Abstract
Spore formation in Saccharomyces cerevisiae occurs via the de novo synthesis of the prospore membrane during the second meiotic division. Prospore membrane formation is triggered by assembly of a membrane-organizing center, the meiotic outer plaque (MOP), on the cytoplasmic face of the spindle pole body (SPB) during meiosis. We report here the identification of two new components of the MOP, Ady4p and Spo74p. Ady4p and Spo74p interact with known proteins of the MOP and are localized to the outer plaque of the SPB during meiosis II. MOP assembly and prospore membrane formation are abolished in spo74Δ/spo74Δ cells and occur aberrantly in ady4Δ/ady4Δ cells. Spo74p and the MOP component Mpc70p are mutually dependent for recruitment to SPBs during meiosis. In contrast, both Ady4p and Spo74p are present at SPBs, albeit at reduced levels, in cells that lack the MOP component Mpc54p. Our findings suggest a model for the assembled MOP in which Mpc54p, Mpc70p, and Spo74p make up a core structural unit of the scaffold that initiates synthesis of the prospore membrane, and Ady4p is an auxiliary component that stabilizes the plaque.
Sporulation in Saccharomyces cerevisiae is a specialized form of cell division in which a single diploid cell produces four haploid spores within the cytoplasm of the original mother cell (10). The limiting membranes of the spores are synthesized de novo during meiotic segregation of nuclear DNA via the redirection of the secretory pathway (23). Golgi-derived vesicles that are destined for the plasma membrane during vegetative growth become targeted to the four spindle pole bodies (SPBs), the functional equivalents of centrosomes in higher eukaryotes, during sporulation. Fusion of these vesicles creates four discrete membrane compartments, the prospore membranes, which surround the nuclei upon the completion of meiosis to generate spores.
Prospore membrane synthesis is initiated during the transition from the first to the second meiotic division. In yeast, both meiotic divisions occur within a single, continuous nuclear envelope, in which the SPBs are embedded. The cytoplasmic face of each SPB, termed the outer plaque, expands in early meiosis II and becomes a site for docking and fusion of secretory vesicles (20). Formation of the prospore membrane occurs via the fusion of these vesicles to form a flattened sac that abuts the meiotic outer plaque (MOP). Each prospore membrane grows toward the center of the spindle during anaphase II and engulfs the adjacent lobe of the nucleus. As nuclear division occurs at the end of meiosis II, each prospore membrane fuses with itself to enclose a haploid nucleus within two continuous membranes. Spore wall material is deposited into the lumen between the two new membranes to produce a mature spore (19).
Regulation of SPB function during meiosis is critical for spore formation. The morphological expansion of the outer plaque during sporulation reflects a shift in its primary role from the anchoring of cytoplasmic microtubules to the initiation of prospore membrane synthesis. This modification of the outer plaque is required for prospore membrane formation (1, 5, 16). The change in morphology and function of the outer plaque during sporulation is due to an alteration in its protein composition. Spc72p, a component of the outer plaque during mitotic growth that binds to the gamma-tubulin complex of cytoplasmic microtubules, disappears from SPBs during meiosis and is replaced by the meiosis-specific components Mpc54p and Mpc70p/Spo21p (15, 16). Mpc54p, Mpc70p, and the constitutive outer plaque components Cnm67p and Nud1p/Spc94p are the four known proteins of the MOP (1, 16). Cnm67p, Mpc54p, and Mpc70p are predicted to have extended conformations with amino- and carboxy-terminal globular domains separated by central coiled-coil regions and are required for expansion of the outer plaque and formation of prospore membranes (1, 16, 31). The role of Nud1p in assembly of the MOP is more difficult to assess because this protein is essential for vegetative growth, but the interaction of Nud1p with Cnm67p, Mpc54p, and Mpc70p in the two-hybrid assay suggests that it may function as a docking protein for other outer plaque components (8, 16, 41).
Certain environmental stimuli or genetic perturbations that affect assembly of the MOP result in formation of nonsister dyads, two-spore asci in which one haploid nucleus from each meiosis II spindle has been packaged. For example, the removal of the carbon source after initiation but prior to completion of meiosis leads to modification of only two outer plaques and the production of nonsister dyads (5, 26). Nonsister dyads can also result from mutations that cause reduced levels of structural components of the MOP or affect their recruitment to the SPB (1, 7, 13, 40). In contrast, dyads caused by mutations that affect subsequent steps in sporulation, such as growth of prospore membranes or synthesis of spore wall components, contain spores in which two meiotic products have been packaged at random (9, 21, 25).
The identification of proteins with potential roles in assembly of the MOP and formation of the prospore membrane has been facilitated by genome-wide analyses (4, 14, 28, 29, 39). The results from such large-scale analyses have been used to elucidate the specific functions of several proteins involved in these processes (1, 21, 25). Other genes that have null phenotypes or transcription profiles comparable to those of components that mediate modification of the outer plaque and synthesis of the prospore membrane have yet to be analyzed intensively.
In this study, we have characterized the products of two meiotically induced genes, ADY4 and SPO74, involved in sporulation. We find that Ady4p and Spo74p are components of the MOP that are required for normal prospore membrane formation. MOPs and prospore membranes are absent in spo74Δ/spo74Δ cells and display heterogeneous defects in ady4Δ/ady4Δ cells. Quantitative analysis of fluorescence from green fluorescent protein (GFP) fusions during meiosis revealed that loss of either Mpc70p or Spo74p abolishes recruitment of the other to SPBs and that Ady4p is present at SPBs at reduced levels in cells that lack either Mpc54p, Mpc70p, or Spo74p. Our findings suggest that Mpc54p, Mpc70p, and Spo74p assemble at the SPB during meiosis into the ordered structure that initiates formation of the prospore membrane and that Ady4p is an ancillary component of the MOP that stabilizes the plaque.
MATERIALS AND METHODS
Yeast strains and methods.
