Abstract
Ycf1p, a member of the yeast multidrug resistance-associated protein (MRP) subfamily of ATP-binding cassette proteins, is a vacuolar membrane transporter that confers resistance to a variety of toxic substances such as cadmium and arsenite. Ycf1p undergoes a PEP4-dependent processing event to yield N- and C-terminal cleavage products that remain associated with one another. In the present study, we sought to determine whether proteolytic cleavage is required for Ycf1p activity. We have identified a unique region within lumenal loop 6 of Ycf1p, designated the loop 6 insertion (L6ins), which appears to be necessary and sufficient for proteolytic cleavage, since L6ins can promote processing when moved to new locations in Ycf1p or into a related transporter, Bpt1p. Surprisingly, mutational results indicate that proteolytic processing is not essential for Ycf1p transport activity. Instead, the L6ins appears to regulate substrate specificity of Ycf1p, since certain mutations in this region lower cellular cadmium resistance with a concomitant gain in arsenite resistance. Although some of these L6ins mutations block processing, there is no correlation between processing and substrate specificity. The activity profiles of the Ycf1p L6ins mutants are dramatically affected by the strain background in which they are expressed, raising the possibility that another cellular component may functionally impact Ycf1p activity. A candidate component may be a new full-length MRP-type transporter (NFT1), reported in the Saccharomyces Genome Database as two adjacent open reading frames, YKR103w and YKR104w, but which we show here is present in most Saccharomyces strains as a single open reading frame.
Many proteins are initially synthesized as precursors that undergo proteolytic processing events to generate their mature form. Such proteins include the vacuolar proteases and mating pheromones in Saccharomyces cerevisiae, which require proteolytic cleavage to generate biologically active molecules (22, 41). Posttranslational processing can also cleave certain transcription factors, such as sterol regulatory element-binding protein (SREBP) or Notch, from a membrane tether so that they can function in the nucleus (8). In other instances, the role of processing is less clear. Presenilin, a component of the γ secretase required for the proteolytic processing of the amyloid precursor protein, is a multispanning membrane protein that itself is proteolytically cleaved into two fragments (50, 51). Although both the N- and C-terminal cleavage products of presenilin have been shown to be necessary for activity, it is uncertain whether processing is required for activity (27, 34).
Ycf1p, a member of the yeast ATP-binding cassette (ABC) transporter superfamily, also undergoes proteolytic processing and is the only known example of an ABC transporter that is cleaved (31, 47). Ycf1p localizes to the vacuolar membrane, where it functions as a glutathione conjugate transporter to detoxify the cell of a variety of compounds, including the heavy metals cadmium and arsenite (16, 28, 29, 43, 47). We have previously shown that the cleavage of Ycf1p depends on the master vacuolar protease, Pep4p, and therefore is likely to occur upon arrival of Ycf1p at the vacuolar membrane (31). Ycf1p is comprised of a typical ABC core region (MSD1, NBD1, L1, MSD2, and NBD2) and an additional N-terminal extension (MSD0 and L0) that is a diagnostic feature of certain members of the multidrug resistance-associated protein (MRP) subfamily to which Ycf1p belongs (see Fig. 2A). The posttranslational cleavage of Ycf1p liberates two fragments, one containing the N-terminal extension plus a small segment of the C-terminal core, and the other containing the remainder of the C-terminal core (31, 49). It does not appear that the fragments created by posttranslational processing represent inactive degradation products since the cleavage products interact and both contain regions that have been shown to be critical for Ycf1p function (12, 13, 31).
FIG. 2.
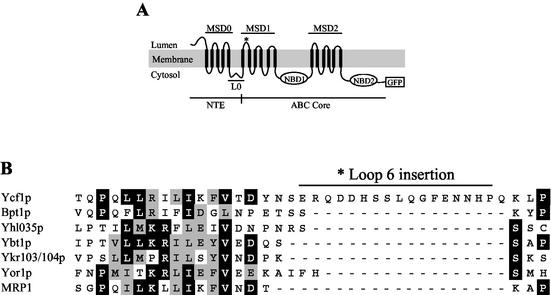
Among the yeast MRPs, Ycf1p contains a unique insertion within loop 6 of MSD1. (A) The proposed topology of Ycf1p is shown. The N-terminal extension (NTE) contains a membrane-spanning domain (MSD0) and a cytosolic linker region (L0). The ABC core region contains two membrane-spanning domains (MSD1 and MSD2) and two nucleotide-binding domains (NBD1 and NBD2), and is separated by a linker (L1) between NBD1 and MSD2 (for simplicity sake, L1 is omitted in this and subsequent figures). The proposed cleavage site within loop 6 is indicated by an asterisk. The GFP coding sequence, fused immediately before the stop codon of Ycf1p, is indicated. (B) The lumenally disposed loop 6 (L6; residues 303 to 341) of Ycf1p was aligned to the five other yeast MRP subfamily members (Bpt1p, Yhl035p, Ybt1p, Ykr103/104p, and Yor1p) and its closest human homologue, MRP1, by using CLUSTALW software (46). Black boxes represent amino acid identity, and gray boxes indicate amino acid similarity as determined with the Boxshade server (www.ch.embnet.org/software/box_form.html). The unique L6 insertion in Ycf1p corresponds to residues 321 to 337 and is absent in all of the other proteins as indicated by the gap in this region.
