Abstract
After the process of DNA barcoding has become well advanced in a group of organisms, as it has in the economically important fungi, the question then arises as to whether shorter and literally more barcode-like DNA segments should be utilized to facilitate rapid identification and, where applicable, detection. Through appropriate software analysis of typical full-length barcodes (generally over 500 base pairs long), uniquely distinctive oligonucleotide ‘microcodes’ of less than 25 bp can be found that allow rapid identification of circa 100–200 species on various array-like platforms. Microarrays can in principle fulfill the function of microcode-based species identification but, because of their high cost and low level of reusability, they tend to be less cost-effective. Two alternative platforms in current use in fungal identification are reusable nylon-based macroarrays and the Luminex system of specific, colour-coded DNA detection beads analysed by means of a flow cytometer. When the most efficient means of rapid barcode-based species identification is sought, a choice can be made either for one of these methodologies or for basic high-throughput sequencing, depending on the strategic outlook of the investigator and on current costs. Arrays and functionally similar platforms may have a particular advantage when a biologically complex material such as soil or a human respiratory secretion sample is analysed to give a census of relevant species present.
Keywords: macroarrays, species identification, barcoding, fungi
1. Introduction
The concept of DNA barcoding emerged from a background of molecular phylogenetic analysis (Hebert et al. 2003). Scientists working with fungi, like those working with other microorganisms, were attracted to molecular phylogenetics very early in its history. This was partly because the organisms they dealt with were morphologically enigmatic, and thus biosystematically intractable, and partly because most of the organisms could readily be grown in culture, facilitating centralized DNA sequence comparison of strains sampled all over the world (e.g. O'Donnell et al. 1998, Kurtzman 1994, Scorzetti et al. 2002). Comparative biosystematics by nature demands the comparison of homologies—folk wisdom about comparing apples to apples and not to oranges applies here—and therefore there was a rapid emergence of a very small number of easily sequenced gene loci that could be roughly used as comparison standards at different taxonomic levels. For the most part, the nuclear ribosomal internal transcribed spacer (ITS) regions and the variable D1/D2 domains within the 28S ribosomal subunit were used at the species and generic levels. The 28S subunit and the 18S ribosomal subunit were utilized for taxonomic levels above the genus. In a few fungal groups where these ribosomal regions were clearly shown to provide inadequate resolution, one or two additional housekeeping gene loci such as translation elongation factor 1α (EF-1) and β-tubulin were also brought into play. Though the appropriateness of this strong concentration on a few individual genes was occasionally questioned—there were legitimate concerns, for example, about individual gene trees being mistaken for species phylogenetic trees (Taylor et al. 2000)—what this intuitive comparative standardization accomplished was to introduce basic DNA barcoding to fungal microbiology long before the expression was coined.
It should be noted that there was no tradition in mycology of using the mitochondrial cytochrome oxidase subunit I gene (cox1) used for most DNA barcoding of animals (Hebert et al. 2003). Preliminary results with Penicillium, however, indicate that it may be of interest and should be investigated further (K. Seifert, A. Lévesque, unpublished data).
For several years now, many thousands of fungal ribosomal sequences as well as a significant representation of EF-1 and other sequence types have been available in public repositories. Most fungal groups of any economic or practical importance are already represented to some degree. A significant proportion of the fungi represented in sequence databases are misidentified (de Hoog & Horré 2002, Hawksworth 2004, Lévesque & de Cock 2004, Kopchinskiy et al. 2005), and many more are unvouchered (Crous 2002) and thus stripped of their biological context. Nonetheless, careful sifting of the data for sequences connected to accessible, nomenclaturally significant isolates (ex-type strains, biosystematically well studied strains, etc.) allows assembly of a useful preliminary barcode set for numerous fungal taxonomic and ecological groups. The question then becomes how most efficiently to press this information, plus any comparable new sequences investigators wish to contribute, into service to accomplish one of the major aims of DNA barcoding, that is, facilitating identification. It should be noted that most twentieth century molecular identification approaches that relied on comparison of nucleic acid or protein band migration rates were vulnerable to coincidence (non-homologous bands migrating at essentially the same rate) and other factors limiting their resolution. The very sharp specificity of sequences has thus become the unquestioned molecular identification gold standard in recent years. The application of sequencing remains preliminary in insufficiently studied fungal groups where sequence types are not yet known to correspond to well-delimited species or other biosystematically or ecologically relevant units, but in many groups, sequences can now be straightforwardly used for species identification, especially when specialized databases are consulted (e.g. the Fusarium Database, http://fusarium.cbio.psu.edu/). Investigators trying to devise effective routine species identification systems for fungi that have already been barcoded soon find that they are at ‘a fork in the road’, in that one of two strategies must be followed in sequence-based identification. Since both strategies can be conceived of as subtypes of DNA barcoding, let us call them basic barcode identification—the use of whole gene or large partial gene amplicons in sequence identification—and microcoding, the use in identification of oligonucleotide sequences that are often not much longer than PCR primers. Both strategies are becoming increasingly rapid and cost-effective over time. Some strengths and weaknesses of these approaches are discussed here. Though these approaches are mainly discussed in the context of current examples involving economically important, culturable fungi, parallel considerations are expected to become salient in work with many other groups of organisms, including other fungal groups, as these groups reach the state where a significant amount of primary barcoding work has already been accomplished.
