Abstract
DNA barcoding is a promising approach to the diagnosis of biological diversity in which DNA sequences serve as the primary key for information retrieval. Most existing software for evolutionary analysis of DNA sequences was designed for phylogenetic analyses and, hence, those algorithms do not offer appropriate solutions for the rapid, but precise analyses needed for DNA barcoding, and are also unable to process the often large comparative datasets. We developed a flexible software tool for DNA taxonomy, named TaxI. This program calculates sequence divergences between a query sequence (taxon to be barcoded) and each sequence of a dataset of reference sequences defined by the user. Because the analysis is based on separate pairwise alignments this software is also able to work with sequences characterized by multiple insertions and deletions that are difficult to align in large sequence sets (i.e. thousands of sequences) by multiple alignment algorithms because of computational restrictions. Here, we demonstrate the utility of this approach with two datasets of fish larvae and juveniles from Lake Constance and juvenile land snails under different models of sequence evolution. Sets of ribosomal 16S rRNA sequences, characterized by multiple indels, performed as good as or better than cox1 sequence sets in assigning sequences to species, demonstrating the suitability of rRNA genes for DNA barcoding.
Keywords: DNA barcoding, molecular taxonomy, cox1, 16S rRNA, species recognition
1. Introduction
Recent work suggested that a DNA-based identification system can aid the resolution of the vast diversity of life with its millions of species (Tautz et al. 2003). Hebert et al. (2003a,b) proposed that a DNA barcoding system for animal life could best be based upon sequence diversity in the 5′ section of the mitochondrial gene cytochrome oxidase subunit I (cox1). Although DNA variation has long been used successfully for the identification and classification of microorganisms (Rosello-Mora & Amann 2001), scepticism against this approach for more complex taxa has been expressed. Two primary objections have focused on (i) the concern that DNA sequence differences among closely related species will often be too small to allow their discrimination (Mallet & Willmot 2003) and (ii) the fact that present strategies and programs suffer from difficulties of aligning sequences of different lengths (Lipscomb et al. 2003), especially in automated large-scale analyses. Hebert et al. (2003b) argued against the mitochondrial 12S and 16S rRNA genes as standard DNA barcoding markers because the presence of multiple insertions and deletions in these genes pose potential problems due to difficulties and ambiguities in their alignment. This problem would apply as well to the nuclear 28S rRNA and internal transcribed spacer genes (ITS). On the other hand, a variety of arguments have been voiced and evidence has been brought forth to suggest that 28S (Tautz et al. 2003; Markmann & Tautz 2005), ITS (Blaxter 2003) and 16S rRNA (Vences et al. 2005b) could be valuable taxonomic markers in the framework of a large-scale DNA barcoding system.
One of the most promising applications of DNA barcoding is the molecular identification of often phenotypically disparate life-history stages in taxonomic, ecological, behavioural and conservation studies. Animal juvenile or larval morphology is often distressingly uniform among different species such as in fish larvae. Larvae might, on the other hand, be radically different in morphology from the adult (e.g. in frogs or holometabolous insects), and some species such as parasites might even display a complex variety of larval, semi-adult and adult stages. Reliable field identification in these taxa requires extensive expertise, and even then can be impossible. Furthermore, any classical taxonomic method to estimate population numbers and monitor trends is costly and time-intensive. Recent developments in non-invasive genetic sampling techniques in combination with DNA barcoding provide a practical alternative for such research.
In this study we present a flexible software tool for DNA taxonomy named TaxI (available at http://www.evolutionsbiologie.uni-konstanz.de/Software). This tool is based on pairwise sequence divergences between query and reference sequences that can be defined by the user. We test the suitability of this program for the identification of larval and juvenile stages of organisms with two datasets, fish larvae from Lake Constance, Central Europe, and juvenile land snails, and compare the performance of two proposed markers, cox1 and 16S rRNA, in DNA barcoding.
2. Material and methods
(a) Molecular datasets and methods
Thirty 16S rRNA fish reference sequences were used in this study representing most fish species that occur in Lake Constance (Eckmann & Rösch 1998). These sequences were obtained from GenBank (accession numbers given in table 1).