In the present study we identify a 17-amino-acid insertion in loop 6 of MSD1, designated the loop 6 insertion (L6ins), and show that this region is necessary and sufficient to promote proteolytic cleavage. A major goal of the present study was to determine whether the proteolytic cleavage of Ycf1p modulates transport function. Notably, mutational analysis of L6ins indicates that the activity of Ycf1p is independent of its posttranslational processing status. Instead, we found that mutations within L6ins can affect Ycf1p substrate specificity, even though the L6ins lies on the lumenal face of the vacuolar membrane where the substrate is released after its transport. Surprisingly, we detected dramatic differences in resistance to toxic substances of two different laboratory strains both bearing the same L6ins mutation. These data suggest that an additional component present in some strains, but absent in others, may be important for Ycf1p activity. We report here the discovery of a new full-length yeast MRP-type transporter gene (NFT1) comprised of two adjacent open reading frames (ORFs), YKR103w and YKR104w, that represents a candidate gene which may account for the observed strain differences.
MATERIALS AND METHODS
Yeast strains, media, and growth conditions.
Yeast strains used in the present study are listed in Table 1. The strains containing chromosomal forms of wild-type and mutant ycf1-GFP were generated by the standard two-step integration/excision method. Each of the integrating plasmids (pSM1817, pSM1826, pSM1853, pSM1859, pSM1860, pSM1863, pSM1864, and pSM1865) was digested with AvrII and then transformed into SM3851, a ycf1Δ derivative of strain SM1058, and SM4719, a ycf1Δ::KanMX4 derivative of strain BY4741. Transformants for each strain were selected on SC-Ura plates. Excisants, leaving behind wild-type and mutant gene replacements, were selected on 5-fluoroorotic acid and confirmed by Southern blot analysis and PCR. The expression of wild-type and mutant forms of Ycf1p-green fluorescent protein (GFP) in each of the strains was confirmed by Western blot analysis (see Fig. 4B and C). The resulting strains are listed as integrants in the strain table (Table 1). All yeast transformations were performed as described previously (11).
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| BY4741 | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 | 7a |
| F744 | MATa | Σ1278b derivative from G. Fink via J. Boeke |
| MS10 | MATaura3-52 leu2-3,112 ade2-101 | M. Rose |
| MS1907 | MATaura3-52 leu2-3,112 ade2-101 pep4::LEU2 | M. Rose |
| NKY276 | MATalys2 ura3 ho::LYS2 | SK1 derivative from N. Kleckner via J. Boeke |
| SM1058 | MATatrp1 leu2 ura3 his4 can1 | 32 |
| SM3794 | MATatrp1 leu2 ura3 his4 can1 bpt1Δ::KanMX4 | 38 |
| SM3851 | MATatrp1 leu2 ura3 his4 can1 ycf1Δ | 38 |
| SM4516 | SM3851 [2μ URA3] | Transformant of SM3851 with pSM217 |
| SM4517 | SM3851 [2μ URA3 YCF1] | Transformant of SM3851 with pSM1752 |
| SM4518 | SM3851 [2μ URA3 YCF1-GFP] | Transformant of SM3851 with pSM1753 |
| SM4522 | SM3794 [2μ URA3 BPT1-GFP] | Transformant of SM3794 with pSM1490 |
| SM4523 | MS10 [2μ URA3 YCF1-GFP] | Transformant of MS10 with pSM1753 |
| SM4524 | MS1907 [2μ URA3 YCF1-GFP] | Transformant of MS1907 with pSM1753 |
| SM4527 | MS10 [2μ URA3] | Transformant of MS10 with pSM217 |
| SM4528 | MS1907 [2μ URA3] | Transformant of MS1907 with pSM217 |
| SM4539 | MS10 [CEN URA3 YCF1-GFP] | Transformant of MS10 with pSM1762 |
| SM4540 | MS1907 [CEN URA3 YCF1-GFP] | Transformant of MS1907 with pSM1762 |
| SM4543 | SM3851 [2μ URA3 ycf1(ΔL6)-GFP] | Transformant of SM3851 with pSM1775 |
| SM4590 | SM3794 [2μ URA3 BPT1::L6-GFP] | Transformant of SM3794 with pSM1784 |
| SM4643b,c | MATatrp1 leu2 ura3 his4 can1 YCF1-GFP | Integrant in SM3851 using pSM1817 |
