Phylogenet. Evol. 2000;17:129–132. doi: 10.1006/mpev.2000.0737. 10.1006/mpev.2000.0737 [DOI] [PubMed] [Google Scholar]
- Ruiz-Garcia M, Alvarez D. RFLP analysis of mtDNA from six platyrrhine genera: phylogenetic inferences. Folia Primatol. 2003;74:59–70. doi: 10.1159/000069998. 10.1159/000069998 [DOI] [PubMed] [Google Scholar]
- Saunders G.W. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Phil. Trans. R. Soc. B. 2005;360:1879–1888. doi: 10.1098/rstb.2005.1719. 10.1098/rstb.2005.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen V, Reeves G. A Plea for DNA banking. Science. 2004;304:1445. doi: 10.1126/science.304.5676.1445b. 10.1126/science.304.5676.1445b [DOI] [PubMed] [Google Scholar]
- Singer S.S, Schmitz J, Schwiegk C, Zischler H. Molecular cladistic markers in New World monkey phylogeny (Platyrrhini Primates) Mol. Phylogenet. Evol. 2003;26:490–501. doi: 10.1016/s1055-7903(02)00312-3. 10.1016/S1055-7903(02)00312-3 [DOI] [PubMed] [Google Scholar]
- Smith M.A, Fisher B.L, Hebert P.D.N. DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar. Phil. Trans. R. Soc. B. 2005;360:1825–1837. doi: 10.1098/rstb.2005.1714. 10.1098/rstb.2005.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 1998. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4. [Google Scholar]
- Szmulewicz M.N, Andino L.M, Reategui E.P, Woolley-Barke T, Jolly C.J, Disotell T.R, Herrera R.J. An Alu insertion polymorphism in a baboon hybrid zone. Am. J. Phys. Anthropol. 1999;109:1–8. doi: 10.1002/(SICI)1096-8644(199905)109:1<1::AID-AJPA1>3.0.CO;2-X. 10.1002/(SICI)1096-8644(199905)109:1%3C1::AID-AJPA1%3E3.0.CO;2-X [DOI] [PubMed] [Google Scholar]
- Thalmann O, Hebler J, Poinar H.N, Paabo S, Vigilant L. Unreliable mtDNA data due to nuclear insertions: a cautionary tale from analysis of humans and other great apes. Mol. Ecol. 2004;13:321–335. doi: 10.1046/j.1365-294x.2003.02070.x. 10.1046/j.1365-294X.2003.02070.x [DOI] [PubMed] [Google Scholar]
- Vandeberg J.L, Williams-Blangero S. Strategies for using nonhuman primates in genetic research on multifactorial diseases. Lab. Anim. Sci. 1996;46:146–151. [PubMed] [Google Scholar]
- Vandeberg J.L, Williams-Blangero S. Advantages and limitations of nonhuman primates as animal models in genetic research on complex diseases. J. Med. Primatol. 1997;26:113–119. doi: 10.1111/j.1600-0684.1997.tb00042.x. [DOI] [PubMed] [Google Scholar]
- Warren K.S, et al. Speciation and intrasubspecific variation of Bornean orangutans, Pongo pygmaeus pygmaeus. Mol. Biol. Evol. 2001;18:472–480. doi: 10.1093/oxfordjournals.molbev.a003826. [DOI] [PubMed] [Google Scholar]
- Wu S, Schmidt T.R, Goodman M, Grossman L.I. Molecular evolution of cytochrome c oxidase subunit I in primates: is there coevolution between mitochondrial and nuclear genomes? Mol. Phylogenet. Evol. 2000;17:294–304. doi: 10.1006/mpev.2000.0833. 10.1006/mpev.2000.0833 [DOI] [PubMed] [Google Scholar]
- Xu X, Arnason U. The mitochondrial DNA molecule of Sumatran orangutan and a molecular proposal for two (Bornean and Sumatran) species of orangutan. J. Mol. Evol. 1996;43:431–437. doi: 10.1007/BF02337514. [DOI] [PubMed] [Google Scholar]