2. Basic barcode identification
Rapid development in sequencer and DNA handling technologies has made it ever more practical to routinely sequence whole-gene or large partial-gene amplicons as an identification technique. A front-running and well-known example in the field of medical mycology was provided by Pryce et al. (2003), who showed that all but a very small number of medically important fungal species growing in culture could be identified by full-length ITS sequences obtained within 24 h and costing less than AUS$ 10 (∼US$ 7.50 or € 6.25) per sequence inclusive of materials and labour. Unlike the many PCR-restriction fragment length polymorphism techniques developed in the immediately preceding period of technological development, this allowed a large number of species from a very broad taxonomic range to be handled in a maximally uniform way. Some years prior to this publication, important US reference centres for basidiomycetous (Fell et al. 2000) and ascomycetous yeasts (Kurtzman 1994) as well as Fusarium species (O'Donnell et al. 1998) had already altered procedures to identify (or, in the case of poorly resolved taxa, cluster) essentially all incoming isolates by means of well-chosen sequence regions. This was made possible in part by the use of high-throughput sequencing apparatus and robotics.
The main obstruction to using a uniform basic barcoding procedure for rapid identification of fungi is that it is a major challenge to find DNA extraction and PCR procedures, as well as primer pairs, that will reliably yield suitable high-quality, long sequence reads in rapid cycle sequencing with a wide range of fungi. This tends to be true even for loci such as ITS with highly conserved flanking regions, theoretically reliably amplified by ‘universal primers.’ Buried in the seemingly minor variations in many mycological papers' materials and methods sections are the results of months of struggle with isolates that at first seemed completely resistant to ‘universal’ sequencing or even to PCR. Currently, it is seldom possible to predict which fungi will prove difficult to sequence, and this means that there is no way as yet to make a priori adjustments to streamline basic barcode identification in such groups. There are a few clues available. Anecdotally, it can be suggested that slow-growing filamentous fungi with dense colonies, especially when these colonies are heavily pigmented, appear to be especially likely to generate cycle sequencing problems. Recently, for example, repeated, concerted and technically varied attempts by two very competent molecular mycology laboratories to obtain ribosomal sequences for the dark, slow-growing new species Oidiodendron fimicola (Rice & Currah in press) were unsuccessful, and the species ultimately had to be described based on its phenotype. Other groups of fungi where sequencing is difficult may not offer any obvious clue to this state of affairs. Even when isolates have phylogenetic affinity with easily sequenced groups, this may not be predictive of success in normal barcoding procedures. For example, in the present first author's laboratory, we have recently dealt with members of a group of Acremonium species, including A. ochraceum and A. bacillisporum, that largely withstood ITS and 28S sequencing attempts involving various primers and purification techniques (ranging from high-cost affinity columns to primitive cetyltrimethylammonium bromide procedures). It was ultimately found that they could be sequenced once 3% dimethylsulfoxide was added to the amplification step of PCR (Demeke & Adams 1992). Meanwhile, successful 18S sequences done over 2 years earlier without problem had already revealed that the recalcitrant species were a close sister group, or perhaps even an internal clade, related to the uniformly easily sequenced clade containing Acremonium strictum and A. kiliense (Bills et al. 2004). The members of the easily sequenced group and the difficult sister group were very similar in phenotype except that the latter produced conidia not in the mucoid clumps typical of the former group, but rather in chains, signalling a switch to airborne conidial dissemination. This switch likely involved at least one major change in fungal biochemistry, namely the additional production of hydrophobic anti-desiccation substances, but at present, we cannot directly attribute difficulty in sequencing to this factor. Perhaps in future, however, links may be discovered between cycle sequencing difficulties and certain readily discernible phenotypic, ecological or chemical properties. Authors are advised to briefly document such difficulties in their own studies, and to look for any patterns that could be used in predicting the occurrence of similar problems in other fungi.
