Abstract
Large-scale DNA barcoding projects are now moving toward activation while the creation of a comprehensive barcode library for eukaryotes will ultimately require the acquisition of some 100 million barcodes. To satisfy this need, analytical facilities must adopt protocols that can support the rapid, cost-effective assembly of barcodes. In this paper we discuss the prospects for establishing high volume DNA barcoding facilities by evaluating key steps in the analytical chain from specimens to barcodes. Alliances with members of the taxonomic community represent the most effective strategy for provisioning the analytical chain with specimens. The optimal protocols for DNA extraction and subsequent PCR amplification of the barcode region depend strongly on their condition, but production targets of 100K barcode records per year are now feasible for facilities working with compliant specimens. The analysis of museum collections is currently challenging, but PCR cocktails that combine polymerases with repair enzyme(s) promise future success. Barcode analysis is already a cost-effective option for species identification in some situations and this will increasingly be the case as reference libraries are assembled and analytical protocols are simplified.
Keywords: museum specimens, DNA isolation, PCR, species identification, taxonomy, cox1
1. Assembling DNA barcodes: the challenge
DNA barcoding promises fast, accurate species identifications by focusing analysis on a short standardized segment of the genome (Hebert et al. 2003). Several studies have now established that sequence diversity in a ∼650 bp region near the 5′ end of the mitochondrial cytochrome oxidase subunit I (cox1; also referred to as COI) gene provides strong species-level resolution for varied animal groups including birds (Hebert et al. 2004b), fishes (Ward et al. 2005), springtails (Hogg & Hebert 2005), spiders (Barrett & Hebert 2005) and moths (Hebert et al. 2003; Janzen et al. 2005). These early results have provoked larger-scale barcoding efforts and global projects for fishes and birds have now been initiated (Marshall 2005). These projects represent the first wave in a series of initiatives which will demand the capability to assemble barcodes rapidly and cost-effectively. As one looks further to the future, the need for substantial analytical capacity looms. For example, an effort to barcode the 1.7 million described species (Hawksworth 1995) would require the assembly of some 20 million barcodes, given a target of about 10 barcodes per species. This total will rise fivefold if barcode coverage is desired for all 10 million eukaryote species (e.g. Hammond 1992), producing a sequence library of 65 billion base pairs, approximately twice the current size of GenBank (April 2005). This task could be completed within a decade by establishing 50 core laboratories, each producing 200 000 barcode records per year. When viewed from the perspective of major genomic facilities, some of which generate more than 50 million sequences a year, the production goals for barcode facilities may seem modest. However the business of generating barcodes is complex; each record represents a sequence derived from a specimen that had to be collected, archived and databased.
In the balance of this paper, we direct most of our attention to an evaluation of how the primary steps in the analytical chain extending from specimens to barcode records can be optimized, scaled up and economized. Because the single most critical step to achieve high production involves a move from protocols based on single specimens to those compatible with 96-well format, we only consider methods compliant with this approach.
2. Specimens
(a) Sourcing specimens
Specimens are the raw material for any barcode facility. This need can be met most easily by sequencing all specimens encountered, but because of the lognormal distribution of species abundance (May 1975), most of the resultant sequences will derive from a few common species. Collaborations with taxonomists represent a far more effective strategy for provisioning the analytical chain with specimens (Janzen et al. 2005; Smith et al. 2005; Ward et al. 2005). With this approach, it is feasible to assemble a library of sequences that provides both broad species coverage and similar sampling intensity across species (e.g. 10 barcodes each). Moreover, sample sizes can be increased in cases where complexities, such as cryptic species, are encountered in the first pass (Hebert et al. 2004a; Janzen et al. 2005).
We have adopted the TrakMates micro-plate system (Matrix Technologies, Hudson, NH, USA) to force the organization of specimen shipments into the blocks of 96 needed for the later stages of analysis. One micro-plate holds 96 vials (94 specimens, two controls), each uniquely barcoded on the bottom of the vial. These barcoded vials can be rapidly scanned, aiding the tracking of specimens as they enter the analytical chain. Aside from an organized flow of specimens to the barcode facility, there is a critical need for the firm connection of specimens to their collaterals. To facilitate this, we have developed a spreadsheet that organizes key specimen information. We have, as well, developed web-based software to both organize the specimen information and to connect each barcode sequence with its source specimen (see below).
(b) Preservation/handling
Whenever possible, animal specimens should be killed and preserved in a DNA-friendly fashion (freezing, cyanide and ethanol). Even brief exposure to agents that damage DNA, such as ethyl acetate or formaldehyde, should be avoided (Prendini et al. 2002). While fresh or freshly frozen tissues are ideal for analysis, DNA in dried specimens ordinarily remains easily analysed for 5 years, although degradation rises as time passes. Specimens preserved in absolute ethanol are easily analysed when young, but acidification soon degrades their DNA unless it is regularly replaced or buffered. As a general principle, barcode analysis should follow collection as soon as possible, but delays of a few months will cause little problem.
To minimize external or cross-contamination, all tissue samples should be handled on a clean working surface and all instruments should be acid or flame sterilized before handling a new specimen. When using 96-well plates for tissue assembly, particular care must be taken when adding samples to avoid cross-contamination between wells.