| SM4644b,c | MATatrp1 leu2 ura3 his4 can1 ycf1(ΔL6)-GFP | Integrant in SM3851 using pSM1826 |
| SM4648 | SM3851 [2μ URA3 ycf1(ΔL6, L12::L6)-GFP] | Transformant of SM3851 with pSM1832 |
| SM4656 | SM3851 [2μ URA3 ycf1(ΔL6-1)-GFP] | Transformant of SM3851 with pSM1833 |
| SM4657 | SM3851 [2μ URA3 ycf1(ΔL6-2)-GFP] | Transformant of SM3851 with pSM1834 |
| SM4658 | SM3851 [2μ URA3 ycf1(ΔL6-3)-GFP] | Transformant of SM3851 with pSM1835 |
| SM4672 | SM3851 [2μ URA3 ycf1(L6-2→A)-GFP] | Transformant of SM3851 with pSM1850 |
| SM4678 | SM3851 [2μ URA3 ycf1(L6-2→L6-1)-GFP] | Transformant of SM3851 with pSM1851 |
| SM4680 | SM3851 [2μ URA3 ycf1(L6-2→E)-GFP] | Transformant of SM3851 with pSM1854 |
| SM4697b,c | MATatrp1 leu2 ura3 his4 can1 ycf1(L6-2→L6-1)-GFP | Integrant in SM3851 using pSM1853 |
| SM4717b,c | MATatrp1 leu2 ura3 his4 can1 ycf1(L6-2→A)-GFP | Integrant in SM3851 using pSM1859 |
| SM4718b,c | MATatrp1 leu2 ura3 his4 can1 ycf1(L6-2→E)-GFP | Integrant in SM3851 using pSM1860 |
| SM4719a | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 ycf1::KanMX4 | Research Genetics (Huntsville, Ala.) |
| SM4720b,d | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 YCF1-GFP | Integrant in SM4719 using pSM1817 |
| SM4721b,d | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 ycf1(ΔL6)-GFP | Integrant in SM4719 using pSM1826 |
| SM4722b,d | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 ycf1(L6-2→L6-1)-GFP | Integrant in SM4719 using pSM1853 |
| SM4723b,d | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 ycf1(L6-2→A)-GFP | Integrant in SM4719 using pSM1859 |
| SM4724b,d | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 ycf1(L6-2→E)-GFP | Integrant in SM4719 using pSM1860 |
| SM4726b,d | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 ycf1(ΔL6-1)-GFP | Integrant in SM4719 using pSM1863 |
| SM4727b,d | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 ycf1(ΔL6-2)-GFP | Integrant in SM4719 using pSM1864 |
| SM4728b,d | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 ycf1(ΔL6-3)-GFP | Integrant in SM4719 using pSM1865 |
| SM4729b,c | MATatrp1 leu2 ura3 his4 can1 ycf1(ΔL6-1)-GFP | Integrant in SM3851 using pSM1863 |
| SM4730b,c | MATatrp1 leu2 ura3 his4 can1 ycf1(ΔL6-2)-GFP | Integrant in SM3851 using pSM1864 |
| SM4731b,c | MATatrp1 leu2 ura3 his4 can1 ycf1(ΔL6-3)-GFP | Integrant in SM3851 using pSM1865 |
| SM4757 | SM3851 [2μ URA3 ycf1(ΔL6, L12::L6-2)-GFP] | Transformant of SM3851 with pSM1876 |
A derivative of the strain BY4741, provided by Research Genetics via J. Boeke (Johns Hopkins University School of Medicine, Baltimore, Md.).
Strains constructed by the standard two-step integration-excision method by using the designated plasmid as described in Materials and Methods.
Derived from SM3851, a derivative of the strain SM1058.
Derived from SM4719, a derivative of the strain BY4741.
FIG. 4.
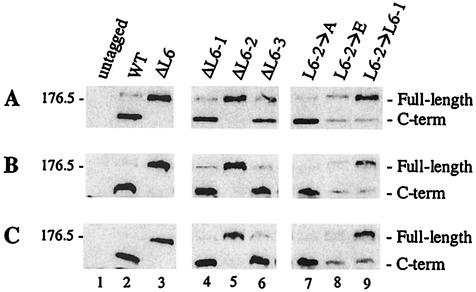
Mutations within the L6ins have variable effects on Ycf1p processing. The steady-state protein level of wild-type and mutant Ycf1p-GFP expressed from multicopy plasmids in SM1058 (A) or from the chromosome in the SM1058 (B) and BY4741 (C) strain backgrounds was determined by immunoblot analysis as in Fig. 1A, except that panels B and C contain 0.8 OD600 cell equivalents per lane. Ycf1p-GFP was detected by using either mouse anti-GFP monoclonal antibodies (A) or rabbit anti-GFP polyclonal antibodies (B and C). The sequence for each mutant is shown in Table 3. The strains used in lanes 1 to 9 were SM4517, SM4518, SM4543, SM4656, SM4657, SM4658, SM4672, SM4680, and SM4678 (A); SM1058, SM4643, SM4644, SM4729, SM4730, SM4731, SM4717, SM4718, and SM4697 (B); and BY4741, SM4720, SM4721, SM4726, SM4727, SM4728, SM4723, SM4724, and SM4722 (C), respectively.