In some cases, isolates that are problematic in cycle sequencing can be sequenced via cloning procedures. In the case of our A. ochraceum-complex isolates, however, the problem interfering with cycle sequencing appeared also to obstruct cloning; such findings are not uncommon in our experience. In any case, cloning requires a specially licensed laboratory in many countries (e.g. The Netherlands) and may be held in low favour because of its relative inconvenience and, in certain circumstances, its vulnerability to random selection of paralogous gene forms that may be present at low copy number.
Fungal identification by basic barcoding is also vulnerable to the common molecular phylogenetic problem, seen in many groups of organisms, that ‘one size does not fit all’ in molecular sequence identification. Some predominantly phytopathogenic groups like Gibberella (anamorphs: Fusarium subgenus Liseola), perhaps influenced by the constant generation of variation that is of advantage in pathogenicity (Brasier 2000), and, partially in consequence of this, by relatively rapid speciation, show poor resolution at the species level in concertedly evolving multicopy loci such as ITS; in some cases they have also developed paralogous forms that complicate both cycle sequencing and cloning procedures (O'Donnell & Cigelnik 1997). In such cases, recourse must be made to single-copy genes such as EF-1. These in turn then must occasionally be optimized for primer choice and PCR techniques, as flanking regions and topologies may not be as highly conserved as in the ribosomal coding regions. Single-copy genes that work well in some fungal groups may be troublesome in others.
Development of a uniform barcode standard for species identification in fungi is thus hindered by the variation in the evolutionary ages of species in different groups. Given that there can be no rules in biology for how quickly or slowly functional species may evolve, construction of a uniform barcode identification procedure may be regarded as a theoretical impossibility. In some fungi, for example, minor changes in the mating type loci, followed by inbreeding, may be sufficient to initiate a newly separated sexual lineage intersterile with its parental forerunner. This lineage may rapidly emerge as a new biological species (Aanen et al. 2000) that will inexorably diverge evolutionarily from its forerunner even though, early in its evolutionary history, the sequences of its housekeeping genes are mainly unchanged from the ancestral type even in the introns and spacers. The real possibility of such events occurring merely entails that barcode identification may need to follow nested protocols to deal with known rapidly evolving species groups, or that investigators may need to do a preliminary glance at morphology in order to make the correct selection of barcode genes to examine for members of particular fungal groups. Much like the image of a barcode that is over 500 characters long, however, these contingencies do tend to detract from the simple vision of a tricorder-like reading of fungal biodiversity (Godfray & Knapp 2004; If the term ‘tricorder’ is unfamiliar, it is the fictional device in the science fiction drama series Star Trek that can be pointed at life forms to immediately read their species identity and physiological properties).
To a certain extent, the PCR and cloning inhibition problems discussed above also affect techniques based on use of microcodes, and the inability of any single gene region to serve as an identifying standard for all fungi applies a fortiori when gene regions are represented by small subregions. With microcode procedures, however, there is a built-in expectation that organisms will be batched in advance into particular groups or that analytical platforms will be tailored to specific taxonomic or ecological circumstances, so this is not generally perceived as a disadvantage. If the difficulties in using basic barcoding in rapid identification can be optimistically summarized with the phrase, ‘some customization may be necessary for certain species groups’, then it is clear that microcoding procedures, by nature, cannot make a better offer. For the most part, they can only become relatively advantageous by being faster, cheaper or more convenient to handle than full barcoding techniques. The circumstances in which they might achieve these advantages are not yet well resolved. Nonetheless, these techniques appear to offer a potential for very rapid handling, and there has been considerable interest in their development despite the increasing convenience of routine whole-gene sequencing.