Standard S. cerevisiae genetic methods and media were used (30). Strains are listed in Table 1. Diploid strains with AN, MND, and NY designations were derived from SK1 haploids AN117-4B and AN117-16D and are isogenic with each other, diploid strains with MYA designation are derived from a separate SK1 stock strain and are isogenic with each other, and YCJ4 is not of SK1 ancestry. AN117-4B, AN117-16D, AN120, YCJ4, and MYA strains have been described previously (12, 24, 29). The following chromosomal insertions were made in AN117-4B and AN117-16D, and the resulting strains were mated to create AN279, AN282, MND57, MND58, and NY541, respectively: GFP-HIS3MX6 at the 3′ end of the ADY4 open reading frame (ORF), GFP-HIS3MX6 at the 3′ end of the SPO74 ORF, HIS3MX6 replacing the ADY4 ORF, HIS3MX6 replacing the SPO74 ORF, and HIS3MX6 replacing the MPC54 ORF. SnaBI-cut pRS304 was integrated into one allele of trp1 in MND57 to create MND59 and into an unknown site in AN117-4B, which was then mated to the spo74::HIS3MX6 derivative of AN117-16D to create MND61.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| AN117-4B | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 | 24 |
| AN117-16D | MATahis3Δ/SK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 | 24 |
| YCJ4 | MATaADE2::pGAL-URA3 canR gal4 gal80 his3 leu2 LYS2::Lexop-lacZ trp1 ura3 | 12 |
| AN120 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura/ura3 | 24 |
| AN161 | MATa/MATα ARG4/arg4-NspI cnm67Δ::HIS3MX6/cnm67Δ::HIS3MX6 his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 | 1 |
| AN180 | MATa/MATα ARG4/arg4-Nsp1 his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 mpc70Δ::HIS3MX6/mpc70Δ::HIS3MX6 RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 | 1 |
| AN279 | MATa/MATα ADY4::GFP-HIS3MX6/ADY4::GFP-HIS3MX6 ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 | This study |
| AN282 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 RME1/rme1Δ::LEU2 SPO74::GFP-HIS3MX6/SPO74::GFP-HISS3MX6 trp1::hisG/trp1::hisG ura3/ura3 | This study |
| MND57 | MATa/MATα ady4Δ::HIS3MX6/ady4Δ::HIS3MX6 ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 | This study |
| MND58 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 RME1/rme1Δ::LEU2 spo74Δ::HIS3MX6/spo74Δ::HIS3MX6 trp1::hisG/trp1::hisG ura3/ura3 | This study |
| MND59 | MATa/MATα ady4Δ::HIS3MX6/ady4Δ::HIS3MX6 ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 RME1/rme1Δ::LEU2 trp1/trp1::pRS304-TRP1 ura3/ura3 | This study |
| MND61 | MATa/MATα ARG4/arg4-Nsp1 his3/his3 hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 RME1/rme1Δ::LEU2 SPO74/spo74Δ::HIS3MX6 trp1::hisG/trp1::hisG ura3/ura3 pRS304-TRP1 at unknown locus | This study |
| MYA-1898 | MATa/MATα his3/his3 HO/HO LEU2::pURA3-tetR-GFP/LEU2::pURA3-tetR-GFP lys2/lys2 REC8::HA3-URA3/REC8::HA3-URA3 spo74Δ::HIS3MX6/spo74Δ::HIS3MX6 trp1/trp1URA3::tetO224/URA3::tetO22 | 29 |
| MYA-1993 | MATa/MATα ady4Δ::HIS3MX6/ady4Δ::HIS3MX6 his3/his3 HO/HO LEU2::pURA3-tetR-GFP/LEU2::pURA3-tetR-GFP lys2/lys2 REC8::HA3-URA3/REC8::HA3-URA3 trp1/trp1URA3::tetO224/URA3::tetO22 | 29 |
| MYA-2047 | MATa/MATα his3/his3 HO/HO LEU2::pURA3-tetR-GFP/LEU2::pURA3-tetR-GFP lys2/lys2 mpc70Δ::HIS3MX6/mpc70Δ::HIS3MX6 REC8::HA3-URA3/REC8::HA3-URA3 trp1/trp1URA3::tetO22/URA3::tetO22 | 29 |
| MYA-2089 | MATa/MATα his3/his3 HO/HO LEU2::pURA3-tetR-GFP/LEU2::pURA3-tetR-GFP lys2/lys2 REC8::HA3-URA3/REC8::HA3-URA3 trp1/trp1 URA3::tetO224/URA3::tetO22 | 29 |
| NY541 | MATa/MATα ARG4/arg4-Nsp1 his3ΔSK/his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 mpc54Δ::HIS3/MX6/mpc54Δ::HIS3MX6 RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 | H. Tachikawa |
| NY556 | MATa/MATα ARG4/arg4-Nsp1 his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 MPC54::GFP-HIS3MX6/MPC54::GFP-HIS3MX6 RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 | H. Tachikawa |
Gene insertions and replacements were performed by transformation of strains with PCR-generated DNA cassettes and verified by PCR (18). The sequences of oligonucleotide primers used to amplify insertion cassettes by PCR are listed in Table 2. The cassettes used to replace ADY4, MPC54, and SPO74 in MND57, NY541, and MND58, respectively, were amplified from pFA6a-HIS3MX6 with the following pairs of primers: ANO299-ANO283, HT76-HT77, and ANO297-ANO292. The cassettes used to insert the GFP gene into the 3′ ends of ADY4 and SPO74 in AN279 and AN282, respectively, were amplified from pFA6a-yEGFP(S65T)-HIS3MX6 with the primer pairs ANO282-ANO283 and ANO291-ANO292.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| ANO282 | TTG CCG GAC ATT TTG TCA CGT ATA TAT CAA ACC TTT ACT GAT CCC TCC AAT CGG ATC CCC GGG TTA ATT AA |
| ANO283 | GAA CCC CAG ATG CCA AAA AAC CTA AAA TAT AAA AAT AGA GTA CAA GTT ATG TGA ATT CGA GCT CGT TTA AAC |
| ANO291 | AAT AGA ATG CCC AGA AGA AGT AAT GAA GAA ATG TAT AGC TGG TCA CGT AAT CGG ATC CCC GGG TTA ATT AA |
| ANO292 | TAT GCA AGA CAT GTA TAT ATG ATT AGT AGT TTT TTT TAT TTA CTT TCA TTG AGA ATT CGA GCT CGT TTA AAC |
| ANO297 | ACT CTT CCC GGT TAT GTT GTA TTG GCA AGG AAA GAG CTG AAT TGG TTG AGT GCG GAT CC CGG GTT AAT TAA |
| ANO298 | GTT CTT TCT AGA TAG GTA TGT GCG ATG AGG |
| ANO299 | ACA TAT ATG CCC TAT TTA TTG ATC ATA GTC ACA AAT TGT CGG CTC ACG TAT ACG GAT CCC CGG GTT AAT TAA |
| ANO300 | GTT CTT TCT AGA TAT CTT CGG ATG CAA GGG |
| HT66 | GAA GAA TTC AGA TCT ATA TTA CCC TGT TAT CC |
| HT76 | TGA ACT GAC TAA GAT AAG TTG TTA GTC TCT GTA ACT TCT CGT ATA AGG CCC GGA TCC CCG GGT TAA TTA A |
| HT77 | TCT TAA ACA TAA TGT CTT TTT AAT GTT TGT AGG TTA GGT TAT AAA CCT TAG AAT TCG AGC TCG TTT AAA C |
| HT99 | GAA GAA CTC GAG TAC TTC GTT AAC CGC GTC CGT CTC |
| MNO115 | GGG GGG GAT CCC AGT ATG GAT AAT ATT TTA AAG |
| MNO116 | GGG GGG CTC GAG TCA CTT GAT GGA GTG CGG |
| MNO117 | GGG GGG GAT CCC TAC TCT TCT ATC TGT GGT AAC |
| MNO119 | GGG GGG GAT CCC CAA TAC AAT GTC GAT ACG CAA GC |
| MNO120 | GGG GGG CTC GAG TCA GCT CAT AAT CGA TGC GGT G |
| MNO121 | GGG GGG GAT CCC AAA AGT CAA AAT GAC AAA ATT TTG |
| MNO122 | GGG GGG CTC GAG TCA TAA TTG CGC TTC TTT TTC |
| MNO123 | GGG GGG GAT CCC AAA TAT GCA AAA TTA GCT G |
| MNO124 | GGG GGG CTC GAG TCA TTC TTT AAA TAA AGA GAA C |
| MNO125 | GGG GGG GAT CCC CGG TTC TTA CTG CCA AGT G |
| MNO126 | GGG GGG CTC GAG TCA ATA ACA TGT CAG AAT ATT AG |
| MNO170 | GGG GGG CTC GAG AGA TCT ATA TTA CCC TGT TAT CC |
| MNO173 | TTG CCG GAC ATT TTG TCA CG |
| MNO174 | AGA ATT GGG ACA ACT CCA GTG |
| MNO177 | GCG GCG CTC GAG TTA ATT GGA GGG ATC AGT AAA |
| MNO178 | GCG GCG GAT CCC ATG GGA GCT GGC ACT CTT |
| MNO179 | GCG GCG GGG ATC CGT ATG GGA GCT GGC ACT CTT |
| MNO180 | GCG GCG CTC GAG TCA ATT ACG TGA CCA GCT ATA |
| MNO181 | GCG GCG GCA GAT CTC ATG AAT AAA GAC GTC GAC |
| MNO182 | GCG GCG GAG ATC TGT ATG AAT AAA GAC GTC GAC |
| MNO183 | GCG GCG GAT CCC ATG CCA GAA GAT ACA AGC |
| MNO184 | GCG GCG GGG ATC CGT ATG CCA GAA GAT ACA AGC |
| MNO185 | GCG GCG CTC GAG TCA ATT TGT AAC GTG TTC ATT |
Cells were induced to sporulate in liquid medium as described previously (25). Cells were cultured to saturation in either yeast extract-peptone-dextrose or synthetic complete medium lacking the appropriate nutrient, cultured overnight to mid-log phase in YPA, and transferred to 2% potassium acetate at a concentration of 3 × 107 cells/ml (optical density at 600 nm of ∼2.0).
Plasmids.