Plate and liquid drop-out media were prepared as previously described (32). The plates used for the spot tests were prepared by adding the indicated concentrations of CdSO4 or AsNaO2 (Sigma, St. Louis, Mo.) to the minimal plate medium immediately prior to pouring the plates. A fresh batch of cadmium plates was made for each experiment, since toxicity tended to vary somewhat from batch to batch and to decrease with age of the plates.
To examine growth inhibition by toxic compounds, cells were grown overnight to saturation in minimal medium and then subcultured at a 1:500 dilution in minimal medium and grown overnight at 30°C to an optical density at 600 nm (OD600) of ca. 1.0. This overnight culture was diluted to an OD600 of 0.1, which in turn was diluted in 10-fold increments. Aliquots (5 μl) of each 10-fold dilution were spotted onto SC-Ura or SC plates containing the indicated concentration of CdSO4 or AsNaO2 and incubated for 4 to 6 days at 30°C.
Plasmid constructions and DNA sequence analysis.
Plasmids used in the present study are listed in Table 2. To analyze single copy (CEN) Ycf1p with GFP fused to the C terminus, we constructed the plasmid pSM1761 (CEN LEU2 YCF1-GFP) by recombinational cloning (33). A PvuI restriction fragment from pSM1753 (2μ URA3 YCF1-GFP) (38) was recombined into BamHI-XhoI-digested pRS315 (CEN LEU2) (39). A URA3 version of pSM1761 was constructed by recombinational cloning of a PvuI restriction fragment from pSM1761 with BamHI-XhoI-digested pRS316 (CEN URA3) (39), yielding pSM1762 (CEN URA3 YCF1-GFP).
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant genotypea | Reference or source |
|---|---|---|
| pJAW50 | 2μ TRP1 YCF1 | 48 |
| pJAW53 | YIp 5′1018nt-YCF1-hisG-URA3-hisG-3′1356nt-YCF1 | 48 |
| pRS306 | YIp URA3 | 39 |
| pRS315 | CEN LEU2 | 39 |
| pRS316 | CEN URA3 | 39 |
| pSM217 | 2μ URA3 | 10 |
| pSM1490 | 2μ URA3 BPT1-GFP | 38 |
| pSM1752 | 2μ URA3 YCF1 | 38 |
| pSM1753 | 2μ URA3 YCF1-GFP | 38 |
| pSM1761 | CEN LEU2 YCF1-GFP | This study |
| pSM1762 | CEN URA3 YCF1-GFP | This study |
| pSM1775 | 2μ URA3 ycf1(ΔL6)-GFP | This study |
| pSM1783 | 2μ URA3 BPT1::AgeI-GFP | This study |
| pSM1784 | 2μ URA3 BPT1::L6-GFP | This study |
| pSM1785 | 2μ URA3 YCF1-GFP + (3′ region) | This study |
| pSM1817 | YIp URA3 YCF1-GFP | This study |
| pSM1825 | 2μ URA3 ycf1(ΔL6)-GFP + (3′ region) | This study |
| pSM1826 | YIp URA3 ycf1(ΔL6)-GFP | This study |
| pSM1831 | 2μ URA3 ycf1(ΔL6, L12::NotI)-GFP | This study |
| pSM1832 | 2μ URA3 ycf1(ΔL6, L12::L6)-GFP | This study |
| pSM1833 | 2μ URA3 ycf1(ΔL6-1)-GFP | This study |
| pSM1834 | 2μ URA3 ycf1(ΔL6-2)-GFP | This study |
| pSM1835 | 2μ URA3 ycf1(ΔL6-3)-GFP | This study |
| pSM1850 | 2μ URA3 ycf1(L6-2→A)-GFP | This study |
| pSM1851 | 2μ URA3 ycf1(L6-2→L6-1)-GFP | This study |
| pSM1853 | YIp URA3 ycf1(L6-2→L6-1)-GFP | This study |
| pSM1854 | 2μ URA3 ycf1(L6-2→E)-GFP | This study |
| pSM1859 | YIp URA3 ycf1(L6-2→A)-GFP | This study |
| pSM1860 | YIp URA3 ycf1(L6-2→E)-GFP | This study |
| pSM1863 | YIp URA3 ycf1(ΔL6-1)-GFP | This study |
| pSM1864 | YIp URA3 ycf1(ΔL6-2)-GFP | This study |
| pSM1865 | YIp URA3 ycf1(ΔL6-3)-GFP | This study |
| pSM1876 | 2μ URA3 ycf1(ΔL6, L12::L6-2)-GFP | This study |
YIp denotes yeast integrating plasmid.