(c) The importance of archival specimens
Natural history museums and herbaria maintain most of the world's known biodiversity within their collections. In some groups, species coverage may be nearly complete. For example, museums hold nearly 10 million bird specimens (Roselaar 2003), assuring deep coverage for most of the 10 000 known species. The analysis of museum specimens could enable rapid growth in barcode coverage (Janzen et al. 2005). Unfortunately, they are generally poor targets for analysis because of DNA degradation due to hydrolysis and oxidation (Lindahl 1993), exposure to ultraviolet light (Eglinton & Logan 1991) and preservation agents such as formaldehyde (Schander & Halanych 2003). Methods used to retrieve DNA from museum specimens typically aim to isolate DNA with high efficiency (Junqueira et al. 2002). Because many copies of the mitochondrial genome are present in each cell, its component genes, such as cox1, represent optimal targets for analysis in archival specimens. However, because the template DNA is degraded, few amplicons longer than 300–400 bp can be obtained from specimens more than a decade old (Su et al. 1999; Junqueira et al. 2002; Rohland et al. 2004). When degradation is particularly severe, one common strategy involves the amplification of less than 100 bp DNA fragments (Goldstein & Desalle 2003). In such cases, obtaining a DNA barcode will require the concatenation of several short sequences (i.e. Su et al. 1999).
3. DNA isolation
(a) Different strategies
Methods for DNA isolation fall into two broad categories: DNA release and DNA extraction. DNA release protocols aim to rapidly release DNA into solution, making it accessible for downstream applications such as PCR. Release-based methods also enable DNA isolation from samples without their physical disruption. In this case, the entire specimen can be removed after DNA isolation, allowing the retention of a voucher in cases where this would not otherwise be possible. Release methods are, however, not very sensitive and do not produce high purity DNA suitable for long-term storage (e.g. more than 1 year). By contrast, DNA extraction methods aim to purify DNA, often by binding it to a membrane (e.g. silica) or by chemical fractionation. Some classical methods, such as phenol/chloroform extractions (Sambrook et al. 1989), are not attractive because they are time consuming and involve toxic materials. The type and condition of specimens is a key factor in selecting a DNA isolation method. For fresh or recently collected tissue, a release-based DNA extraction usually provides sufficient DNA for barcoding. However, for archival material, more sensitive approaches should be used. Because little DNA is needed for barcode analysis, the amount of tissue used in DNA isolation is usually minute. Figure 1 shows four typical tissue samples for barcode analysis.
Figure 1.
Typical specimen types and sizes used for DNA barcoding analysis as compared to a pencil head. (a), a lepidopteran leg; (b), a Daphnia; (c), a feather; (d), muscle tissue.
(b) Comparing DNA isolation techniques
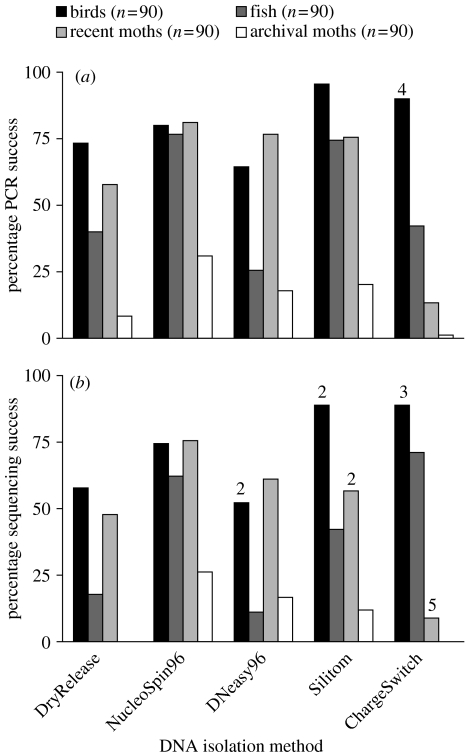
In order to determine an optimal procedure for high volume barcoding, we compared five DNA isolation methods on four sets of specimens (birds, fish, recent and archival moths—see Electronic Appendix part 1A for details). The major criterion for the inclusion of methods in this performance test was their capacity for high-throughput analysis, but we also considered cost and sensitivity. These methods included an artisanal (=homemade) DNA release method, called DryRelease, which employs Chelex resin as a DNA release agent (Walsh et al. 1991). We also examined three DNA extraction methods that use silica to bind DNA: Silitom, an artisanal method based on the protocols of Elphinstone et al. (2003) and Boom et al. (1990), NucleoSpin96 tissue kit (Machery-Nagel, Düren, Germany) and DNeasy96 tissue kit (QIAGEN, Hilden, Germany). Finally, we tested a DNA extraction method that uses magnetic beads to bind DNA: ChargeSwitch Forensic kit (Invitrogen, Carlsbad, California, USA; see Electronic Appendix part 1B and 2A for details of these methods).
The effectiveness of these five DNA isolation methods was compared by testing their success in PCR amplification (using visual inspection on an agarose gel; see Electronic Appendix part 1F for details) of the full-length (∼650 bp) cox1 barcode with primer sets specific for each taxonomic group (see Electronic Appendix part 1E for details) (figure 2; table 1). Overall, the NucleoSpin96 kit was most effective, producing more than 75% success for three of the groups of specimens, and 31% for the most difficult group (archival moths). However, it was not always the best: the Silitom and ChargeSwitch methods produced higher success for bird samples. Interestingly, the DNeasy96 kit was less effective than the NucleoSpin96 kit, despite their very similar methodologies. This difference was particularly striking for fishes where the NucleoSpin96 kit delivered three times as many successful amplifications. The ChargeSwitch method produced the most variable results with 90% PCR success for birds, but only 13% and 1% for recent and archival moths, respectively.