Plasmids made for this study are listed in Table 3. All plasmids created in this study except pRS424-ADY4-GFP were made in three steps: PCR amplification of the sequence of interest from a plasmid or genomic DNA template, digestion of the PCR product at restriction sites within the PCR primers, and subcloning of the digested PCR product into restriction sites in the polylinker of a standard yeast vector. To make pRS424-ADY4-GFP, two partially overlapping PCR products with a common PacI site were amplified, each product was digested at the PacI site and at a second restriction site within the primer away from the region of overlap, and the two PCR products were simultaneously subcloned into pRS424. Oligonucleotide primers, template DNAs, and restriction sites used for subcloning are listed in Table 3.
TABLE 3.
PCR primers, templates, and restriction sites used for plasmid construction
| Plasmid (position) | Oligonucleotide primers
|
Primer restriction endonuclease sites
|
Template | Vector restriction endonuclease sites
|
|||
|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | ||
| pEG202-ADY4 (1-493) | MNO182 | MNO177 | Bg/II | XhoI | pRS424-ADY4-GFP | BamHI | XhoI |
| pEG202-MPC54 (1-465) | MNO184 | MNO185 | BamHI | XhoI | pRS314-MPC54-GFP | BamHI | XhoI |
| pEG202-SPO74 (1-413) | MNO179 | MNO180 | BamHI | XhoI | pRS424-SPO74-GFP | BamHI | XhoI |
| pGADGH-ADY4 (1-493) | MNO181 | MNO177 | BglII | XhoI | pRS424-ADY4-GFP | BamHI | XhoI |
| pGADGH-MPC54 (1-465) | MNO183 | MNO185 | BamHI | XhoI | pRS314-MPC54-GFP | BamHI | XhoI |
| pGADGH-MPC70 (1-200) | MNO115 | MNO120 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (1-300) | MNO115 | MNO122 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (1-400) | MNO115 | MNO124 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (1-500) | MNO115 | MNO126 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (1-609) | MNO115 | MNO116 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (101-609) | MNO117 | MNO116 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (201-609) | MNO119 | MNO116 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (301-609) | MNO121 | MNO116 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (401-609) | MNO123 | MNO116 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-MPC70 (501-609) | MNO125 | MNO116 | BamHI | XhoI | pRS316-SPO21 | BamHI | XhoI |
| pGADGH-SPO74 (1-413) | MNO178 | MNO180 | BamHI | XhoI | pRS424-SPO74-GFP | BamI | XhoI |
| pRS314-MPC54-GFP | HT99 | MNO170 | XbaI | XhoI | Genomic DNA of NY556 | SpeI | XhoI |
| pRS314-SPO74-GFP | ANO298 | HT66 | XbaI | EcoRI | Genomic DNA of AN282 | SpeI | EcoRI |
| pRS424-ADY4-GFP | Genomic DNA of AN279 | SpeI | XhoI | ||||
| ADY4 ORF | ANO300 | MNO174 | XbaI | PacI | |||
| GFP ORF | MNO173 | MNO170 | PacI | XhoI | |||
The following plasmids used in this study have been described elsewhere: 1-66 (38); pFA6a-HIS3MX6 (18); pFA6a-yEGFP-HIS3MX6 and pGADGH-MPC70 (25); pMK169, pMK183, pMK184, pSM613, and pSM614 (8); and pRS316-SPO21 (1). The following plasmids were used to express the indicated fusion proteins for two-hybrid assays: 1-66, GAD-Mpc70p133-609; pMK169, GAD-Spc42p1-363; pMK183, GAD-Cnm67p386-580; pMK184, LexA-Cnm67p386-580; pSM613, GAD-Nud1p405-852; and pSM614, LexA-Nud1p405-852. pRS424-G20 contains a chimeric gene that consists of a fragment of SPO20 fused to the GFP gene (H. Nikanishi, unpublished data.)
Fluorescence microscopy.
Preparation of cells for fluorescence microscopy and collection and analysis of images were performed essentially as described elsewhere (25). For visualization of naturally fluorescent proteins only, cells were fixed in 4% formaldehyde for 5 min, washed with phosphate-buffered saline (PBS) (130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4), and mounted with Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole) (Vector, Burlingame, Calif.). For simultaneous visualization of tubulin and naturally fluorescent proteins, cells were fixed for 10 to 15 min in 4% formaldehyde and processed for immunofluorescence as described elsewhere (25). Tubulin was visualized by using a 1:10 dilution of MAS 078b primary antibody (Harlan, Sussex, United Kingdom) and a 1:400 dilution of goat anti-mouse Alexa 546 (Molecular Probes, Eugene, Oreg.) secondary antibody, which cross-reacts with the MAS 078 rat monoclonal antibody. Fluorescence images were collected with a Zeiss Axioplan 2 microscope and Zeiss AxioCam HRm digital camera, and Axiovision 3.0.6 software was used for deconvolution and quantitation. Figures were prepared by using Photoshop 6.0 (Adobe Software, San Jose, Calif.) and Canvas 5.0.2 (Deneba Software).
Analysis of fluorescence intensity of GFP fusions for SPB components was performed as follows. Cells transformed with plasmids encoding GFP fusions were fixed after 6 to 8 h in 2% potassium acetate as described above, and the number of GFP foci at the spindle poles were analyzed in 50 meiosis II cells from each culture. Quantification of fluorescence intensity was performed on Z-series collections of raw (i.e., not deconvolved) images. A circle corresponding to a radius of 0.3 μm was drawn around each SPB, and the average pixel intensity in that circle from the brightest Z section was taken as the total value. The average pixel intensity of a same-sized circle that did not overlap an SPB was taken for each cell as the background value, and the specific signal was calculated as the background signal subtracted from the total signal for each SPB. The specific signal of ≥15 SPBs was quantified for each combination of strain (including the wild type) and GFP fusion. The mean specific signal for a given GFP fusion in each mutant was compared to the mean specific signal for that GFP fusion in wild-type cells, which was defined as 100%, to calculate the relative signal intensity. The following strains and plasmids were used: strains AN120 (wild type), MND57 (ady4Δ/ady4Δ), AN161 (cnm67Δ/cnm67Δ), NY541 (mpc54Δ/mpc54Δ), AN180 (mpc70Δ/mpc70Δ), and MND58 (spo74Δ/spo74Δ) and plasmids pRS424-ADY4-GFP, pRS314-MPC54-GFP, pSB33 (MPC70-GFP), and pRS314-SPO74-GFP.
Electron microscopy.
Cells were prepared for standard transmission electron microscopy as described elsewhere (37). Cells were prepared for transmission immunoelectron microscopy essentially as described previously (11) with the following slight modifications. Cells from sporulating cultures were harvested, fixed for 1 h in PBS containing 2% paraformaldehyde and 0.1% glutaraldehyde, washed in PBS, resuspended in PBS containing 8% sucrose, and stored overnight on ice. The cells were then rapidly frozen by high-pressure freezing (BAL-TEC HPM-010; Technotrade International, Manchester, N.H.) and freeze-substituted at −90°C in 0.2% glutaraldehyde plus 0.01% uranyl acetate in acetone for 96 h in an EM AFS device (Leica, Vienna, Austria). The cells were warmed over 22.5 h to −45°C and then infiltrated with HM20 resin over a period of 4 days. The cells were flat embedded under UV light at −45°C in HM20 for 3 days and then warmed to room temperature over a 24 h period. Embedded cells were sectioned and immunostained as follows: sections were (i) floated on blocking buffer (0.02% Tween 20, 0.8% bovine serum albumin, and 0.1% fish gelatin in 1× PBS) for 1 h, (ii) immunostained with anti-GFP overnight at 4°C as described previously (42), (iii) rinsed with PBS plus 0.1% Tween 20 three times, (iv) incubated with 10-nm-diameter colloidal gold (BB International, Cardiff, United Kingdom) for 2 h at room temperature, (v) rinsed and fixed with 1.0% glutaraldehyde for 5 min, (vi), stained with aqueous uranyl acetate and lead citrate, and (vii) imaged in a Philips CM-10 electron microscope.
β-Galactosidase assays.