To create mutations within the Ycf1p processing site (Table 3), the region within L6 between amino acids 321 and 337 was deleted and mutated as follows. The entire region was deleted by recombinational cloning of a YCF1 PCR product, generated from an oligonucleotide containing an engineered deletion of amino acids 321 to 337 (designated ΔL6), into AgeI-digested pSM1753, thereby yielding pSM1775 (2μ URA3 ycf1ΔL6-GFP). Three smaller deletions were created within this region by recombinational cloning of various YCF1 PCR products containing engineered deletions of amino acids 321 to 326 (ΔL6-1), 327 to 332 (ΔL6-2), or 333 to 337 (ΔL6-3) into AgeI-digested pSM1775 (for the ΔL6-1 mutant) or into AgeI-digested pSM1753 (for the ΔL6-2 and ΔL6-3 mutants). The resulting plasmids were pSM1833 (2μ URA3 ycf1ΔL6-1-GFP), pSM1834 (2μ URA3 ycf1ΔL6-2-GFP), and pSM1835 (2μ URA3 ycf1ΔL6-3-GFP).
TABLE 3.
Summary of sequences and phenotypes of Ycf1p variants in SM1058 strain background
| Row | L6ins sequencea
|
Ycf1p allele | Phenotypec
|
||||
|---|---|---|---|---|---|---|---|
| L6-1 | L6-2 | L6-3 | Processingb | Cdr | Asr | ||
| 1 | ERQDDH | SSLQGF | ENNHP | WT | + | + | + |
| 2 | ERQDDH | SSLQGF | ENNHP | WT (+GFP) | + | + | − |
| 3 | ------ | ------ | ----- | ΔL6 | − | + | − |
| 4 | ------ | SSLQGF | ENNHP | ΔL6-1 | + | + | − |
| 5 | ERQDDH | ------ | ENNHP | ΔL6-2 | − | + | − |
| 6 | ERQDDH | SSLQGF | ----- | ΔL6-3 | + | +/− | + |
| 7 | ERQDDH | AAAAAA | ENNHP | L6-2→A | + | +/− | + |
| 8 | ERQDDH | EEEEEE | ENNHP | L6-2→E | +/− | +/− | + |
| 9 | ERQDDH | ERQDDH | ENNHP | L6-2→L6-1 | −/+ | +/− | + |
The sequences correspond to mutations within the L6ins of Ycf1p (amino acids 321 to 337). Deletions are indicated by a dash (-).
The processing phenotypes (scored as +, +/−, −/+, and −) are based on the immunoblot analysis in Fig. 4.
The cadmium (Cdr) and arsenite (Asr) resistance phenotypes reflect the results shown in Fig. 5.
The entire L6-2 region from amino acids 327 to 332 was replaced with various residues as shown in Table 3 and as described below. The L6-2 region was replaced with six alanines or with a duplication of the amino acids in the L6-1 region (ERQDDH) by recombinational cloning of YCF1 PCR products containing the engineered mutations into AgeI-digested pSM1753. The resulting plasmids were pSM1850 (2μ URA3 ycf1L6-2→A-GFP) or pSM1851 (2μ URA3 ycf1L6-2→L6-1-GFP). The L6-2 region was also replaced with six glutamates by recombinational cloning of a YCF1 PCR product containing the engineered mutation into AgeI-digested pSM1850, thereby yielding pSM1854 (2μ URA3 ycf1L6-2→E-GFP).
To move the region required for Ycf1p cleavage from loop 6 to loop 12 of Ycf1p, first a NotI site was inserted after amino acid 984 in pSM1775 to yield pSM1831 (2μ URA3 ycf1ΔL6, L12::NotI-GFP). Next, the region from amino acids 321 to 337 was amplified from pJAW50 (2μ TRP1 YCF1) (48) and cloned into NotI-digested pSM1831 by homologous recombination, generating pSM1832 (2μ URA3 ycf1ΔL6, L12::L6-GFP). In the resulting plasmid, the region required for Ycf1p cleavage is deleted from its original location and transferred to the region between amino acids 984 and 985. A smaller region of the cleavage site (L6-2; residues 327 to 332) was also transferred to loop 12 by recombinational cloning to generate pSM1876 (2μ URA3 ycf1ΔL6, L12::L6-2-GFP).
The region required for Ycf1p cleavage site was also transferred to the analogous region in loop 6 of BPT1. First, an AgeI restriction site was inserted after amino acid 325 in pSM1490 (2μ URA3 BPT1-GFP) (38) to yield pSM1783 (2μ URA3 BPT1::AgeI-GFP). Next, the region from amino acids 321 to 337 in YCF1 was amplified from pJAW50 and cloned into AgeI-digested pSM1783 by homologous recombination, generating pSM1784 (2μ URA3 BPT1::L6-GFP). In the resulting plasmid, the potential Ycf1p cleavage site has been transferred to the region between amino acids 325 and 326 in Bpt1p.