Figure 2.
Evaluation of DNA isolation methods for high volume DNA barcoding analysis in different types of specimens. Five DNA isolation methods were compared for the amplification of full-length (∼650 bp) cox1 barcode sequence, by examining (a), % PCR success and (b), % sequencing success. Numbers on columns indicate cases of contamination.
Table 1.
Comparison of the overall performance of DNA isolation methods.
| method | category | % sensitivitya | ageb | ease of use | time/96 samples (h) (technician, total) | contaminationc | storage potentiald | price per sample (US$)e |
|---|---|---|---|---|---|---|---|---|
| DryRelease | release | 44.7 | 2 | easy | 1.5, 20 | 0 | low | 0.27 |
| NucleoSpin96 | extract | 66.7 | 15 | moderate | 2.5, 21.5 | 0 | moderate | 1.90 |
| DNeasy96 | extract | 45.8 | 4 | moderate | 2.5, 21.5 | 2 | moderate | 2.11 |
| Silitom | extract | 66.1 | 8 | moderate | 3.0, 21.5 | 4 | moderate | 0.32 |
| ChargeSwitch | extract | 36.7 | 2 | easy | 3.5, 5.0 | 8 | moderate | 1.25 |
Average percentage PCR success, on an agarose gel, for all samples tested.
Maximum age of samples with positive PCR result.
Number of sequencing contaminations across all 90 specimens.
One year at −20°C.
Based on 2005 US list prices.
All PCR reactions were sequenced to ascertain their performance in delivering both a clean cox1 sequence and one that derived from the presumptive source specimen (Electronic Appendix part 1G provides sequencing protocol). In most cases, a small percentage of the visible PCR products failed to generate a clean sequence, but the differences between extraction methods were small (figure 2). The only exception involved the ChargeSwitch method for fishes where the number of sequences obtained was higher than the number of visible PCR products (figure 2).
The strong performance of silica-based approaches makes them appropriate for high-throughput barcoding, especially when work is focused on the analysis of small tissue samples. Substantial cost savings (80%) can be realized by the use of an artisanal protocol such as Silitom rather than commercial kits. Moreover, when samples are young or well-preserved a simple release-based artisanal protocol such as DryRelease could represent the optimal approach in many cases (table 1). We note that the amount of tissue used in the DryRelease protocol, where potential PCR inhibitors in the tissue are not removed, can greatly influence the results. As a consequence, success rates can often be substantially increased by optimizing the amount of tissue at the initiation of a study and we have done this for many large-scale projects.
None of the negative controls (six in each set of 96 samples for a total of 24 per method) produced an amplification product, except the ChargeSwitch analysis on birds, where four of six negative controls showed an amplicon (figure 2). However, we did observe more cases where the PCR product did not derive from the correct specimen. The ChargeSwitch kit showed the highest number of such contaminated sequences (five and three in recent moths and birds, respectively), but four sequence contaminations were observed in Silitom isolations (two in both recent moths and birds) and two in the DNeasy96 amplifications, both from birds. These results suggest that some protocols are more sensitive to contamination than others, and that the risk of its occurrence is greatest when one is working with tissues that yield relatively large amounts of DNA. Our results further suggest that the ChargeSwitch method, where the DNA is attached to magnetic beads in liquid phase, is particularly sensitive to handling errors leading to contamination, especially when the protocol is performed in 96-well format. By contrast, the two kits (NucleoSpin96, DNeasy96) where the DNA is bound to a silica gel membrane inside a column offer more security.
(c) Which DNA isolation method is best for archival specimens?
In our studies, the NucleoSpin96 kit produced the highest amplification success for the full-length barcode region of cox1 in archival moths (31%) followed by Silitom (20%), DNeasy96 kit (18%), DryRelease (8%) and ChargeSwitch kit (1%; figure 2). These results make it clear that silica-based methods should be used for DNA isolation from archival specimens.
4. PCR amplification
(a) Primer design is critical for high success
Before starting a barcode project on any new taxonomic group, it is essential to test the performance of existing primers on fresh specimens from a range of species in the target group. If one or two current primer sets do not deliver more than 95% amplification success for the test assemblage, primer redesign should be undertaken. Our past studies on varied taxonomic assemblages have shown that minor adjustments in primer sequences can have a large impact on barcode recovery. Primer reconfiguration begins by aligning all available sequences for the target taxonomic group. Subsequent adjustments in sequence to maximize matches have enabled the development of effective primer sets (more than 95% amplification across species) for large taxonomic assemblages, such as Lepidoptera (Janzen et al. 2005), birds (Hebert et al. 2004b) and fish (Ward et al. 2005). In most cases, effectively complete barcode recovery for all species in a group can be achieved with two sets of non-degenerate primers. Primers with degenerate positions or modified bases such as inosine (which can form base pairs with all four nucleotides) can help with recalcitrant groups where variable nucleotide positions across taxa compromise amplification (Candrian et al. 1991). Using primers with degenerate positions may also reduce the chance of preferential amplification of nuclear pseudogenes (Sorenson et al. 1999). Many software packages are available to aid primer design, but we recommend Primer3 (Rozen & Skaletsky 2000) for designing non-degenerate primers and Codehop (Rose et al. 2003) for degenerate primers.