β-Galactosidase assays were performed as described previously (25). Transformants of YCJ4 were cultured overnight at 30°C on a Whatman 50 filter on the surface of a plate of synthetic complete medium lacking His and Leu, and the filter was immersed in liquid N2 for 10 s and then incubated at 30°C in Z buffer containing 0.1% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and 0.027% β-mercaptoethanol. At least three independent transformants for each combination of constructs were analyzed. None of GAD fusions interacted with LexA alone, but LexA-Spo74p1-413 gave moderate induction of β-galactosidase expression in the absence of a GAD fusion. The following LexA fusions were used as positive controls for interactions with GAD fusions: Cnm67p386-580 for Spc42p1-363 and Nud1p405-852 for Mpc70p and Nud1p405-852.
Analysis of segregation of centromere-linked markers in dyads.
Centromere linkage values of 11 and 9.5 centimorgans for ARG4 and RME1, respectively, in strain AN120 were calculated by analysis of 253 nonsister dyads formed in response to environmental conditions, and these values were used to calculate expected frequencies for dyad classes resulting from packaging of nonsister meiotic nuclei. All segregants from dyads were competent to mate with either MATa or MATα strains and thus were inferred to be haploid.
RESULTS
Ady4p and Spo74p are localized to SPBs during meiosis.
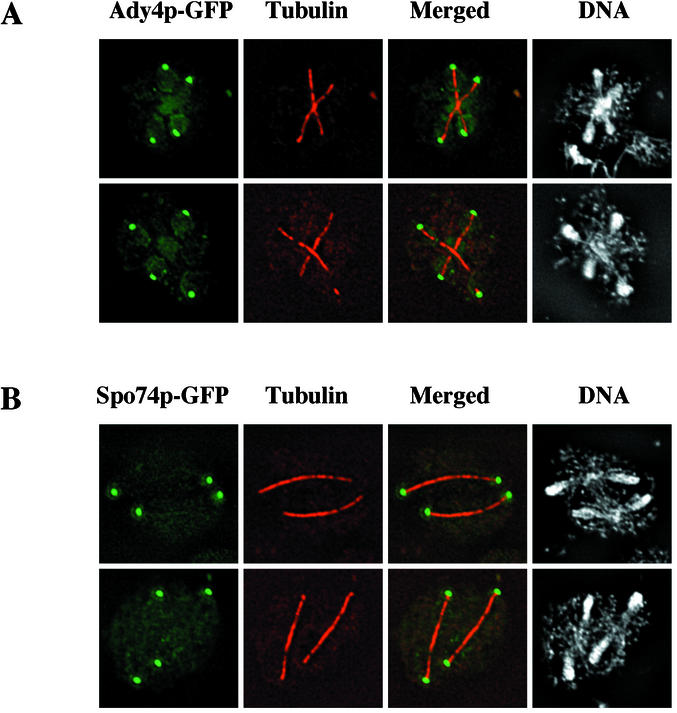
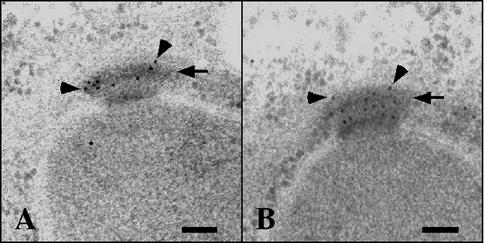
To identify novel proteins with potential roles in prospore membrane formation, the subcellular localization of the products of genes that are induced during meiosis and required for normal ascus formation was analyzed (29). Strains with GFP fused to the 3′ ends of ADY4, AMA1, SPO73, SPO74, and SPO77 were individually constructed and examined by fluorescence microscopy. The two of these gene products that produced functional, visible GFP fusions, Ady4p and Spo74p, formed foci at the ends of the meiotic spindles (Fig. 1). Spo74p-GFP could be easily detected and showed the same distribution when expressed homozygously from its natural locus or from a centromeric plasmid. Ady4p-GFP could be visualized consistently only when expressed from a multicopy plasmid but showed a similar distribution when expressed homozygously from its natural locus or from a centromeric plasmid. Analysis of Ady4p and Spo74p by immunoelectron microscopy revealed that both proteins were localized to the meiotic outer plaque of the SPB (Fig. 2). The localization patterns of Ady4p and Spo74p are similar to those that have been described for the MOP components Mpc54p and Mpc70p (1, 16).
FIG. 1.
Localization of Ady4p-GFP and Spo74p-GFP by fluorescence microscopy. Sporulating AN120 cells transformed with either pRS424-ADY4-GFP (top panels) or pRS314-SPO74-GFP (bottom panels) were processed for immunofluorescence microscopy with an antibody to tubulin and DAPI to visualize DNA. Natural fluorescence from GFP fusions is shown in green, immunofluorescent signal from antitubulin antibody is shown in red, and fluorescent signal from DAPI-stained DNA is shown in gray. Merged column shows overlaid images of GFP and tubulin fluorescences. Images of two cells from each strain are shown.
FIG. 2.
Localization of Ady4p-GFP and Spo74p-GFP by electron microscopy. Sporulating cells from strains AN279 (ADY4-GFP/ADY4-GFP) (A) and AN282 (SPO74-GFP/SPO74-GFP) (B) were processed for electron microscopy, and sections were stained with primary antibodies to GFP and gold-conjugated secondary antibodies. Arrows point to MOPs, and arrowheads point to selected gold particles. Bars, 100 nm.
Ady4p and Spo74p interact with components of the MOP in the two-hybrid assay.
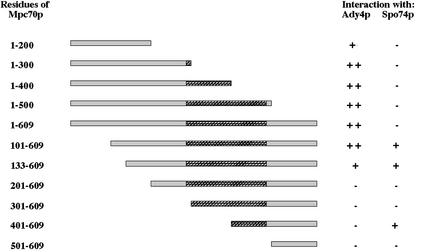
To examine whether Ady4p and Spo74p interact with known components of the SPB, these proteins were tested for the ability to bind various fusions of SPB proteins in the two-hybrid assay. A LexA fusion of Ady4p interacted with GAD fusions containing sequences from Cnm67p, Mpc70p, and Nud1p (Table 4). A LexA fusion of Spo74p interacted with GAD fusions containing sequences from Mpc70p and itself, and a GAD fusion of Spo74p interacted with a LexA fusion of Mpc54p (Table 4). The regions of Mpc70p that interact with Ady4p and Spo74p were mapped by using a series of fusions that contain the Gal4p activation domain and different regions of Mpc70p. Residues 100 to 300 of Mpc70p were required for maximal binding to Ady4p, whereas residues 401 to 609 were sufficient to mediate the interaction with Spo74p (Fig. 3). These results indicate that Ady4p and Spo74p interact with components of the MOP.
TABLE 4.
Two-hybrid interactions with Ady4p and Spo74p
| GAD fusion | Interaction with LexA Fusion:
|
||
|---|---|---|---|
| Ady4p1-493 | Mpc541-465 | Spo74p1-413 | |
| Ady4p1-493 | − | − | − |
| Cnm67p386-580 | + | NDa | − |
| Mpc541-465 | − | + | − |
| Mpc70pb | + | + | + |
| Nud1p405-852 | + | ND | − |
| Spc42p1-363 | − | ND | − |
| Spo74p1-413 | − | + | + |
ND, not determined.
GAD fusions containing various regions of MPC70p were tested for interactions; see Fig. 3 for details.
FIG. 3.
Schematic of GAD-Mpc70p fusions and their ability to interact with LexA-Ady4p and LexA-Spo74p. Residues of Mpc70p included in fusion proteins are indicated at the left, and stippled regions represent coiled-coil domains of Mpc70p.
ady4Δ/ady4Δ and SPO74/spo74Δ mutants accumulate random and nonsister dyads, respectively.