To analyze chromosomal ycf1 mutants, we constructed yeast integrating plasmids (YIp) to use for generating two-step gene replacements. First, the 1,092-bp region downstream of the YCF1 ORF was amplified from pJAW53 (48) and cloned into NaeI-digested pSM1753 by recombinational cloning, generating pSM1785 (2μ URA3 YCF1-GFP + 3′ region), which is identical to pSM1753 except for the added 3′ downstream region. Next, a KpnI-BamHI fragment from pSM1785, containing YCF1-GFP, was subcloned into the same sites of pRS306 (YIp URA3) (39), yielding pSM1817 (YIp URA3 YCF1-GFP). The plasmid pSM1825 (2μ URA3 ycf1ΔL6-GFP + 3′ region) was generated by subcloning an AgeI-KpnI fragment from pSM1775 into the same sites of pSM1785. As described above, a KpnI-BamHI fragment from pSM1825, containing ycf1ΔL6-GFP, was subcloned into the same sites of pRS306, yielding pSM1826 (YIp URA3 ycf1ΔL6-GFP). Each of the smaller loop 6 deletions (ΔL6-1, ΔL6-2, and ΔL6-3) and the L6-2 mutations (L6-2→A, L6-2→L6-1, and L6-2→E) were also subcloned into integrating plasmids. AgeI-KpnI fragments from pSM1833, pSM1834, pSM1835, pSM1850, pSM1851, and pSM1854 were subcloned into the same sites of pSM1817 to generate pSM1863 (YIp URA3 ycf1ΔL6-1-GFP), pSM1864 (YIp URA3 ycf1ΔL6-2-GFP), pSM1865 (YIp URA3 ycf1ΔL6-3-GFP), pSM1859 (YIp URA3 ycf1L6-2→A-GFP), pSM1853 (YIp URA3 ycf1L6-2→L6-1-GFP), and pSM1860 (YIp URA3 ycf1L6-2→E-GFP), respectively.
All of the deletions and mutations described above were confirmed by DNA sequence analysis. Further details of the plasmid constructions have been described (30) and will be furnished upon request.
To sequence the junction of the ORFs YKR103w and YKR104w, a 626-bp region spanning the two ORFs was amplified by PCR from genomic DNA (colony PCR or purified genomic DNA) with 18mer oligonucleotide primers (24). The PCR products were sequenced by automated DNA sequencing by using the 5′ primer. Sequences of YKR103w/104w and related sequences in other Saccharomyces species were obtained from the Saccharomyces Genome Database (SGD) website (http://genome-www.stanford.edu/Saccharomyces/).
Antibodies.
The rabbit anti-GFP polyclonal antibody was a gift from R. Jensen (Johns Hopkins University School of Medicine, Baltimore, Md.). The mouse anti-GFP monoclonal antibody was purchased from Clontech (Palo Alto, Calif.). The horseradish peroxidase-conjugated secondary antibodies (donkey anti-rabbit immunoglobulin and sheep anti-mouse immunoglobulin) used for immunoblotting were purchased from Amersham Pharmacia Biotech (Piscataway, N.J.).
Immunoblot analysis.
Cell extracts and immunoblots used to detect Ycf1p were prepared as previously described (15) except that samples were either heated at 65°C for 10 min prior to electrophoresis (Fig. 1A and 3B) or not heated at all, in order to minimize aggregation (Fig. 3A and 4). The primary antibodies used were rabbit anti-GFP (1:5,000) or mouse anti-GFP (1:670).
FIG. 1.
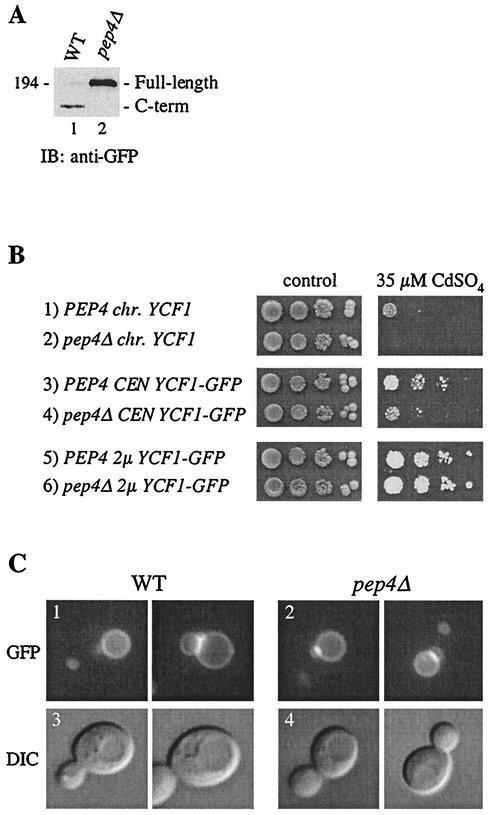
Ycf1p processing and resistance of cells to cadmium is impaired when PEP4 is deleted. (A) The steady-state level of Ycf1p-GFP in wild-type (WT) and pep4Δ strains was examined by immunoblot analysis. Crude yeast cell extracts (0.4 OD600 cell equivalents per lane) were resolved by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis and transferred to nitrocellulose. Ycf1p-GFP was detected by using rabbit anti-GFP polyclonal antibodies. Unprocessed Ycf1p-GFP is indicated as full-length and the C-terminal cleavage product as C-term. A molecular weight marker is indicated. The strains used were SM4523 (lane 1) and SM4524 (lane 2). (B) Cadmium resistance was tested by spotting an aliquot (5 μl) of cells at 0.1 OD600, and serial 10-fold dilutions thereof, onto plates containing no CdSO4 (control) or 35 μM CdSO4, followed by incubation at 30°C for 2 or 5 days, respectively. The strains used in rows 1 to 6 are SM4527, SM4528, SM4539, SM4540, SM4523, and SM4524, respectively. (C) Vacuolar membrane localization of Ycf1p-GFP in wild-type (1 and 3) and pep4Δ (2 and 4) cells by fluorescence microscopy. In the differential interference contrast (DIC) images (panels 3 and 4), the vacuole appears as an indentation. In the corresponding GFP panels (panels 1 and 2), the bright areas of fluorescence correspond to a region where two vacuolar lobes intersect. The strains used were SM4523 (wild type) and SM4524 (pep4Δ).