(b) PCR optimization
An optimized PCR for the barcode region of cox1 should yield a single sharp amplicon, with no more than minor sub-banding when examined on an agarose gel. This can often be achieved by optimizing cycling conditions, especially the annealing temperature, and by altering the concentration of PCR reagents such as magnesium, dNTPs and primers through pilot studies on a few taxonomically divergent members of the target assemblage. Optimization often also dramatically increases amplification success and can eliminate the need for PCR cleanup prior to the sequencing reaction. PCR amplification can also be enhanced with additives such as bovine serum albumin, betaine, DMSO (Abu Al-Soud & Radstrom 2000) and trehalose. Trehalose is especially useful because it acts as a potent PCR enhancer by both lowering the DNA melting temperature and stabilizing Taq polymerase (Spiess et al. 2004). Trehalose can also overcome the effect of PCR inhibitors that are often present in crude DNA extracts (e.g. DNA release methods). Minimalization of the volume of each PCR reaction is also important to reduce reagent use and cost; 10 μl reactions should be employed.
(c) Evaluation of different polymerases
Taq DNA polymerase from Thermus aquaticus (Saiki et al. 1988) is standard for PCR, but a wide variety of other polymerases have higher fidelity or processivity (e.g. Cline et al. 1996). As well, more complex PCR cocktails that include one or more repair enzymes offer new hope for the amplification of degraded DNA (Di Bernardo et al. 2002; Mitchell et al. 2005). Restorase (Sigma-Aldrich, St. Louis, MO, USA) represents one recently introduced commercial enzyme cocktail that couples AccuTaq, a high accuracy polymerase, with a repair enzyme.
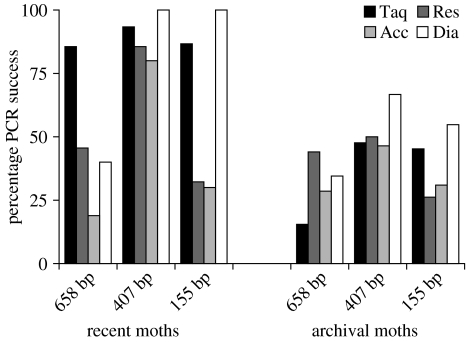
We evaluated the effectiveness of four polymerases on DNA isolated using the NucleoSpin96 kit from two sets of specimens: recent moths (90 samples, six negative controls) and archival moths (84 samples, 12 negative controls). The recent moths were all less than 1 year old, whereas the archival moths included 14 specimens from each of six age groups (2, 4, 8, 16, ∼32, ∼64 years; See Electronic Appendix part 1A for details). We tested amplification of the DNA extract from each specimen using: Taq polymerase, Diamond DNA polymerase (Bioline, Randolph, MA, USA), AccuTaq (Sigma-Aldrich, St. Louis, MO, USA) and Restorase. Each enzyme was used according to the manufacturer's instructions, but the amount of template DNA was constant across all four enzymes (See Electronic Appendix part 1D for details). We tested these enzymes for their ability to amplify the full-length cox1 barcode (658 bp), as well as partial barcode sequences of 407 and 155 bp (see Electronic Appendix part 1E for sequences of primers).
As expected, positive PCR results were much higher for recent than archival specimens (figure 3). For recent moths, the highest overall success was obtained with Taq polymerase (86%, 93% and 87% success for 658, 407 and 155 bp amplicons, respectively). For archival moths, Restorase performed best overall (44%, 50% and 26% success for 658, 407 and 155 bp amplicons, respectively), but the Diamond and Taq polymerases outperformed it for the smallest amplicon. This latter result was not wholly surprising as Restorase is not recommended for the amplification of small targets. Our results indicate that standard Taq polymerase provides both high performance and low cost for specimens whose DNA has not been degraded, while the use of Restorase merits consideration in archival specimens.
Figure 3.
Evaluation of different PCR enzymes for the amplification of cox1 for recent and archival specimens. Four enzymes including Taq polymerase (Taq), Restorase (Res), AccuTaq (Acc) and Diamond polymerase (Dia) were compared for the amplification of full-length and partial cox1 barcodes (658, 407, 155 bp). Results are shown as % PCR success.
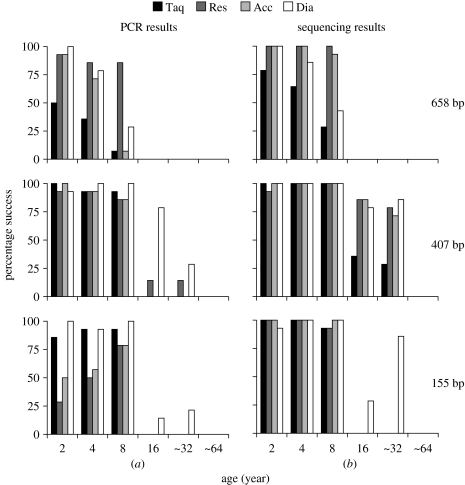
We further compared PCR and sequencing results for different age groups of the archival moths (figure 4). For all four enzymes, success in both PCR and sequencing declined with specimen age for all three amplicons. Restorase delivered the highest PCR success for the full-length product, but none of the enzymes produced 658 bp amplicons from samples older than 8 years (figure 4). For the 407 bp amplicon, all four enzymes performed well, amplifying almost 100% of the samples 8 years and younger. In samples older than 8 years, Taq polymerase showed lower success compared to the other three enzymes (figure 4). Restorase, AccuTaq and Diamond polymerase performed similarly and produced 407 bp amplicons from about 70% of the samples as old as 32 years (figure 4). However, results with Diamond polymerase were inflated by two sequence contaminations. Surprisingly, for the smallest amplicon (155 bp), all four enzymes performed poorly for samples older than 8 years (figure 4).