3. Microcoding (oligonucleotide barcoding)
In recent years, as indicated above, an increasing number of techniques have been deployed that are predicated on the use of distinctive short nucleic acid segments—that is, short DNA barcodes—in rapid identification procedures. Among these are silicon-based microarrays, nylon-membrane-based macroarrays, and the patented Luminex system of DNA-tagged polystyrene beads sorted by flow cytometry. These systems share the feature that fully sequenced genes are studied to find small, thermodynamically stable and non-hairpin-forming areas of high sequence uniqueness that can then be used as fixed single-stranded oligonucleotides on an identification platform. The bound oligonucleotides then anneal specifically to matching, complementary DNA regions in a test solution bearing labelled single-stranded amplicons from one or more unidentified strains. These systems can only be developed as a second step in DNA barcoding procedures for any given group of organisms, because all the relevant basic barcodes need to be assembled before the unique short segments useable as oligonucleotides can be discerned. Because of practical considerations related to how many oligonucleotides can be proof-tested as unique in relation to one another and then attached together in a common test platform, oligonucleotide-based systems so far have been limited to groups not larger than circa 200 test species.
Fungal systems were among the earliest to be studied using silicon-platform microarrays (Shalon et al. 1996); these studies, however, were and mainly remain dedicated to studying broad gene expression patterns among different isolates, and in individual isolates exposed to differing conditions. In theory, species identification by means of microarrays is relatively straightforward. In practice, however, the high cost and non-reusability of current microarrays makes their use cost-effective primarily to answer genomics questions producing far higher quantities of useful data per chip exposure than simply ‘this is species X’. Thus, though microarrays still have potential for use in identifying spectra of species or species-groups occurring together in complex environmental materials, they may not be an optimal technology for the rapid identification of single isolates or for analysis of media, such as bronchoalveolar lavage, at most containing just a small number of fungal species. It should be noted, however, that techniques allowing re-use of microarrays a small number of times (e.g. 1–4 times) are in development (Dolan et al. 2001).
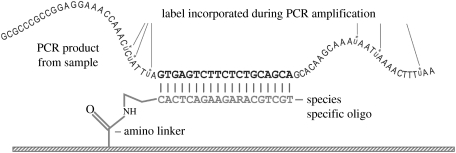
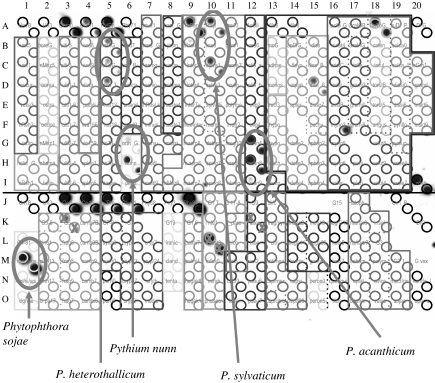
Nylon-membrane-based reverse dot-blot macroarrays are similar in principle to silicon microarrays, but have the advantage that they can be re-used up to 50 times (Fessehaie et al. 2003). This makes it conceivable that they could efficiently be used to identify single isolates, and, in particular, it makes them a prime candidate for analysis of extracted DNA from environmental materials containing a limited number of relevant species. Indeed, such macroarrays are already in commercial production under the name DNA Multiscan in four European countries in relation to detection of important phytopathogens in greenhouse soils (Lievens et al. 2003; http://www.denhaan.nl/ukdna.html). In this technique, a number of different oligonucleotides are simultaneously spotted onto a nylon membrane using a multipin array spotter. The spotted array is then exposed to PCR amplicons labelled with digoxigenin d-UTP. Figure 1 shows the principle of detection of labelled amplicons using specific oligonucleotides amino-linked to the membrane surface. Some of the species detected in substrata analysed using macroarrays may be very difficult to obtain in culture. This may simply be because they tend to be overgrown by more prevalent species that are also more aggressive and faster growing in artificial culture. For example, it was found that macroarrays spotted with oligonucleotides for soilborne Oomycota in the genera Pythium and Phytophthora (as per Lévesque et al. (1998) but with an updated version of the array) facilitated the early detection of the biocontrol agent Pythium nunn in soybean roots early in the growing season (figure 2), when this species was present only at inoculum levels lower than those of several other oomycetous species (Anonymous 2002). Coupled with appropriate techniques for unbiased, proportionate extraction and amplification of DNA of all relevant organisms from substrate material such as soils, this technique does offer a rapid census of populations present in otherwise inscrutable material, giving a genuine foretaste of tricorder-like biodetection.