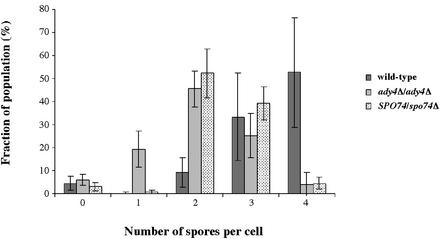
To characterize more thoroughly the roles of ADY4 and SPO74 in sporulation, the effects of null mutations in these genes on the distribution of ascal types was analyzed. In wild-type cultures, the majority of cells formed asci with three or four spores (Fig. 4). The spo74Δ/spo74Δ mutant, as reported previously, failed to sporulate (29; unpublished data), but SPO74/spo74Δ cells sporulated efficiently and formed mostly dyads and triads (Fig. 4). The accumulation of dyads due to heterozygosity of SPO74 is similar to the gene dosage effect observed for the outer plaque components MPC70 and MPC54 (1, 40; unpublished data). In ady4Δ/ady4Δ cultures, most cells formed spores, but the predominant ascal type was a dyad, which is consistent with the previous description of this mutant (Fig. 4) (29).
FIG. 4.
Distribution of ascal types in wild-type, ady4Δ/ady4Δ, and SPO74/spo74Δ cells. Cells from strains AN120 (wild type), MND57 (ady4Δ/ady4Δ), and MND61 (SPO74/spo74Δ) were cultured overnight in 2% potassium acetate, and 200 cells per culture were analyzed. The data shown are averages and standard deviations from at least three independent experiments.
To determine whether the dyads produced in ady4Δ/ady4Δ and SPO74/spo74Δ cells are nonsisters, segregation of the centromere-linked markers ARG4 and RME1 was analyzed. In the SPO74/spo74Δ mutant, the observed distributions of dyad types for both ARG4 and RME1 were close to the values predicted for nonsister dyads and were significantly different (P < 0.001) from the expected frequencies for random dyads (Table 5). This haplo-insufficient phenotype of SPO74 is also similar to that caused by loss of one copy of MPC70, in which dyads that are formed are exclusively nonsisters (1, 40). In contrast, the observed distributions of dyad types for both ARG4 and RME1 in the ady4Δ/ady4Δ mutant were significantly different (P < 0.001) from the values predicted for nonsister dyads and were close to the expected frequencies for random dyads (Table 5). The production of nonsister dyads in SPO74/spo74Δ cells suggests that Spo74p is a MOP component required for formation of prospore membranes, whereas the accumulation of random dyads in ady4Δ/ady4Δ cells represents a novel mutant phenotype for a sporulation-specific SPB component.
TABLE 5.
Segregation of ARG4 and RME1 in ady4Δ/ady4Δ and SPO74/spo74Δ dyads
| Dyad type | Expected frequency (%)
|
Observed frequency (%)
|
|||||
|---|---|---|---|---|---|---|---|
| Random | Nonsister
|
ady4Δ/ady4Δa
|
SPO74/spo74Δb
|
||||
| ARG4 | RME1 | ARG4 | RME1 | ARG4 | RME1 | ||
| +/+ | 16.7 | 5.5 | 4.8 | 15.1 | 16.1 | 6.5 | 0.9 |
| +/− | 66.7 | 8.9 | 90.5 | 73.1 | 62.3 | 8.7 | 95.3 |
| −/− | 16.7 | 5.5 | 4.8 | 11.8 | 21.5 | 6.5 | 3.7 |
Ninety-three dyads of MND59 (ady4Δ/ady4Δ ARG4/arg4 RME1/rme1Δ::LEU2) were analyzed.
One hundred seven dyads of MND61 (ARG4/arg4 RME1/rme1Δ::LEU2 SPO74/spo74Δ) were analyzed.
Prospore membrane formation is defective in ady4Δ/ady4Δ and spo74Δ/spo74Δ cells.
To determine whether Ady4p and Spo74p promote prospore membrane formation, the number and morphology of prospore membranes in mutants that lack these proteins were analyzed. Cells that express a prospore membrane marker, a fusion of GFP and an amino-terminal fragment of Spo20p (H. Nakanishi and A. M. Neiman, unpublished data), were induced to undergo meiosis and analyzed by fluorescence microscopy. In wild-type cultures, most cells in meiosis II had three or four prospore membranes of comparable size and shape, and nearly all prospore membranes in postmeiotic cells (>99%) surrounded a nucleus (Fig. 5 and Table 6). In spo74Δ/spo74Δ cultures, the GFP fusion could be detected in some cells as cytoplasmic spots, but no prospore membranes were observed in cells at any stage of meiosis (Fig. 5). The absence of prospore membranes in spo74Δ/spo74Δ cells is similar to the phenotype caused by loss of either MPC54 or MPC70 and demonstrates that Spo74p is essential for initiation of prospore membrane synthesis (1, 16).
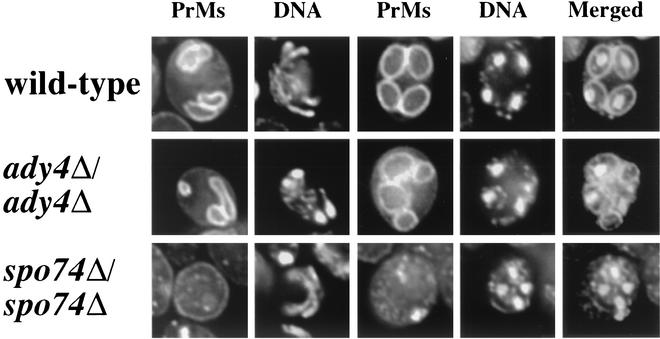
FIG. 5.
Prospore membrane morphology in wild-type, ady4Δ/ady4Δ, and spo74Δ/spo74Δ cells. Cells from strains AN120 (wild type), MND57 (ady4Δ/ady4Δ), and MND58 (spo74Δ/spo74Δ) that had been transformed with pRS424-G20 were induced to enter meiosis, fixed, stained with DAPI, and analyzed by fluorescence microscopy. Prospore membranes (PrMs) were visualized by using the fusion protein of GFP and an amino-terminal fragment of Spo20p, and DNA was visualized with DAPI. PrM and DNA images from one meiosis II cell (left two columns) and PrM, DNA, and merged images from one postmeiotic cell (right three columns) from each strain are shown.
TABLE 6.
Prospore membrane formation in ady4Δ/ady4Δ cells
| Relevant genotype | No. of cells with the following no. of prospore membranes per cella:
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Wild type | 0 | 2 | 32 | 66 |
| ady4Δ/ady4Δ | 26 | 40 | 26 | 10 |
One hundred meiosis II cells with at least one visible prospore membrane from strains AN120 (wild type) and MND57 (ady4Δ/ady4Δ) transformed with pRS424-G20 were analyzed.
Prospore membranes were present in ady4Δ/ady4Δ cells but varied in morphology and number per cell. Among ady4Δ/ady4Δ cells in meiosis II, cells with two prospore membranes represented the predominant class and cells with one or three prospore membranes each made up about a quarter of the population (Table 6). In the ady4Δ/ady4Δ mutant, cells in meiosis II often displayed heterogeneity in the size and morphology of individual prospore membranes, and 16% of prospore membranes in postmeiotic cells had failed to capture nuclei (Fig. 5). These results suggest that Ady4p may facilitate growth of prospore membranes.
MOPs are defective in ady4Δ/ady4Δ and spo74Δ/spo74Δ cells.
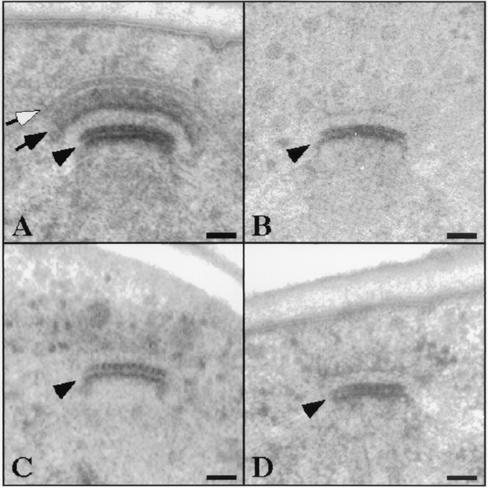
To determine whether modification of the outer plaque and anchoring of the prospore membrane occur normally during meiosis in ady4Δ/ady4Δ and spo74Δ/spo74Δ mutants, SPBs in these cells were examined by electron microscopy. In wild-type cells, the MOP appeared as an arc composed of two electron-dense layers of equal thickness (Fig. 6A). The prospore membrane in wild-type cells was closely apposed to the outer layer of the MOP and extended laterally from the MOP parallel to the nuclear envelope (Fig. 6A). In spo74Δ/spo74Δ cells, only a trace amount of MOP material could be detected in meiosis II, and prospore membranes were absent (Fig. 6C and D). The absence of prospore membranes and most outer plaque material from the SPBs of spo74Δ/spo74Δ cells is similar to the phenotype caused by loss of either MPC54 or MPC70 and provides further evidence that Spo74p is a MOP component required for prospore membrane synthesis (Fig. 6B) (1, 16).