Figure 4.
Effectiveness of four PCR enzymes for the amplification of cox1 in archived specimens of varied ages. Taq polymerase (Taq), Restorase (Res), AccuTaq (Acc) and Diamond polymerase (Dia) were compared for the amplification of full and partial cox1 barcodes (658, 407,155 bp) in archival moth specimens from six age groups (2, 4, 8, 16, ∼32, ∼64 years). Results are shown as (a), % PCR success and (b), % sequencing success.
Direct sequencing of all PCR reactions revealed an interesting result: sequences were sometimes recovered from samples where no PCR product was evident on the agarose gel. This was particularly the case for samples 8 years and older. For example, agarose gels revealed only two of 14 positive PCR products in Restorase amplification of 32 year old moths, but we obtained 11 sequences (with no sign of contamination) from the same reactions (figure 4). Visualization of PCR products on agarose requires a product concentration of at least 1–2 ng μl−1 (Sambrook et al. 1989; White & Wu 2001), while capillary sequencers are known to be more sensitive. This fact suggests that all PCR products from archival specimens should be sequenced.
(d) Archival specimens and DNA repair
AccuTaq is the polymerase present in the Restorase enzyme blend so a comparison of results using Restorase versus those using AccuTaq can indicate if the repair mechanism in Restorase aids barcode recovery from archival specimens. We found that Restorase produced more PCR positives on agarose gels for both the 658 and 407 bp amplicons than AccuTaq (figure 4). However, we found no difference once these samples were sequenced (figure 4). This result suggests that Restorase aids PCR yield, perhaps by repairing template damage, but that the effect is small. In earlier experiments with Restorase, we were able to amplify full-length 658 bp cox1 barcodes from moths up to 70 years old. However, this success required extensive optimizations that are not time- or cost-effective when the goal is high production rates. However, in the case of extremely rare or extinct species, this capacity could be valuable.
5. Screening PCR products
When working on a new taxonomic group or on specimens where PCR success is uncertain, it is helpful to screen PCR reactions for product. This has traditionally been a laborious task involving gel casting and the loading of individual reactions onto the gel. We have explored two options to accelerate this process: microfluidic devices and pre-cast agarose gels. Microfluidic devices ‘sip’ small volumes of the PCR reaction from each of the 96 wells on a plate and then run electrophoresis on a very small scale to determine both the size and concentration of the PCR product (e.g. Greiner et al. 2004). Unfortunately, current devices have several limitations for DNA barcoding; they are expensive (more than $100K, €80K), have high operating costs and are relatively slow. These disadvantages are not offset by any gain in sensitivity: the detection limits for these systems mirror those that can be achieved through agarose gels. Pre-cast agarose gels represent a second option: they are fast, require little capital investment (less than $1K, €0.8K) and have modest operating cost. We regularly employ the E-Gel 96 system (Invitrogen, Carlsbad, CA, USA) to screen PCR products, but similar gels are manufactured by several other suppliers.
6. Sequencing
(a) Sequencing reaction optimization
Sequencing reactions employ standard chemistry, but reactions can be run in low volume format with diluted sequencing mix (i.e. BigDye; Applied Biosystems, Foster City, CA, USA) without compromising sequencing success or quality. By employing a 10 μl reaction volume containing 0.25 μl BigDye (1/16 of standard reaction), the cost of each sequencing reaction can be substantially reduced. Before the reaction product is submitted for sequencing, it must be cleaned up. There are a variety of solutions for this step and several are scalable to very large production rates. Ethanol precipitation and magnetic bead protocols are widely used by major genomic facilities, but column-based approaches are also effective. Any high volume DNA barcoding facility requires access to one or more capillary sequencers, such as the ABI 3730 DNA Analyser (Applied Biosystems, Foster City, CA, USA). Based on a bidirectional sequencing each barcode record represents two ‘reads’ (see below). Operating seven days a week, a single ABI 3730 can generate just 200K reads or 100K barcode records per year, setting a production threshold for the facility.
(b) Sequence assembly and edit
A bidirectional sequencing strategy has the advantage of enabling the use of automated sequence assembly software to both assign quality values like PHRED scores (Ewing et al. 1998) for each base position and produce a consensus barcode sequence from the reads. It also enhances the quality of the final barcode and ensures its compliance with the minimum read length (i.e. 550 bp) needed to gain barcode designation (by avoiding signal deterioration that often occurs at the end of the reads). Manual inspection and editing of the barcode sequence at the electropherogram level are still required to validate sequence quality and to check for possible polymorphic sites. Their presence, which is often overlooked by sequence assembly software, can indicate the co-amplification of nuclear pseudogenes (Bensasson et al. 2001) along with the authentic mitochondrial sequence. Several software packages are available for visualizing, editing and assembling sequences. Sequencher (Gene Codes Corporation, Ann Arbor, MI, USA) and Seqscape (Applied Biosystems, Foster City, CA, USA) are the most popular commercial software options and include features such as internal basecallers, automatic alignment, contig assembly and trimming of sequences.