Table 1.
Fish reference sequences used in this study and obtained from GenBank given by species and accession number.
| Order | family | species | accession numbers |
|---|---|---|---|
| Anguillifomes | Anguillidae | Anguilla anguilla | AJ244828.1 |
| Cypriniformes | Balitoridae | Barbatula barbatula | DQ077974 |
| Cobitidae | Cobitis taenia | AJ247080.1 | |
| Cyprinidae | Abramis brama | AJ247067.1 | |
| Alburnus alburnus | AJ247063.1 | ||
| Barbus barbus | AJ247065.1 | ||
| Blicca bjoerkna | AJ247064.1 | ||
| Carassius carassius | AJ247070.1 | ||
| Chondrostoma nasus | AJ247047.1 | ||
| Cyprinus carpio | NC001606.1 | ||
| Gobio gobio | AJ247068.1 | ||
| Leuciscus cephalus | AJ247054.1 | ||
| Leuciscus leuciscus | AJ247052.1 | ||
| Leuciscus souffia agassizi | AJ247079.12 | ||
| Phoxinus phoxinus | AJ247062.1 | ||
| Rhodeus sericeus | AJ247086.1 | ||
| Rutilus rutilus | AJ247045.1 | ||
| Scardinius erythrophtalmus | AF215479.1 | ||
| Tinca tinca | AJ247053.1 | ||
| Esociformes | Esocidae | Esox lucius | AF060446.1 |
| Gadiformes | Lotidae | Lota lota | AP004412.1 |
| Gasterosteiformes | Gasterosteidae | Gasterosteus aculeatus | AF355030.1 |
| Perciformes | Percidae | Gymnocephalus cernuus | AY141443.1 |
| Perca fluviatilis | AY141442.1 | ||
| Salmoniformes | Salmonidae | Coregonus lavaretus | NC002646.1 |
| Oncorhynchus mykiss | AF312573.1 | ||
| Salmo trutta | X77564.1 | ||
| Salvelinus alpinus | AJ319820.1 | ||
| Thymallus thymallus | AY430237.1 | ||
| Scopaeniformes | Cottidae | Cottus gobio | AP004442.1 |
Snail reference sequences representing the Mediterranean family Hygromiidae (23 species) and the Helicidae s.l. (39 species including the previous 23 Hygromiidae species) were taken from a previous study (Steinke et al. 2004; GenBank accession numbers AY546342–546381 for 16S rRNA and AY546262–AY546301 for COI=cox1).
The fish larvae and juveniles (seven putative species) and juvenile snail samples (five putative species) were identified by classical morphological methods. Following a proof of principle approach we only used samples of those specimens that could be reliably identified. Sequences were obtained by preparing total DNA extracts from tissue samples with a proteinase K digestion followed by sodium chloride extractions and ethanol precipitations. Published ‘universal’ primers were subsequently used to amplify a 420 bp fragment of the 16S rRNA gene (16Sar-L and 16Sbr-H by Palumbi et al. 1991) in fish and snail specimens and a 520 bp fragment of the cox1 gene (LCO1490 and HCO2198 by Folmer et al. 1994) in juvenile snails. Polymerase chain reaction (PCR)-amplifications were performed according to standard protocols on a GeneAmp 9700 thermocycler (Applied Biosystems). The PCR products were purified using the QiaQuick spin columns extraction kit (Qiagen), sequenced in both directions with the BigDye termination reaction chemistry, and determined on an ABI 3100 Automatic Capillary Sequencer (Applied Biosystems).
(b) Data analysis using TaxI
TaxI is a program for Windows platforms to compute pairwise distances among sequences after their pairwise alignment. Data input is in fastA format, a simple and widely used file format (Pearson & Lipman 1988). TaxI can process multiple files containing a single sequence each or single files with multiple sequences (aligned or unaligned). Hundreds of query sequences can be analysed in one program run. As depicted in the process flowchart in figure 1, the program considers all possible pairs among query and reference files. All these pairs are then aligned using the T-Coffee algorithm (Notredame et al. 2000). T-Coffee performs with high accuracy even if long internal deletions require a method that is able to deal with local similarity. Following the alignment, sequence divergence is determined for each pair by dividing the number of different nucleotides by the total number of nucleotides examined in the alignment. All alignment positions with gaps are excluded from distance computation (complete deletion).