FIG. 3.
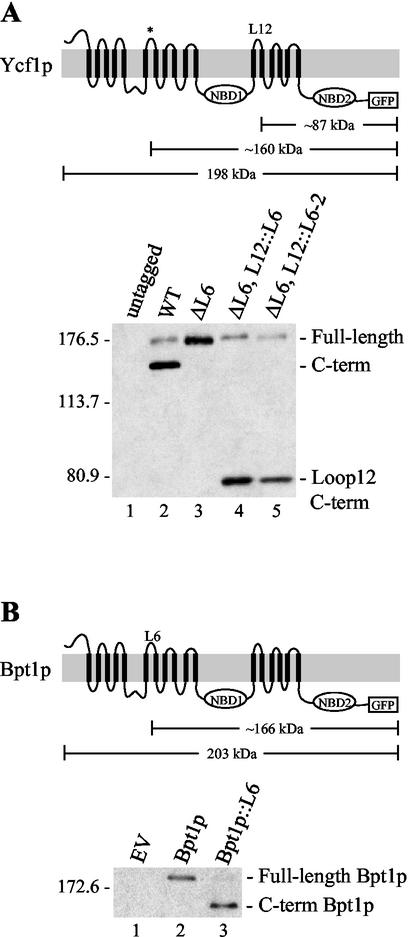
The L6ins is necessary and sufficient for processing within loops 6 and 12 of Ycf1p and within loop 6 of Bpt1p. Schematics (top parts of panels) are shown for Ycf1p (A) and Bpt1p (B). The location of the proposed native cleavage site (*) in Ycf1p and the engineered cleavage sites in Ycf1p L12 and Bpt1p L6 are indicated, as well as the predicted molecular weight of the C-terminal fragments resulting from cleavage in these regions. Immunoblots show the steady-state level of wild-type and mutant Ycf1p-GFP (A) and Bpt1p-GFP (B). Ycf1p-GFP and Bpt1p-GFP were detected by using anti-GFP monoclonal and polyclonal antibodies, respectively. Unprocessed (full-length) and C-terminal cleavage products (C-term) are indicated. Panel A, lanes 1 to 5: SM4517, SM4518, SM4543, SM4648, and SM4757, respectively. Panel B, lanes 1 to 3: SM4516, SM4522, and SM4590, respectively. Abbreviations: WT, wild type; ΔL6ins, L6 insertion deleted (amino acids 321 to 337); ΔL6ins, L12::L6ins (L6ins deleted and amino acids 321 to 337 transferred to L12 of Ycf1p between amino acids 984 and 985); ΔL6ins, L12::L6-2 (L6ins deleted and amino acids 327 to 332 transferred to L12 of Ycf1p between amino acids 984 and 985); EV (empty vector); Bpt1p::L6, Ycf1p amino acids 321 to 337 inserted into Bpt1p between amino acids 325 and 326.
Fluorescence microscopy.
To examine the localization of Ycf1p-GFP, cells were grown overnight to saturation in minimal medium and then subcultured at a 1:1,000 dilution in minimal medium and grown overnight at 30°C to an OD600 of ca. 0.7. Log-phase cells were examined at ×100 magnification on poly-lysine-coated slides by using a Zeiss Axioskop microscope equipped with fluorescence and Nomarski optics (Zeiss, Thornwood, N.Y.). Images were captured with a Cooke charge-coupled device camera and IP Lab spectrum software (Biovision Technologies, Inc., Exton, Pa.).
RESULTS
Is proteolytic processing of Ycf1p required for its ability to confer resistance to cadmium?