7. Barcode of life data systems
Large-scale DNA barcoding projects will create a substantial number of sequence records that must each link to a voucher specimen, as well as to its collateral data. These records need to be organized and analysed. In addition, for the barcode database to be useful for species identification, it must be searchable by sequence, as well as by species name and higher taxonomic categories. The Barcode of Life Data Systems (BOLD), see http://www.barcodinglife.org, provides varied support for a large-scale barcode project. It is an online repository for cox1 sequences as well as a workbench for barcode analysis that includes three components: a laboratory information management system (LIMS), a data management and analysis system (DMAS), and a species identification engine.
(a) Laboratory information management system
The assembly and storage of hundreds of thousands of barcode records requires a LIMS to ensure the accurate tracking of all specimens passing through the multi-step analytical chain. Commercial LIMS are available, but they typically cost $50–100K (€40–80K) per site license, and they lack some key functionality required to support the DNA barcoding initiative.
(b) Data management and analysis system
The DMAS of BOLD provides support for both the warehousing and analysis of barcode records. It includes a simple interface enabling the upload of new sequences to password-protected projects. Its web-based delivery allows work to proceed simultaneously in different labs while being managed from a centralized location, improving communication and preventing data loss or duplication. Moreover, because the DMAS includes information on where each specimen was collected, where it is currently deposited, copies of sequence traces, and high-resolution photographs of each specimen, it allows straightforward traceability of the data stream back to the source. The DMAS was designed to operate at a global scale, ideally supported by mirror sites based at facilities active in barcode analysis.
The DMAS also includes a suite of analysis tools that allow processing or visualization of data. Sequence records, which can be submitted via a simple interface, are automatically aligned. Specimen pages are created automatically from the user-defined data, including an automated plot of GIS coordinates on high-resolution, multi-scale geographic maps. BOLD also includes programs for assembling and exporting neighbour-joining trees (Saitou & Nei 1987), which include colour coding to indicate taxonomic affiliation or other user-defined parameters, as well as tools for specimen display. Finally the DMAS includes an interface that allows the bulk submission of barcode records to GenBank.
(c) Species identification engine
The first step in creating a DNA-based species identification system lies in the assembly of a comprehensive barcode sequence library. The second step involves the development of an effective engine for the comparison and matching of sequences from new specimens to the barcode library. The species identification engine, BOLD-ID, includes a simple user interface to allow cox1 sequences to be entered into a search field and automatically compared against the existing dataset. BOLD-ID makes use of a combination of Local Alignment Search Tool (BLAST; Altschul et al. 1990) and hidden Markov models based on a global protein alignment for the cox1 gene, which increase both the speed and accuracy of the matching procedure. Using this algorithm, BOLD returns a probability-based match profile indicating the likely identity of the source species. Links to the species page provide additional information about it (e.g. photographs) that can be useful in confirming the identification. Aside from identifying single specimens, BOLD-ID also performs batch identifications on 96-well plates of samples. The current version of BOLD-ID is optimized for cox1 gene. However, future versions will include the capability to analyse barcode data from other genes or non-coding regions because barcoding systems in some groups (i.e. plants) will use such data (Chase et al. 2005; Kress et al. 2005).
8. Prospects for high volume DNA barcoding
Few molecular taxonomy and evolution laboratories process more than a few thousand specimens a year, but the assembly of a comprehensive barcode library will require, as noted earlier, 100-fold higher production rates. In one sense, the protocols described in this paper are unproven because no barcoding facility has yet achieved this production target. However, we are confident, based on our own experience (table 2), that these protocols will allow the 100K goal to be achieved (see Electronic Appendix part 2 for routine protocols). We emphasize that there is no single optimized protocol if varied types of specimens are being analysed. For example, our work on recently collected North American Lepidoptera employed the DryRelease protocol for DNA isolation, followed by PCR recovery of the full-length barcode. By contrast, results on Costa Rican Lepidoptera, aged from 1 to 28 years, were greatly improved by using the NucleoSpin96 kit for DNA isolation. Moreover, when a full-length cox1 barcode could not be recovered (mainly in samples more than 10 years old), additional PCRs were used to obtain 400 and 350 bp barcode sequences that were concatenated to produce the full-length sequence. These two examples provide a sense of the methodological flexibility that is critical to achieve high success while minimizing costs. While our work has been mainly focused on animals, we expect that barcode analysis of other organisms, such as plants, will require substantial protocol changes, particularly in the isolation of DNA and in the choice of a target genomic barcode region (Kress et al. 2005).
Table 2.
DNA barcodes generated in various projects.
| project | sample | storage condition | specimens age range (year) | DNA isolation | barcodes generateda | % successb |
|---|---|---|---|---|---|---|
| Lepidoptera of North America | leg | dried | 1–3 | DryRelease | 6510 | 99 |
| Lepidoptera of the ACGc | leg | dried | 1–28 | NucleoSpin96 | 2419 | 80 |
| birds of North America | muscle, liver, feather | ethanol, dried, DMSO | 0–38 | DryRelease | 1782 | 74 |
| fishes of Australia | muscle | ethanol | 1–15 | DryRelease | 913 | 96 |
Number of barcode sequences generated as of April 7, 2005.
Percentage successful DNA barcoding from total number of specimens tested.
Area de Conservacion Guanacaste in northwestern Costa Rica.