FIG. 6.
SPB morphology in spo74Δ/spo74Δ cells. Cells from strains MYA-2089 (wild type) (A), MYA-2047 (mpc70Δ/mpc70Δ) (B), and MYA-1898 (spo74Δ/spo74Δ) (C and D) were induced to enter meiosis and processed for electron microscopy. Open arrow, prospore membrane in wild-type cell; closed arrow, MOP in wild-type cell; arrowheads, central plaques. Bars, 50 nm.
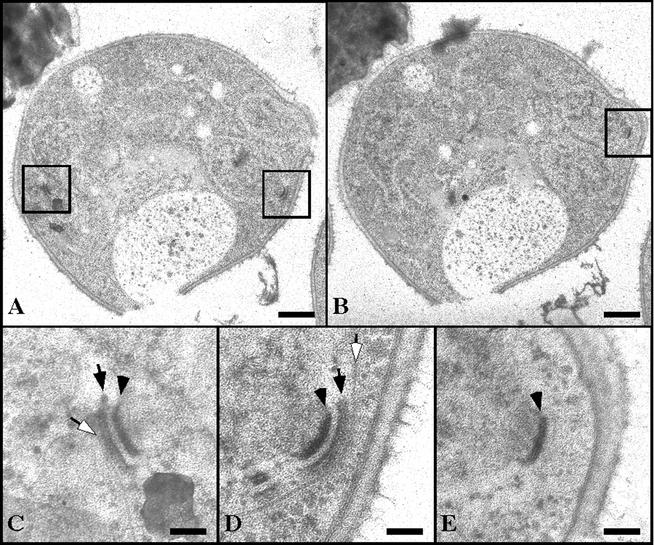
In ady4Δ/ady4Δ cells, SPBs in meiosis II displayed heterogeneous defects. Of 17 SPBs in ady4Δ/ady4Δ cells that were analyzed, 13 had normal-looking MOPs and prospore membranes, 2 displayed defects in attachment of the prospore membrane to the MOP, and 2 showed deficiencies in MOP material. Significantly, morphological differences could be detected among SPBs within the same ady4Δ/ady4Δ cell (Fig. 7). Of the three SPBs that could be identified in a single ady4Δ/ady4Δ cell in meiosis II, one appeared to be normal (Fig. 7D), another displayed detachment of the prospore membrane from the MOP (Fig. 7C), and the third entirely lacked a prospore membrane and the electron-dense layers of the MOP (Fig. 7E). These findings suggest that Ady4p may facilitate growth of the prospore membrane by stabilizing the MOP.
FIG. 7.
SPB morphology in an ady4Δ/ady4Δ cell. Cells from strain MYA-1993 (ady4Δ/ady4Δ) were induced to sporulate and processed for electron microscopy. (A and B) Low-magnification images from different sections of the same cell. (C) High-magnification image of the boxed region on the left side of panel A. (D) High-magnification image of the boxed region on the right side of panel A. (E) High-magnification image of the boxed region of panel B. Open arrows, prospore membranes; closed arrows, MOPs; arrowheads, central plaques. Bars, 500 nm (A and B) and 100 nm (C, D, and E).
Components of the MOP are interdependent for localization to the SPB.
To elucidate the molecular basis of MOP assembly, recruitment to SPBs was individually analyzed for each of the meiosis-specific components of the outer plaque in mutants that lack one of the other components. Strains with homozygous deletions of ADY4, MPC54, MPC70, or SPO74 were transformed with plasmids that contained functional, GFP-tagged versions of the same genes, and plasmids with ADY4-GFP and SPO74-GFP were also introduced into a strain that lacks the constitutive outer plaque component CNM67. Cells were induced to enter meiosis, and the intensity of fluorescent foci at SPBs of cells in meiosis II was analyzed. The results are shown in Table 7. To verify that quantification of the fluorescent signal from SPB foci reflects the amount of a given GFP fusion present at SPBs, fluorescent foci at SPBs during meiosis II were also analyzed in cells in which MPC70-GFP was expressed either homozygously (MPC70-GFP/MPC70-GFP) or heterozygously (MPC70-GFP/MPC70) from its natural locus. The mean fluorescent signal from SPBs in MPC70-GFP/MPC70 cells was 30% of the value from MPC70-GFP/MPC70-GFP cells (data not shown), indicating that the intensity of the fluorescent signal from SPB foci is an approximate representation of the amount of a given GFP fusion present at the SPB.
TABLE 7.
SPB fluorescence of GFP fusion proteins in MOP mutants
| Relevant genotype | Relative intensity of GFP foci (% of signal from wild type)a with the following GFP fusion:
|
|||
|---|---|---|---|---|
| Ady4p | Mpc54p | Mpc70p | Spo74p | |
| ady4Δ/ady4Δ | NDb | 94 ± 21 | 100 ± 49 | 93 ± 57 |
| cnm67Δ/cnm67Δ | —c | ND | ND | — |
| mpc54Δ/mpc54Δ | 16 ± 32 | ND | 46 ± 28 | 39 ± 43 |
| mpc70Δ/mpc70Δ | 4 ± 12 | 42 ± 23 | ND | — |
| spo74Δ/spo74Δ | <1 ± 4 | 18 ± 16 | — | ND |
Fluorescent signals (mean ± standard deviation) were quantified as described in Materials and Methods.
ND, not determined.
—, no fluorescent signal at SPBs.
Analysis of recruitment of MOP components to SPBs in outer plaque mutants revealed various interdependencies of these proteins for assembly of the MOP. First, both Ady4p and Spo74p were absent from SPBs in cnm67Δ/cnm67Δ cells, providing additional evidence that these proteins are components of the outer plaque. Another functional relationship revealed by this analysis is that localization of Mpc70p and Spo74p to SPBs was mutually dependent; the deletion of one eliminated recruitment of the other to SPBs. A third set of interactions that emerged from this analysis is that loss of either Mpc54p, Mp70p, or Spo74p caused a decrease in the recruitment of the other three MOP components to SPBs. Ady4p was most affected by deletion of these components; the loss of any one of these proteins resulted in a ≥84% decrease in the amount of Ady4p present at SPBs. The amount of Mpc54p present at SPBs was also decreased significantly by the loss of either Mpc70p or Spo74p, and reciprocally, the loss of Mpc54p caused a significant decrease in the amount of Mpc70p and Spo74p at SPBs. In contrast, the loss of Ady4p had no discernible effect on the recruitment to SPBs of Mpc54p, Mpc70p, or Spo74p. These results indicate that the MOP is formed by the independent recruitment to the SPB of Ady4p and Mpc54p and the coordinated recruitment of Mpc70p and Spo74p and that the latter two recruitment events are required for MOP assembly.
DISCUSSION
During meiosis in S. cerevisiae, the outer plaque of the SPB becomes a site of de novo membrane synthesis. Expansion of the outer plaque via an alteration in its molecular composition creates a protein scaffold on which formation of the prospore membrane occurs. In this study we have identified two proteins, Ady4p and Spo74p, that are involved in the meiosis-specific modification of the outer plaque and formation of the prospore membrane.
Functions of Ady4p and Spo74p.
Our analysis of SPO74 reveals many similarities between the product of this gene and the meiosis-specific outer plaque components Mpc54p and Mpc70p. First, each of these proteins is localized to spindle poles during meiosis II, and the dependence of this localization on Cnm67p has been demonstrated for Mpc70p and Spo74p (1) (Table 7). Second, heterozygous loss of MPC54, MPC70, or SPO74 leads to accumulation of nonsister dyads (1, 40; M. E. Nickas and A. M. Neiman, unpublished data) (Fig. 4; Table 5). Third, homozygous loss of each of these genes prevents expansion of the outer plaque during meiosis and abolishes formation of the prospore membrane (1, 16) (Fig. 5 and 6). These findings suggest that Spo74p, like Mpc54p and Mpc70p, is a primary structural component of the MOP.