Figure 1.
Processing scheme for documentation of information processing.
The evolutionary distances that are computed from DNA sequence data are primarily estimates of the number of nucleotide substitutions per site (d) between two sequences. There are many methods for estimating evolutionary distances, depending on the pattern of nucleotide substitutions, which might best account for back mutations among more distantly related sequences (see Nei 1987; Gojobori et al. 1990). In TaxI, the user can choose between six methods: Jukes-Cantor distance (Jukes & Cantor 1969), Kimura 2-parameter distance (Kimura 1980), Tajima-Nei distance (Tajima & Nei 1984), Tamura distance (Tamura 1992), Tamura-Nei distance (Tamura & Nei 1993), LogDet (Lockhart et al. 1994). All possible pairwise distances are saved in a matrix and the lowest d is calculated with a simple sort routine for every multiple hit correct method chosen. The output files are in basic ASCII format and contain evolutionary distances and other information related to the data and delimited by tabs. Most spreadsheet programs (e.g. Microsoft Excel) allow editing those files.
In our test datasets, all juvenile sample sequences (table 2) were tested against the reference sequences with all possible distance methods. The percentage of correct matches was calculated to test the accuracy of the method. Twenty-eight newly determined 16S rRNA fish sequences (GenBank accession numbers DQ077946–DQ077973) were tested against the Lake Constance reference data set containing fish species occurring in that lake (Eckmann & Rösch 1998).
Table 2.
Tested juvenile samples, their morphological assignment and the sequences used.
| n individuals | morphological ID | 16S rDNA | cox1 |
|---|---|---|---|
| fish sequences | |||
| 2 | Anguilla anguilla (larvae) | x | — |
| 2 | Lota lota (larvae) | x | — |
| 5 | Barbatula barbatula (juvenile) | x | — |
| 6 | Gasterosteus aculeatus (juvenile) | x | — |
| 6 | Rutilus rutilus (juvenile) | x | — |
| 6 | Perca fluviatilis (juvenile) | x | — |
| 1 | Phoxinuns phoxinus (juvenile) | x | — |
| Snail sequences | |||
| 4 | Candidula unifasciata | x | x |
| 5 | Cochlicella acuta | x | x |
| 5 | Helicopsis striata | x | x |
| 3 | Trichia sericea | x | x |
| 3 | Trochoidea geyeri | x | x |
Twenty 16S rRNA and cox1 juvenile snail sequences were determined morphologically to belong to five different species and were tested against two data sets respectively. In the latter case sequences were tested against a second dataset containing 16 more species at a more-inclusive taxonomic level to test the performance of TaxI in a situation with considerable amounts of sequence divergence.
3. Results
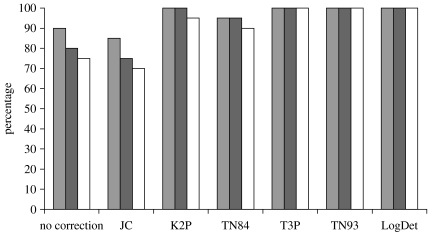
In the 16S rRNA fish dataset all 28 query sequences were correctly assigned to seven species under all possible model regimes. The divergence value of the query sequences to the most similar lineage was between 0 and 2.7% and the mean divergence to the complete reference dataset lies between 15.5 and 26.9%. Query sequences of the snail dataset, using uncorrected distances, were in all cases assigned to the correct genus and in 90% to the right species. Exceptions were Candidula unifasiciata juveniles that grouped most closely with a morphological distinct sister species (Candidula spadae). In all cases, there was 0–16.8% sequence divergence between the test taxon and the lineage in the profile that was most similar to it (table 3), which was in any case lower than the mean divergence to the complete reference dataset (19.6–25.9%). Eighteen out of the 20 juvenile snail specimens were successfully classified using 16S rRNA sequences and 16 were assigned to the correct species using cox1. The use of Tamura-, Tamura-Nei- and LogDet-distances in calculating snail classifications resulted in 100% success of species recognition (figure 2) whereas other substitutions models performed worse, especially with cox1 sequences. The identification using cox1 sequences was more reliable in the taxonomically less inclusive dataset, i.e. when available sequences for the comparison were a priori more closely related to the query sequence, hence required more precise prior taxonomic knowledge.