Figure 1.
Annealing of labelled amplicon with a specific oligonucleotide that is linked to a solid surface via an amino linker.
Figure 2.
Macroarray exposed to Pythium- and Phytophthora-specific nuclear ribosomal internal transcribed spacer-region amplicons obtained from a DNA sample extracted from field-grown soybean roots (T. Barasubiye, Agriculture and Agri-Food Canada, unpublished). Dark spots appear where specific oligonucleotides have captured labelled amplicons corresponding to various species present on the roots. The array is covered by a mask indicating the identity of the various spots seen. ‘X’ symbols are placed over known cross-reacting oligonucleotides.
The selection of appropriate microcodes can be greatly facilitated by the use of software that can go through a large, multi-species alignment (or even an unaligned collection) of full-length barcodes and determine the best signature microcode regions that also are free of thermodynamic and conformational problems. Success in such automated microcode selection has been reported by Seifert & Lévesque (2004) with the software package Signature Oligo, which has been used, for example, to select useable diagnostic oligonucleotides from sequences of mycotoxigenic Aspergillus, Fusarium and Penicillium species. Oligonucleotides can be selected specific to any clustering level in sequence dendrograms, not only allowing the detection of variant organisms not represented by individual species-level microcodes, but also providing a level of redundancy serving as a control against false-positive results (e.g. if the Fusarium thapsinum microcode gives a positive signal in macroarray analysis, the overarching general Fusarium subgenus Liseola microcode should also be positive). A second software package, Array Designer 1.1, is used to ensure that the oligonucleotides selected have a suitable melting temperature (55 °C at 6×SSC) and lack dimers and hairpins. It can also be used to BLAST (Altschul et al. 1990) a batch file of microcodes against GenBank to monitor their uniqueness. As Seifert & Lévesque (2004) state, ‘it is surprising how often 18–22 bp oligonucleotides have no BLAST matches apart from their own sequences’. Occasional random matches that are found tend to have no practical consequence, Seifert and Lévesque explain, because such coincidences have an extremely high likelihood of involving parts of completely different gene regions from organisms considered unrelated to the groups under consideration in the arrays. Even if the fortuitously coinciding DNA were present in a sample being analysed by a microcode-based technique, it would not give a positive result because the gene containing it would not amplify with the PCR primers used.
A similar principle for the selection and use of unique microcodes is embodied in the Luminex system (Spiro et al. 2000, Diaz & Fell 2004). In Luminex, however, sensitivity is enhanced by attaching the synthesized oligonucleotide segments to polystyrene microspheres that can be read, one by one, in a flow cytometer. Each oligonucleotide is partnered with one of 100 types of differently coloured microspheres; each of the 100 colours available is identifiable when probed with a red laser. The amplicons from the test material are tagged with a fluorescent marker readable with a green laser. In using the system to identify, for example, a fungal culture, the 100 types of differently coloured beads partnered with 100 different oligonucleotides are exposed to the test solution of single-stranded, fluorescently tagged amplicons derived from the culture. The amplicon then, at the correct temperature, anneals with the unique complementary species-specific oligonucleotide that is attached to just one of the microsphere types. It may anneal with only that oligonucleotide or, depending on the design of the test, it may also anneal with another, hierarchically supervening oligonucleotide such as that specific for the relevant fungal genus. As the microspheres are channelled through the flow cytometer, they are simultaneously read by two lasers, the red one classifying the microspheres and the green one showing which microspheres bear the annealed DNA tagged with the fluorescent marker. Putting these two data sets together, the identification of the species involved becomes clear.
The use of microcode-based oligonucleotides for species identification in the Luminex system requires the same selection for uniqueness and thermodynamic and conformational appropriateness that must be used for microarray and macroarray technologies. One disadvantage of all such systems is that this process must be carefully done, and then the identification platforms must be thoroughly tested for sensitivity and specificity using all species and other genetic types that are intended to be detected. In particular, cross-reactions among oligonucleotides for closely related species may be difficult to preclude based on computer analysis alone, and occasionally, an oligonucleotide will need to be replaced with a more robustly specific one, or the distinction of two closely related taxa may need to await further testing (Diaz & Fell 2004).