Our analysis of ADY4 demonstrates that it encodes a component of the MOP that is not essential for assembly of this structure. Ady4p is localized to SPBs during meiosis II in a Cnm67p-dependent manner, but expansion of outer plaques and formation of prospore membranes can occur in the absence of Ady4p (Fig. 1, 2, 5, and 7; Tables 6 and 7). Mutations of components involved in assembly of the MOP that incompletely block sporulation result in formation of nonsister dyads, which is thought to be a consequence of the failure to modify daughter SPBs (1, 40). Dyads formed by ady4Δ/ady4Δ cells, however, are random, indicating that the role of Ady4p in MOP structure and/or function is distinct from those of Mpc54p, Mpc70p, and Spo74p.
The formation of prospore membranes that fail to capture nuclei, observed in ady4Δ/ady4Δ cells, has also been described for cells that lack Cnm67p, another outer plaque component (1). Despite this similarity, however, the phenotypic defects in ady4Δ/ady4Δ and cnm67Δ/cnm67Δ cells are distinct. The majority of SPBs in ady4Δ/ady4Δ cells have recognizable MOPs that give rise to prospore membranes (Fig. 7 and Table 6). In contrast, the SPBs in cnm67Δ/cnm67Δ cells lack MOPs entirely, and no prospore membranes can be detected in the majority of these cells (1). When prospore membranes do form in cnm67Δ/cnm67Δ cells, they are synthesized in an Mpc70p-dependent manner in the cytoplasm, unassociated with nuclei (1). Thus, it has been proposed that prospore membranes in cnm67Δ/cnm67Δ cells are synthesized on MOP-like structures that are not bound to SPBs.
Another mutant in which prospore membranes fail to capture nuclei that may shed more light on the function of Ady4p is the spo20Δ mutant. As in ady4Δ/ady4Δ cells, prospore membranes in spo20Δ/spo20Δ cells are initially synthesized on MOPs associated with SPBs in early meiosis II but become detached from SPBs at later stages (23). ady4Δ/ady4Δ and spo20Δ/spo20Δ cells also display a similar distribution in the number of prospore membranes per cell (24) (Table 6). SPO20 encodes a sporulation-specific protein required for fusion of vesicles to promote growth of the prospore membrane (23). The similarity of ady4Δ/ady4Δ and spo20Δ/spo20Δ mutant phenotypes raises the possibility that Ady4p may play a role in vesicle fusion at the SPB during prospore membrane synthesis.
An alternate possibility for the function of Ady4p is that it preserves the structural integrity of the assembled MOP. A role for Ady4p in stabilizing the MOP is consistent with the defects in the morphology of MOPs and prospore membranes observed in ady4Δ/ady4Δ cells. The absence of Ady4p could result in a stochastic probability of MOP disassembly during meiosis II and produce the variable abnormalities observed in ady4Δ/ady4Δ cells. For example, MOPs that disassemble shortly after formation would not form prospore membranes at all, whereas MOPs that disassemble later in meiosis II would produce prospore membranes that become detached from SPBs and fail to capture nuclei. Those MOPs that remain intact in ady4Δ/ady4Δ cells could give rise to viable spores. Moreover, the observation that dyads formed in ady4Δ/ady4Δ cells are random is consistent with the idea that MOP disassembly occurs by chance. Further support for a role for Ady4p in promoting MOP stability comes from preliminary three-dimensional tomographic analysis of meiotic SPBs in ady4Δ/ady4Δ cells, which reveals ultrastructural defects in the MOP that are not readily apparent in standard thin-section transmission electron micrographs (C. Schwartz and A. M. Neiman, unpublished data).
Model of meiotic outer plaque assembly.
The results presented here and those of previous studies suggest that the four meiosis-specific components of the outer plaque are recruited to the SPB via the independent binding to Nud1p of Ady4p, Mpc54p, and Mpc70p. First, Ady4p, Mpc54p, and Mpc70p are localized to SPBs during meiosis II in the absence of either of the other two (16) (Table 7). Further support for this idea is the finding that Ady4p, Mpc54p, and Mpc70p interact with Nud1p in the two-hybrid assay (16) (Table 4). Finally, the observation that Mpc70p and Spo74p are mutually dependent for localization to SPBs suggests that recruitment of Spo74p is mediated by Mpc70p (Table 7). The failure of Mpc70p to localize to SPBs in the absence of Spo74p may indicate that Spo74p regulates the function of Mpc70p or forms a subcomplex with it prior to recruitment to the SPB.
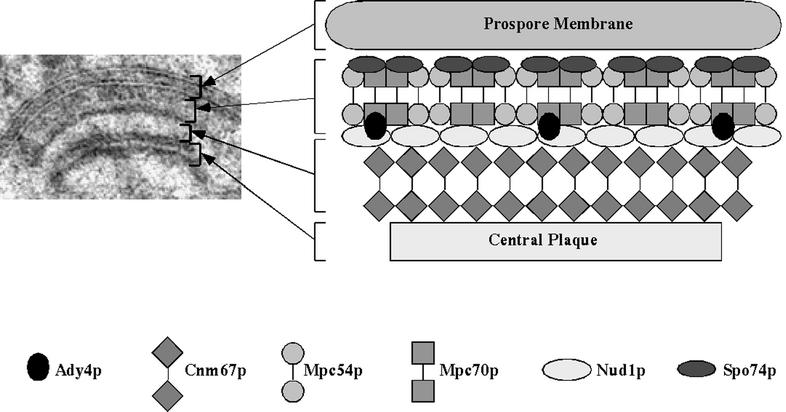
The findings presented here and elsewhere can be incorporated into a structural model of the MOP at the SPB (Fig. 8). Several observations suggest that Mpc54p, Mpc70p, and Spo74p assemble into a repeating unit at the SPB that forms the ordered matrix upon which prospore membrane synthesis occurs. First, the phenotypes described above caused by deletion of MPC54, MPC70, and SPO74 are similar. Second, each of these proteins interacts with itself and with the other two in the two-hybrid assay (16) (Fig. 3 and Table 4). Third, the loss of any one of these proteins leads to a decrease in the amount of the other two and of Ady4p present at SPBs (Table 7). In contrast, the phenotypes of ady4Δ/ady4Δ cells suggest that Ady4p is not a primary structural component of the ordered array that promotes synthesis of the prospore membrane. The observation that Ady4p interacts specifically with Mpc70p and Nud1p in the two-hybrid assay suggests that Ady4p may localize to the interface between the constitutive components of the outer plaque and the meiosis-specific matrix, consistent with a potential role for this protein in maintaining the stability of the MOP, as discussed above.
FIG. 8.
Model of the assembled MOP of the SPB. Six polypeptide subunits of the MOP are shown in ordered array between the central plaque and the prospore membrane. Arrows to the electron micrograph of the MOP on the left show layers that correspond to structures in the model.
Our model of MOP assembly gives rise to specific questions about the structure and function of this protein matrix. One important issue is the stoichiometry and orientation of Mpc54p, Mpc70p, and Spo74p in the basic structural unit of the MOP. In other regions of the SPB, coiled-coil proteins such as Spc42p and Cnm67p form tightly packed, organized arrays of parallel (i.e., head-to-head) subunits that create asymmetric layers (3, 31). Thus, a parallel arrangement seems likely for subunits of Mpc54p and Mpc70p within the meiosis-specific layers of the outer plaque. Another critical question is how the MOP promotes homotypic fusion of secretory vesicles to initiate prospore membrane synthesis. Formation of prospore membranes requires Mpc54p, Mpc70p, and Spo74p but can occur, albeit abnormally, in the absence of either Cnm67p or Ady4p (1, 16) (Fig. 5 to 7). Thus, oligomerization of Mpc54p, Mpc70p, and Spo74p during assembly of the MOP matrix may trigger vesicle fusion and drive prospore membrane formation. Finally, how does the MOP anchor the prospore membrane? Ady4p appears to facilitate anchoring of the membrane to the plaque, but neither Ady4p nor any of the other MOP components yet identified has a recognizable membrane-binding domain. Future studies will be focused on addressing these questions.