Table 3.
The table shows the mean sequence divergence (uncorrected) between the test taxon and the reference dataset and the sequence divergence between the test taxon and the lineage in the reference profile that was most similar to it.
| Test taxon | mean divergence to reference dataset (%) | divergence value to most similar lineage (%) | |
|---|---|---|---|
| Anguilla anguilla | 16S rDNA | 19.8 | 1.8 |
| Barbatula barbatula | 16S rDNA | 20.4 | 0.0 |
| Gasterosteus aculeatus | 16S rDNA | 22.4 | 0.2 |
| Lota lota | 16S rDNA | 21.2 | 1.2 |
| Perca fluviatilis | 16S rDNA | 22.1 | 0.1 |
| Phoxinuns phoxinus | 16S rDNA | 26.9 | 1.3 |
| Rutilus rutilus | 16S rDNA | 15.5 | 2.6 |
| Reference datasets | |||
| Hygromiidae/Helicidae s.l. | |||
| Candidula unifasciata | 16S rDNA | 23.6/24.4 | 1.2 |
| cox1 | 19.7/20.2 | 0.0 | |
| Cochlicella acuta | 16S rDNA | 24.2/24.8 | 0.4 |
| cox1 | 21.3/21.4 | 10.7 | |
| Helicopsis striata | 16S rDNA | 21.2/22.7 | 0.9 |
| cox1 | 22.7/23.3 | 15.0 | |
| Trichia sericea | 16S rDNA | 25.9/25.9 | 6.4 |
| cox1 | 20.7/20.8 | 16.8 | |
| Trochoidea geyeri | 16S rDNA | 23.0/22.6 | 10.8 |
| cox1 | 19.6/20.2 | 0.1 | |
Figure 2.
Bar graph of the percentage success in classifying snail species to the correct taxon. Positive 16S rRNA identifications are shown in grey, positive cox1 identifications using the Hygromiidae profile group (23 species) are shown in black, and positive cox1 identifications using the Helicidae s.l. (39 species) profile group are shown in white.
4. Discussion
In this study we developed and tested a new software tool for a DNA identification system based on pairwise sequence divergence under different model regimes. As highlighted by Moritz & Cicero (2004), DNA barcoding or DNA taxonomy focuses on phenetic identification rather than phylogenetic reconstruction, and the software requirements are different in the two cases. Although trees are used for the visualization of results (e.g. Hebert et al. 2003a,b), the tree topology (especially at inclusive phylogenetic levels) is of less concern in DNA barcoding as long as tips are correctly grouped at the relevant taxonomic level (usually species or genera). In the case of gastropod cox1 sequences, recent studies (Medina & Walsh 2000) indicated a high variability of third codon positions and therefore these are suspected to be of limited utility for phylogenetic analyses among closely related species. However, as long as these positions contain information to identify and distinguish among closely related species, they are valuable for DNA barcoding.