Although barcoding can be executed in a decentralized fashion, economies of scale are gained by establishing core facilities. The capital costs involved in creating a facility capable of generating 100K barcodes a year will range from US$0.5–0.8M (€0.4–0.6M) with the higher figure allowing the emplacement of two capillary sequencers. However, much smaller capital investments ($50K, €40K) will allow the creation of facilities capable of generating 100K PCR products that might then be submitted to any sequencing facility for analysis. The generation of 100K barcode records based on bidirectional sequencing will require an operating budget of approximately $0.3M (€0.2M) before salaries. Although such investments will allow an impressive advance on past production levels, it may be insufficient as work moves from the construction of barcode libraries to the routine application of DNA barcodes for rapid, large-scale assessments of biodiversity in conservation biology and other ecological contexts (DeSalle & Amato 2004). Fortunately there are prospects for both further reductions in cost and increases in production. Costs will drop as reaction volumes shrink and microfluidic devices, which employ nanolitre reaction volumes for PCR and sequencing, are under development. There is also the potential for robotic intervention, which when coupled with unidirectional short reads for identifications (as opposed to reference barcodes) could drive production levels to more than 500 000 specimens per year from a single sequencer. In short, the prospects for both the assembly and use of barcode libraries appear bright enough to expect illumination of many key problems in biodiversity science.
Acknowledgments
Funding for this study was provided by the Gordon and Betty Moore Foundation, the Canada Foundation for Innovation, the Ontario Innovation Trust, the Canada Research Chairs Program and NSERC. We thank Daniel Janzen, Scott Miller, Jean-François Landry, John Burns, Tyler Zemlak and Kevin Kerr for providing specimens and Andrey Poltaraus for aid with analytical protocols. We also thank the Sigma-Aldrich R & D group, especially Kevin Kayser, for providing Restorase and Accutaq enzymes and for helpful discussions. Alex Borisenko aided the assembly of graphics for this paper, while Janet Topan oversaw the sequencing. We thank the Consortium for Barcoding Life for facilitating the DNA barcoding initiative.
Footnotes
One contribution of 18 to a Theme Issue ‘DNA barcoding of life’.
Supplementary Material
References
- Abu Al-Soud W, Radstrom P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 2000;38:4463–4470. doi: 10.1128/jcm.38.12.4463-4470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F, Gish W, Miller W, Myers E.W, Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Barrett R, Hebert P.D.N. Identifying spiders through DNA barcodes. Can. J. Zool. 2005;83:481–491. [Google Scholar]
- Bensasson D, Zhang D, Hartl D.L, Hewitt G.M. Mitochondrial pseudogenes: evolution's misplaced witnesses. Trends Ecol. Evol. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. 10.1016/S0169-5347(01)02151-6 [DOI] [PubMed] [Google Scholar]
- Boom R, Sol C.J, Salimans M.M, Jansen C.L, Wertheim-van Dillen P.M, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candrian U, Furrer B, Hofelein C, Luthy J. Use of inosine-containing oligonucleotide primers for enzymatic amplification of different alleles of the gene coding for heat-stable toxin type I of enterotoxigenic Escherichia coli. Appl. Environ. Microbiol. 1991;57:955–961. doi: 10.1128/aem.57.4.955-961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase M.W, Salamin N, Wilkinson M, Dunwell J.M, Kesanakurthi R.P, Haidar N, Savolainen V. Land plants and DNA barcodes: short-term and long-term goals. Phil. Trans. R. Soc. B. 2005;360:1889–1895. doi: 10.1098/rstb.2005.1720. 10.1098/rstb.2005.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline J, Braman J.C, Hogrefe H.H. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. 10.1093/nar/24.18.3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalle R, Amato G. The expansion of conservation genetics. Nat. Rev. Genet. 2004;5:702–712. doi: 10.1038/nrg1425. 10.1038/nrg1425 [DOI] [PubMed] [Google Scholar]
- Di Bernardo G, Del Gaudio S, Cammarota M, Galderisi U, Cascino A, Cipollaro M. Enzymatic repair of selected cross-linked homoduplex molecules enhances nuclear gene rescue from Pompeii and Herculaneum remains. Nucleic Acids Res. 2002;30:e16. doi: 10.1093/nar/30.4.e16. 10.1093/nar/30.4.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eglinton G, Logan G.A. Molecular preservation. Phil. Trans. R. Soc. B. 1991;333:315–327. doi: 10.1098/rstb.1991.0081. discussion 327–328. [DOI] [PubMed] [Google Scholar]
- Elphinstone M.S, Hinten G.N, Anderson M.J, Nock C.J. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Mol. Ecol. Notes. 2003;3:317–320. 10.1046/j.1471-8286.2003.00397.x [Google Scholar]
- Ewing B, Hillier L, Wendl M.C, Green P. Base-calling of automated sequencer traces using phred I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Goldstein P.Z, Desalle R. Calibrating phylogenetic species formation in a threatened insect using DNA from historical specimens. Mol. Ecol. 2003;12:1993–1998. doi: 10.1046/j.1365-294x.2003.01860.x. 10.1046/j.1365-294X.2003.01860.x [DOI] [PubMed] [Google Scholar]