The limited number of species that can be identified by any given array or sphere set may be problematical in some contexts, e.g. in general fungal soil ecology, where more than 100–200 organisms may be potentially significant in a given microhabitat. Of course, use of a second array or sphere set doubles the number of species that can be identified. Similarly, if some species are identified based on ITS sequences and others must be analysed with another locus such as EF-1 in order to establish specificity, either two arrays or sphere sets need to be analysed, or the platforms must be configured with the oligonucleotides corresponding to signature microcode regions in both amplicon types. In connection with latter option, both amplicon types will need to be simultaneously or serially exposed to the detection platforms, increasing the amount of work that needs to be done to obtain an identification. Optimization of multiplex PCR for the two gene regions tested may minimize the extra work involved in such identifications. Macroarrays can be used with labelled whole-cell DNA, so strictly speaking, amplification is not necessary (Trad et al. 2004). Clearly, however, the use of amplicons would improve sensitivity by ensuring that a relatively high quantity of compatible DNA was present for binding with the appropriate complementary oligonucleotide.
It is unclear if the use of microcodes would alleviate the problem, mentioned above in the context of basic barcode identification, of some species being difficult to analyse because difficult-to-remove inhibitors in their cellular chemistry obstruct one or more of the PCR-related steps involved in cycle sequencing. Such organisms often seem to show adequate DNA amplification, and they frequently even yield short sequence runs of varying quality. It is thus possible that microcode platforms may successfully detect and identify them even though the amplicons involved are of inadequate quality for full barcode reading. The matter has not, to our knowledge, been tested.
One feature of microcodes is that, being lower in information content than the full-length barcodes that served as their sources, they tend by nature to be a less sensitive taxonomic indicator than the full barcodes. This may, however, be an advantage in some cases. In a relatively genetically complex species, there may be considerable minor variation within the entire barcode region, but the statistical probability that this variation will have affected the particular small region designated as the microcode is relatively small. Thus the use of microcodes in identification may minimize the potential influence of distracting mutational ‘noise’ and provide users with clear, unambiguous results. By the same token, however, there is always the chance that an unknown and unbarcoded sibling species will be misidentified as its already described sibling based on 100% microcode similarity. With full-length barcodes, the chance is much greater that information distinguishing the unknown species from the known would come to light. Recently, a specialized BLAST search program for Trichoderma species was made available (Kopchinskiy et al. 2005; www.isth.info) based on a sophisticated use of microcodes to increase identification accuracy beyond the level obtainable with standard whole-gene BLAST searching. In part, this was to diminish the influence of long, highly homologous regions that strongly influence sorting in common BLAST procedures (Kopchinskiy et al. 2005). The full potential of microcodes as a diagnostic tool has not yet been circumscribed.
4. Microcoding for organisms other than fungi
Microcoding has mainly been used with culturable organisms, partly because many were sequenced at signature barcoding regions long ago and partly because plenty of material is always available for extraction of the DNA used in microcode test development. Microcodes may be very advantageous, however, in the identification of organisms that are inconvenient for artificial cultivation or husbandry. In application to living organisms, the strengths and weaknesses of these techniques do not differ significantly from those mentioned above in connection with culturable fungi and Oomycetes. Where microcoding methods may have unique value is in the DNA-based identification of preserved specimens, e.g. plant herbarium specimens, with degraded DNA (but see Chase et al. 2005). Presumably in most such degraded DNA, the less-than-25-bp segments used in microcoding are much more likely to remain intact than whole barcodes over 500 bp long. Double-ended ligation of restriction-enzyme-pretreated DNA fragments to DNA linkers serving as templates for PCR amplification, similar to the process used in amplified fragment length polymorphism (AFLP) analysis, could speed the process of identifying preserved specimens. Various related techniques are possible. Proof of principle for the convenience of short signature DNA regions in such situations is provided by the successful identification and strain typing of Mycobacterium spp. strains from Egyptian mummies using the spoligotyping (spacer oligotyping) technique (Zink et al. 2003). This technique is based on membrane array detection of amplicons derived from a series of 34–41 bp spacer regions in the chromosomal DR (direct repeat) locus. Each spacer is separated from neighbouring spacers by a conserved direct repeat region that can serve as a primer binding site for amplicons spanning single short spacers or pairs or triads of adjacent spacers (Kamerbeek et al. 1997). The spoligotyping technique itself is not directly applicable to most non-mycobacterial organisms, as far as is known, but, as one of numerous possible alternative strategies based on similar principles, conserved primers closely flanking signature microcode regions within closely related species groups could be designed.