In phylogenetic analyses, a correct alignment of sequences is of paramount importance but is highly complicated in DNA fragments characterized by multiple insertions and deletions (e.g. Morrison & Ellis 1997). Most methods for maximum parsimony, maximum likelihood and Bayesian analyses require a multiple alignment, which is rather time consuming if hundreds of sequences are used. Furthermore, algorithms for multiple alignments often produce errors in such large datasets (Wheeler et al. 1994). For DNA barcoding purposes, distances need to be compared among sequences. Homologous nucleotide positions must be identified and compared between pairs of sequences, but homology between the separate alignments is less relevant because mutations are not analysed in a phylogenetic context and evolution at particular nucleotide positions needs not to be reconstructed. Although different pairwise alignments may yield different homology statements for one locus, the pairwise alignment approach followed in TaxI is faster than large multiple alignments, and produces an output that is easier to interpret than those of the faster BLAST algorithm (Altschul et al. 1990), particularly when more than one query sequences are subjected to analysis. A major drawback of BLAST is that this algorithm often partitions highly divergent sequences into separate fragments that are then compared separately, and therefore it is not straightforward to obtain a standard measure of overall sequence divergence. TaxI also provides information on alignment length, number of transitions and transversions and the number of introduced gaps for each pairwise comparison to interpret an assignment and to resolve ambiguous findings. Due to the fact that a user can define the reference dataset TaxI is flexible and reduces computational time. We suggest that this program will facilitate the application and wider acceptance of DNA barcoding, and increased practical use will help to identify further tools required for such applications, to be incorporated in this or other software packages.
In ribosomal DNA indels are relatively common because the excision or insertion of a few nucleotides often has little impact on rRNA function. Their alignment and analysis can therefore be particularly difficult. However, this study provides further evidence that even partial 16S rRNA gene fragments are useful in DNA barcoding. In our fish dataset, 16S performed successfully, and in the snail dataset, species identification was more successful using 16S compared to cox1. We have furthermore successfully used TaxI to calculate distances among large numbers of amphibian sequences of the 16S rRNA and cox1 genes, and to identify conspecific and closely related taxa from these datasets (Köhler et al. in press; Vences et al. 2005a).
Helicid land snails pose a challenge for DNA barcoding because they are characterized by extremely high levels of intraspecific mitochondrial DNA divergence (e.g. Thomaz et al. 1996). Strong differences among the reference and query sequences explain why our snail dataset species identification was not in all cases successful. However, a correct identification of all query sequences was achieved using particular models of sequence evolution (figure 2), namely the LogDet, Tamura, and Tamura and Nei models. The latter two correct for multiple hits taking into account substitutional rate differences between nucleotides and inequality of nucleotide frequencies. These methods also distinguish between transversional and transitional substitution rates, which is also the case in the Kimura-2-parameter model misplacing a sequence only in one case. Given the fact that there is a high transition/transversion bias in mitochondrial DNA this indicates that models that consider these two classes of substitutions or calculate additive distances with variable base composition (like LogDet) are most suitable for DNA barcoding in groups of large intraspecific divergences of mitochondrial haplotypes.
Acknowledgments
We would like to thank Elke Hespeler for technical assistance. Support from the Deutsche Forschungsgemeinschaft (DFG) to A.M. and M.V. and from the European Union to W.S. is gratefully acknowledged.
Footnotes
One contribution of 18 to a Theme Issue ‘DNA barcoding of life’.
References
- Altschul S.F, Gish W, Miller W, Myers E.W, Lipman D.J. Basic alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Blaxter M. Counting angels with DNA. Nature. 2003;421:122–124. doi: 10.1038/421122a. 10.1038/421122a [DOI] [PubMed] [Google Scholar]
- Eckmann R, Rösch R. Lake Constance fisheries and fish ecoloy. Arch. Hydrobiol. Spec. Issues Advan. Limnol. 1998;53:285–301. [Google Scholar]