- Greiner M, Carter P, Korn B, Zink D. New approach to complete automation in sizing and quantitation of DNA and proteins by the Automated Lab-on-a-Chip Platform from Agilent Technologies. Nat. Methods. 2004;1:87–89. 10.1038/nmeth1004-87 [Google Scholar]
- Hammond P. Species inventory. In: Groombridge B, editor. Global biodiversity: status of the earth's living resources. Chapman & Hall; London: 1992. [Google Scholar]
- Hawksworth D.L. Cambridge University Press; Cambridge: 1995. Global biodiversity assessment. [Google Scholar]
- Hebert P.D, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D, Penton E.H, Burns J.M, Janzen D.H, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004a;101:14 812–14 817. doi: 10.1073/pnas.0406166101. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D, Stoeckle M.Y, Zemlak T.S, Francis C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004b;2:E312. doi: 10.1371/journal.pbio.0020312. 10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg I.D, Hebert P.D.N. Biological identifications of springtails (Hexapoda: Collembola) from the Canadian arctic, using mitochondrial barcodes. Can. J. Zool. 2005;82:749–754. [Google Scholar]
- Janzen D.H, Hajibabaei M, Burns J.M, Hallwachs W, Remigio E, Hebert P.D.N. Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1835–1845. doi: 10.1098/rstb.2005.1715. 10.1098/rstb.2005.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira A.C, Lessinger A.C, Azeredo-Espin A.M. Methods for the recovery of mitochondrial DNA sequences from museum specimens of myiasis-causing flies. Med. Vet. Entomol. 2002;16:39–45. doi: 10.1046/j.0269-283x.2002.00336.x. 10.1046/j.0269-283x.2002.00336.x [DOI] [PubMed] [Google Scholar]
- Kress J.W, Wurdack K.J, Zimmer E.A.C, Weigt L.A, Janzen D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl Acad. Sci. USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. 10.1073/pnas.0503123102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. 10.1038/362709a0 [DOI] [PubMed] [Google Scholar]
- Marshall E. Taxonomy. Will DNA bar codes breathe life into classification? Science. 2005;307:1037. doi: 10.1126/science.307.5712.1037. 10.1126/science.307.5712.1037 [DOI] [PubMed] [Google Scholar]
- May R.M. Patterns of species abundance and diversity. In: Cody M, Diamond J, editors. Ecology and evolution of communities. Belknap Press of Harvard University Press; Cambridge: 1975. pp. 81–120. [Google Scholar]
- Mitchell D, Willerslev E, Hansen A. Damage and repair of ancient DNA. Mutat. Res. 2005;571:265–276. doi: 10.1016/j.mrfmmm.2004.06.060. [DOI] [PubMed] [Google Scholar]
- Prendini L, Hanner R, DeSalle R. Obtaining, storing and archiving specimens and tissue samples for use in molecular studies. In: DeSalle R, Giribet G, Wheeler W.C, editors. Techniques in molecular evolution and systematics. Birkhaeuser Verlag AG; Basel: 2002. pp. 176–248. [Google Scholar]
- Rohland N, Siedel H, Hofreiter M. Nondestructive DNA extraction method for mitochondrial DNA analyses of museum specimens. Biotechniques. 2004;36:814–816. doi: 10.2144/04365ST05. see also pp. 818–821. [DOI] [PubMed] [Google Scholar]
- Rose T.M, Henikoff J.G, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. 10.1093/nar/gkg524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselaar K. An inventory of major European bird collections. Bull. Br. Ornithol. Clin. 2003;123A:253–337. [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Saiki R.K, Gelfand D.H, Stoffel S, Scharf S.J, Higuchi R, Horn G.T, Mullis K.B, Erlich H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E.F, Maniatis T. Cold Spring Harbor Press; Cold Spring Harbor: 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- Schander C, Halanych K.M. DNA, PCR and formalinized animal tissue—a short review and protocols. Org. Divers. Evol. 2003;3:195–205. [Google Scholar]
- Smith M.A, Fisher B.L, Hebert P.D.N. Barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar. Phil. Trans. R. Soc. B. 2005;360:1825–1834. doi: 10.1098/rstb.2005.1714. 10.1098/rstb.2005.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson M.D, Ast J.C, Dimcheff D.E, Yuri T, Mindell D.P. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol. Phylogenet. Evol. 1999;12:105–114. doi: 10.1006/mpev.1998.0602. 10.1006/mpev.1998.0602 [DOI] [PubMed] [Google Scholar]
- Spiess A.N, Mueller N, Ivell R. Trehalose is a potent PCR enhancer: lowering of DNA melting temperature and thermal stabilization of taq polymerase by the disaccharide trehalose. Clin. Chem. 2004;50:1256–1259. doi: 10.1373/clinchem.2004.031336. 10.1373/clinchem.2004.031336 [DOI] [PubMed] [Google Scholar]
- Su B, Wang Y.X, Lan H, Wang W, Zhang Y. Phylogenetic study of complete cytochrome b genes in musk deer (genus Moschus) using museum samples. Mol. Phylogenet. Evol. 1999;12:241–249. doi: 10.1006/mpev.1999.0616. 10.1006/mpev.1999.0616 [DOI] [PubMed] [Google Scholar]
- Walsh P.S, Metzger D.A, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Ward R.D, Zemlak T.S, Innes B.H, Last P.R, Hebert P.D.N. DNA barcoding Australia's fish species. Phil. Trans. R. Soc. B. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. 10.1098/rstb.2005.1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White H.W, Wu M. Factors affecting quantitation of DNA bands in gels using a charge-coupled device imaging system. Electrophoresis. 2001;22:860–863. doi: 10.1002/1522-2683()22:5<860::AID-ELPS860>3.0.CO;2-D. 10.1002/1522-2683()22:5%3C860::AID-ELPS860%3E3.0.CO;2-D [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.