The microcodes used for identification would first, naturally, need to be demonstrated as sensitive and specific in prior testing with living material of the same species, if this is at all possible (i.e., if the species in question is not extinct). The use of microcoding techniques, however, offers to redeem many DNA-degraded taxonomic specimens that belong, for example, to closely interrelated species complexes where species identification can now only be accomplished using molecular or other in vitro techniques. Such techniques may also allow forensic DNA analysis of materials that contain DNA that has been damaged but not completely destroyed, e.g. lightly burned or dried, heavily pulverized material. Ultimately, with microcode platforms prepared with various selections from complete genome sequence databases, extensive genomic analysis may be possible for materials bearing only fragmented DNA. Such prospects have been widely publicized in connection with microarrays, but recent biosystematic work has shown, as summarized above, that more tractable and cost-effective alternatives may exist.
Footnotes
One contribution of 18 to a Theme Issue ‘DNA barcoding of life’.
References
- Aanen D.K, Kuyper T.W, Mes T.H, Hoekstra R.F. The evolution of reproductive isolation in the ectomycorrhizal Hebeloma crustuliniforme aggregate (Basidiomycetes) in northwestern Europe: a phylogenetic approach. Evol. Int. J. Org. Evol. 2000;54:1192–1206. doi: 10.1111/j.0014-3820.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- Altschul S.F, Gish W, Miller W, Myers E.W, Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Anonymous . Top Crop Manager. 2002. Early season diseases take their toll. Dec. 2002, http://www.topcropmanager.com/6_search/article_e.asp?article=1300. [Google Scholar]
- Bills G.F, Platas G, Gams W. Conspecificity of the cerulenin and helvolic acid producing ‘Cephalosporium caerulens’, and the hypocrealean fungus Sarocladium oryzae. Mycol. Res. 2004;108:1291–1300. doi: 10.1017/s0953756204001297. 10.1017/S0953756204001297 [DOI] [PubMed] [Google Scholar]
- Brasier C.M. The rise of the hybrid fungi. Nature (London) 2000;405:134–135. doi: 10.1038/35012193. 10.1038/35012193 [DOI] [PubMed] [Google Scholar]
- Chase M.W, Salamin N, Wilkinson M, Dunwell J.M, Kesanakurthi R.P, Haidar N, Savolainen V. Land plants and DNA barcodes: short-term and long-term goals. Phil. Trans. R. Soc. B. 2005;360:1889–1895. doi: 10.1098/rstb.2005.1720. 10.1098/rstb.2005.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W. Adhering to good cultural practice (GCP) Mycol. Res. 2002;106:1378–1379. 10.1017/S0953756202227136 [Google Scholar]
- de Hoog G.S, Horré R. Molecular taxonomy of the Alternaria and Ulocladium species from humans and their identification in the routine laboratory. Mycoses. 2002;45:259–276. doi: 10.1046/j.1439-0507.2002.00747.x. 10.1046/j.1439-0507.2002.00747.x [DOI] [PubMed] [Google Scholar]
- Demeke T, Adams R.P. The effects of plant polysaccharides and buffer additives on PCR. Biotechniques. 1992;12:332–334. [PubMed] [Google Scholar]
- Diaz M.R, Fell J.W. High-throughput detection of pathogenic yeasts of the genus Trichosporon. J. Clin. Microbiol. 2004;42:3696–3706. doi: 10.1128/JCM.42.8.3696-3706.2004. 10.1128/JCM.42.8.3696-3706.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan P.L, Wu Y, Ista L.K, Metzenberg R.L, Nelson M.A, Lopez G.P. Robust and efficient synthetic method for forming DNA microarrays. Nucleic Acids Res. 2001;29:E107. doi: 10.1093/nar/29.21.e107. 10.1093/nar/29.21.e107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell J.W, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Fessehaie A, De Boer S.H, Lévesque C.A. An oligonucleotide array for the identification and differentiation of bacteria pathogenic on potato. Phytopathology. 2003;93:262–269. doi: 10.1094/PHYTO.2003.93.3.262. [DOI] [PubMed] [Google Scholar]