- Folmer O, Black M, Heah W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Gojobori T, Moriyama E.N, Ina Y, Ikeo K, Miura T, Tsujimoto H, Hayami M, Yokoyama S. Evolutionary origin of human and simian immunodeficiency viruses. Proc. Natl Acad. Sci. USA. 1990;87:4108–4111. doi: 10.1073/pnas.87.11.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003a;270:313–321. doi: 10.1098/rspb.2002.2218. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Ratnasingham S, deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B. 2003b;270(Suppl. 1):S96–S99. doi: 10.1098/rsbl.2003.0025. 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T.H, Cantor C.R. Evolution of protein molecules. In: Munro H.N, editor. Mammalian protein metabolism. Academic Press; New York: 1969. pp. 21–132. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Köhler J, Vieites D.R, Bonett R.M, García F.H, Glaw F, Steinke D, Vences M. New amphibians and global conservation: A boost in species discoveries in a highly endangered vertebrate group. BioScience. 2005;55:693–696. [Google Scholar]
- Lipscomb D, Platnick N, Wheeler Q. The intellectual content of taxonomy: a comment on DNA taxonomy. Trends Ecol. Evol. 2003;18:65–66. 10.1016/S0169-5347(02)00060-5 [Google Scholar]
- Lockhart P.J, Steel M.A, Hendy M.D, Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol. Biol. Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- Mallett J, Willmott K. Taxonomy: renaissance of Tower of Babel? Trends Ecol. Evol. 2003;18:57–59. 10.1016/S0169-5347(02)00061-7 [Google Scholar]
- Markmann M, Tautz D. Reverse taxonomy: an approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences. Phil. Trans. R. Soc. B. 2005;360:1917–1924. doi: 10.1098/rstb.2005.1723. 10.1098/rstb.2005.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M, Walsh P.J. Molecular systematics of the order Anaspidea based on mitochondrial DNA sequence (12S, 16S, and COI) Mol. Phylogenet. Evol. 2000;15:41–58. doi: 10.1006/mpev.1999.0736. [DOI] [PubMed] [Google Scholar]
- Moritz C, Cicero C. DNA barcoding: promise and pitfalls. PLoS Biol. 2004;2:1529–1531. doi: 10.1371/journal.pbio.0020354. 10.1371/journal.pbio.0020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.A, Ellis J.T. Effects of nucleotide sequence alignment on phylogeny estimation: A case study of 18S rDNAs of apicomplexa. Mol. Biol. Evol. 1997;14:428–441. doi: 10.1093/oxfordjournals.molbev.a025779. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Notredame C, Higgins D.G, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- Palumbi S.R, Martin A, Romano S, McMillian W.O, Stine L, Grabowski G. v.2.0. Department Zoology, Kewalo Marine Laboratory, University of Hawaii; Honolulu: 1991. The simple fools guide to PCR. [Google Scholar]
- Pearson W.R, Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosello-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol. Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. 10.1016/S0168-6445(00)00040-1 [DOI] [PubMed] [Google Scholar]
- Steinke D, Albrecht C, Pfenninger M. Molecular phylogeny and character evolution in the Western Palaearctic Helicidae s.l. (Gastropoda: Stylommatophora) Mol. Phylogenet. Evol. 2004;32:724–734. doi: 10.1016/j.ympev.2004.03.004. 10.1016/j.ympev.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition–transversion and G+C-content biases. Mol. Biol. Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tautz D, Arctander P, Minelli A, Thomas R.H, Vogler A.P. A plea for DNA taxonomy. Trends Ecol. Evol. 2003;18:70–74. 10.1016/S0169-5347(02)00041-1 [Google Scholar]
- Thomaz D, Guiller A, Clarke B. Extreme divergence of mitochondrial DNA within species of pulmonate land snails. Proc. R. Soc. B. 1996;263:363–368. doi: 10.1098/rspb.1996.0056. [DOI] [PubMed] [Google Scholar]
- Wheeler W.C. Sources of ambiguity in nucleic acid sequence alignment. In: Schierwater B, Streit B, Wagner G.P, DeSalle R, editors. Molecular ecology and evolution: approaches and applications. Birkhauser Verlag; Basel: 1994. pp. 323–352. [DOI] [PubMed] [Google Scholar]
- Vences M, Thomas M, Bonett R.M, Vieites D.R. Deciphering amphibian diversity through DNA barcoding: chances and challenges. Phil. Trans. R. Soc. B. 2005a;360:1859–1868. doi: 10.1098/rstb.2005.1717. 10.1098/rstb.2005.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vences M, Thomas M, van der Meijden A, Chiari Y, Vieites D.R. Comparative performance of the 16S rRNA in DNA barcoding of amphibians. Front. Zool. 2005b;2:5. doi: 10.1186/1742-9994-2-5. 10.1186/1742-9994-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]