Biogenesis of membranes on protein scaffolds.
Insight into the molecular mechanism by which the MOP promotes synthesis of the prospore membrane in S. cerevisiae may come from the study of the biogenesis of other membranous organelles that are assembled on protein scaffolds. One such process is formation of cell walls in the syncytial endosperm of Arabidopsis thaliana. During endosperm development, cellularization of the multinucleate endosperm occurs via the de novo synthesis of cell walls between adjacent nuclei (6). These new cell walls are synthesized by the expansion of tubular membranes that are surrounded by a filamentous coat, which may act as a membrane-organizing center in a manner similar to that of the MOP during formation of the prospore membrane (27).
A better-characterized example of de novo membrane synthesis on a protein scaffold is assembly of the Golgi complex. Upon exit from mitosis, the Golgi complex assembles by concurrent formation of membrane cisternae and a matrix composed largely of coiled-coil proteins termed Golgins (17). Under conditions that disrupt transport of Golgi membranes, the Golgi matrix remains intact during interphase and is equally partitioned during mitosis (32, 33) It has thus been proposed that the Golgi matrix acts as a substratum that maintains the structural organization of the membrane cisternae. The Golgi matrix plays two roles in the formation of Golgi stacks that may be analogous to specific functions of the MOP in assembly of the prospore membrane. First, the proteins of the Golgi matrix and associated coiled-coil proteins act as tethers to dock precursor vesicles in proximity to one another, a requisite step for fusion of vesicles into cisternae (22, 35). Second, these proteins anchor Golgi membranes to the matrix to stabilize the cisternae in the stacked arrangement characteristic of this organelle (2, 34, 36).
Assembly of the MOP and synthesis of the prospore membrane may be similar in mechanism to formation of the Golgi complex. It has been proposed that vesicle precursors to the prospore membrane accumulate at the SPB prior to assembly of the MOP (21). Thus, oligomerizaton of the coiled-coil proteins Mpc54p and Mpc70p may trigger fusion of such precursor vesicles by bringing them into proximity in a Golgin-like manner. Subsequently, Mpc54p and Mpc70p may assume another Golgin-like role by anchoring the growing prospore membrane to the assembled MOP. A more precise understanding of the early events of assembly of the MOP and prospore membrane will reveal the extent to which the similarities between this process and biogenesis of the Golgi complex reflect common molecular mechanisms of de novo synthesis of membranes on protein scaffolds.
Acknowledgments
We thank Hideki Nakanishi for providing pRS424-G20 prior to publication; Hiroyuki Tachikawa, Elmar Schiebel, and Jeremy Thorner for providing strains and plasmids; J. Kahana and P. Silver for providing antibodies to GFP; Eileen O'Toole for technical assistance with electron microscopy; and Andrew Staehelin and members of the Neiman lab for helpful discussion.
This work was supported by National Institutes of Health grant GM62154 to A.M.N. and National Institutes of Health Biotechnology Resources grant RR00592 to J. R. McIntosh.
REFERENCES
- 1.Bajgier, B. K., M. Malzone, M. Nickas, and A. M. Neiman. 2001. SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol. Biol. Cell 12:1611-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr, F. A., M. Puype, J. Vandekerckhove, and G. Warren. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91:253-262. [DOI] [PubMed] [Google Scholar]
- 3.Bullitt, E., M. P. Rout, J. V. Kilmartin, and C. W. Akey. 1997. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell 89:1077-10786. [DOI] [PubMed] [Google Scholar]
- 4.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 5.Davidow, L. S., L. Goetsch, and B. Byers. 1980. Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics 94:581-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMason, D. A. 1997. Endosperm structure and development, p. 73-115. In B. A. Larkins and I. K. Vasil (ed.), Cellular and molecular biology of plant seed development. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Deng, C., and W. S. Saunders. 2001. ADY1, a novel gene required for prospore membrane formation at selected spindle poles in Saccharomyces cerevisiae. Mol. Biol. Cell 12:2646-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott, S., M. Knop, G. Schlenstedt, and E. Schiebel. 1999. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA 96:6205-6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esposito, M. S., R. E. Esposoto, and P. B. Moens. 1974. Genetic analysis of two spored asci produced by the spo3 mutant of Saccharomyces. Mol. Gen. Genet. 135:91-95. [DOI] [PubMed] [Google Scholar]
- 10.Esposito, R. E., and S. Klapholz. 1981. Meiosis and ascosore development, p. 211-287. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Giddings, T. H. J., E. T. O'Toole, M. Morphew, D. N. Mastronarde, J. R. McIntosh, and M. Winey. 2001. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 67:27-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inouye, C., N. Dhillon, T. Durfee, P. C. Zambryski, and J. Thorner. 1997. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein-protein interactions. Genetics 147:479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishihara, S., A. Hirata, M. Minemura, S. Nogami, and Y. Ohya. 2001. A mutation in SPC42, which encodes a component of the spindle pole body, results in production of two-spored asci in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:585-595. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knop, M., and E. Schiebel. 1998. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 17:3952-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knop, M., and K. Strasser. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19:3657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linstedt, A. D. 1999. Stacking the cisternae. Curr. Biol. 9:893-896. [DOI] [PubMed] [Google Scholar]
- 18.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 19.Lynn, R. R., and P. T. Magee. 1970. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moens, P. B., and E. Rapport. 1971. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol. 50:344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko, and M. Knop. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20:6946-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura, N., M. Lowe, T. P. Levine, C. Rabouille, and G. Warren. 1997. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell 89:445-455. [DOI] [PubMed] [Google Scholar]
- 23.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neiman, A. M., L. Katz, and P. J. Brennwald. 2000. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155:1643-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickas, M. E., and A. M. Neiman. 2002. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics 160:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto, S., and T. Iino. 1981. Selective abortion of two nonsister nuclei in a developing ascus of the hfd-1 mutant in Saccharomyces cerevisiae. Genetics 99:197-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otegui, M., and L. A. Staehelin. 2000. Syncytial-type cell plates: a novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12:933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 29.Rabitsch, K. P., A. Toth, M. Galova, A. Schleiffer, G. Schaffner, E. Aigner, C. Rupp, A. M. Penkner, A. C. Moreno-Borchart, M. Primig, R. E. Esposito, F. Klein, M. Knop, and K. Nasmyth. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11:1001-1009. [DOI] [PubMed] [Google Scholar]
- 30.Rose, M. D., F. Winston, and P. Hieter (ed.). 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schaerer, F., G. Morgan, M. Winey, and P. Philippsen. 2001. Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol. Biol. Cell 12:2519-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seemann, J., E. Jokitalo, M. Pypaert, and G. Warren. 2000. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407:1022-1026. [DOI] [PubMed] [Google Scholar]
- 33.Seemann, J., M. Pypaert, T. Taguchi, J. Malsam, and G. Warren. 2002. Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science 295:848-851. [DOI] [PubMed] [Google Scholar]
- 34.Short, B., C. Preisinger, R. Korner, R. Kopajtich, O. Byron, and F. A. Barr. 2001. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shorter, J., M. B. Beard, J. Seemann, A. B. Dirac-Svejstrup, and G. Warren. 2002. Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J. Cell Biol. 157:45-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shorter, J., R. Watson, M. E. Giannakou, M. Clarke, G. Warren, and F. A. Barr. 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18:4949-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Straight, P. D., T. H. Giddings, Jr., and M. Winey. 2000. Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell 11:3525-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tu, J., W. Song, and M. Carlson. 1996. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4199-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 40.Wesp, A., S. Prinz, and G. R. Fink. 2001. Conservative duplication of spindle poles during meiosis in Saccharomyces cerevisiae. J. Bacteriol. 183:2372-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wigge, P. A., O. N. Jensen, S. Holmes, S. Soues, M. Mann, and J. V. Kilmartin. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng, X., J. A. Kahana, P. A. Silver, M. K. Morphew, J. R. McIntosh, I. T. Fitch, J. Carbon, and W. S. Saunders. 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146:415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]