- Godfray H.C.J, Knapp S. One contribution of 19 to a theme issue ‘Taxonomy for the twenty-first century’. Phil. Trans. R. Soc. B. 2004;359:559–569. doi: 10.1098/rstb.2003.1457. 10.1098/rstb.2003.1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth D.L. Fungal diversity and its implications for genetic resource collections. Stud. Mycol. 2004;50:9–18. [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B. 2003;270:313–322. doi: 10.1098/rspb.2002.2218. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopchinskiy A, Komoń M, Kubicek C.P, Druzhinina I.S. TrichoBLAST, a multilocus database for Trichoderma and Hypocrea identifications. Mycol. Res. 2005;109 doi: 10.1017/s0953756205233397. 10.1017/S0953756205233397 e-pub in advance of print: http://www.britmycolsoc.org.uk/files/MR_News_June05.pdf. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P. Molecular taxonomy of the yeasts. Yeast. 1994;10:1727–1740. doi: 10.1002/yea.320101306. [DOI] [PubMed] [Google Scholar]
- Lévesque C.A, de Cock A.W.A.M. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 2004;108:1363–1383. doi: 10.1017/s0953756204001431. 10.1017/S0953756204001431 [DOI] [PubMed] [Google Scholar]
- Lévesque C.A, Harlton C.E, de Cock A.W.A.M. Identification of some oomycetes by reverse dot blot hybridization. Phytopathology. 1998;88:213–222. doi: 10.1094/PHYTO.1998.88.3.213. [DOI] [PubMed] [Google Scholar]
- Lievens B, Brouwer M, Vanachter A, Lévesque C.A, Cammue B, Thomma B. Design and development of a DNA array for rapid detection and identification of multiple tomato vascular wilt pathogens. FEMS Microbiol. Lett. 2003;223:113–122. doi: 10.1016/S0378-1097(03)00352-5. 10.1016/S0378-1097(03)00352-5 [DOI] [PubMed] [Google Scholar]
- O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylog. Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- O'Donnell K, Cigelnik E, Nirenberg H.I. Molecular systematics and phylogeography of the Gibberella fujikoroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- Pryce T.M, Palladino S, Kay I.D, Coombs G.W. Rapid identification of fungi by sequencing the ITS1 and ITS2 regions using an automated capillary electrophoresis system. Med. Mycol. 2003;41:369–381. doi: 10.1080/13693780310001600435. 10.1080/13693780310001600435 [DOI] [PubMed] [Google Scholar]
- Rice, A. V. & Currah, R. S. In press. Profiles from Biolog FF plates and morphological characteristics support the recognition of Oidiodendron fimicola sp. nov. Stud. Mycol.53
- Scorzetti G, Fell J.W, Fonseca A, Statzell-Tallman A. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. 10.1016/S1567-1356(02)00128-9 [DOI] [PubMed] [Google Scholar]
- Seifert K.A, Lévesque C.A. Phylogeny and molecular diagnosis of mycotoxigenic fungi. Eur. J. Plant Pathol. 2004;110:449–471. 10.1023/B:EJPP.0000032385.41877.7a [Google Scholar]
- Shalon D, Smith S.J, Brown P.O. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- Spiro A, Lowe M, Brown D. A bead-based method for multiplexed identification and quantitation of DNA sequences using flow cytometry. Appl. Environ. Microbiol. 2000;66:4258–4265. doi: 10.1128/aem.66.10.4258-4265.2000. 10.1128/AEM.66.10.4258-4265.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.W, Jacobson D.J, Kroken S, Kasuga T, Geiser D.M, Hibbett D.S, Fisher M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- Trad S, Allignet J, Frangeul L, Davi M, Vergassola M, Couve E, Morvan A, Kechrid A, Buchrieser C, Glaser P, El-Solh N. DNA macroarray for identification and typing of Staphylococcus aureus isolates. J. Clin. Microbiol. 2004;42:2054–2064. doi: 10.1128/JCM.42.5.2054-2064.2004. 10.1128/JCM.42.5.2054-2064.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink A.R, Sola C, Reischl U, Grabner W, Rastogi N, Wolf H, Nerlich A.G. Characterization of Mycobacterium tuberculosis complex DNAs from Egyptian mummies by spoligotyping. J. Clin. Microbiol. 2003;41:359–367. doi: 10.1128/JCM.41.1.359-367.2003. 10.1128/JCM.41.1.359-367.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]