Ycf1p is proteolytically processed in a PEP4-dependent manner when it reaches the vacuolar membrane (31) (Fig. 1A). Our previous studies indicated that both the N- and the C-terminal cleavage products contain regions that contribute to the biological function of Ycf1p (31). However, the functional significance of the cleavage event per se has not been determined. Here, we examined whether Ycf1p requires proteolytic processing for its activation, or instead, if unprocessed Ycf1p remains functional.
We compared Ycf1p-dependent cadmium resistance in wild-type (PEP4) and pep4Δ strains (Fig. 1B). Because the latter is defective for Ycf1p processing, the ability of full-length unprocessed Ycf1p to confer resistance to cadmium could potentially be assessed in this mutant. Chromosomally expressed Ycf1p confers a low level of resistance to cadmium in a wild-type strain but not in a pep4Δ mutant (Fig. 1B, rows 1 and 2). An analogous difference in cadmium resistance between the wild-type and pep4Δ strains is apparent for strains expressing Ycf1p-GFP from a low-copy (CEN) plasmid (Fig. 1B, rows 3 and 4), although vacuolar localization was similar in both strains (Fig. 1C). These results initially raised the possibility that processing might be essential for the functional activity of Ycf1p. Interestingly, however, overexpression of Ycf1p from a multicopy plasmid (Fig. 1B, rows 5 and 6) suppressed the cadmium sensitivity observed in the pep4Δ strain, even though little or no processing was apparent (Fig. 1A). This result provided the first hint that processing may not, in fact, be a prerequisite for Ycf1p function. It should be noted that the extent of Ycf1p processing is the same in the presence or absence of cadmium (data not shown).
Cleavage of Ycf1p requires a unique 17-amino-acid region of MSD1 that is necessary and sufficient for processing.
To more directly assess the functional requirement for processing, we sought to construct mutations within YCF1 that inhibited processing. Based on the gel mobility and the PEP4-dependent production of the Ycf1p proteolytic products (∼38 and ∼160 kDa) (31), we reasoned that the processing must occur within or near loop 6 (L6) in MSD1, which is lumenally disposed (Fig. 2A). Interestingly, analysis of the L6 region in an alignment between all yeast MRP subfamily members identifies an insertion of ca. 17 amino acids that is unique to Ycf1p and is designated here as the L6 insertion (L6ins) (Fig. 2B). Deletion of the L6ins (ΔL6ins) completely blocks processing of Ycf1p (Fig. 3A, compare lanes 2 and 3). Thus, we conclude that the L6ins is required for proteolytic processing of Ycf1p. Although additional regions of Ycf1p could impact proteolytic processing, this possibility has not yet been further investigated.
To determine whether L6ins is sufficient for processing, we inserted this 17-amino-acid region into a different lumenal loop of Ycf1p. Strikingly, movement of this region within Ycf1p, from L6 in MSD1 to the middle of loop 12 (L12) in MSD2 (ΔL6ins, L12::L6ins), creates a novel processing site within Ycf1p, suggesting that L6ins carries the information necessary for both recognition and cleavage (Fig. 3A, lane 4). Ycf1p is also processed within or near L12 when an even more minimal segment (six amino acids, referred to below as L6-2) from L6ins is transferred to the middle of L12 (Fig. 3A, lane 5).
We sought to determine whether L6ins confers processing when inserted into a different ABC protein, by transferring it to L6 of Bpt1p, the closest yeast homologue of Ycf1p (38, 44). Notably, the normally unprocessed Bpt1p is now cleaved (Fig. 3B, compare lanes 2 and 3). Taken together, the results shown in Fig. 3A and B indicate that the L6ins is necessary and sufficient for proteolytic processing.
Mutations within the L6ins have variable effects on the processing of Ycf1p.
To further define the minimal region required for proteolytic processing, we divided the L6ins into three subregions, L6-1, L6-2, and L6-3 (see Table 3 for residues within these intervals) and generated deletion and substitution mutations in these subregions. Results from this analysis are shown in Fig. 4 and are summarized in Table 3. Immunoblot analysis was used to monitor the processing of mutant versions of Ycf1p expressed either from a multicopy plasmid (Fig. 4A) or from a chromosomally integrated copy of wild-type or mutant GFP-tagged YCF1 (Fig. 4B and C). The integrants were generated by a two-step gene replacement into two different ycf1Δ strain backgrounds (derived from our laboratory strain, SM1058, or from the parental strain for the yeast knockout collection, BY4741). The processing phenotype for each mutant expressed from the chromosome is identical for the two strain backgrounds and essentially identical to that observed when the mutants are overexpressed from multicopy plasmids (Fig. 4, compare B and C to A). Deletion of the first six (ΔL6-1) or the last five (ΔL6-3) amino acids of the L6ins had no effect on Ycf1p cleavage (Fig. 4, lanes 4 and 6). However, deletion of the middle six amino acids (ΔL6-2) severely impaired processing (Fig. 4, lane 5).
To determine whether the specific sequence of the L6-2 region (SSLQGF) is critical for processing, we replaced this subregion with various residues, mutated singly and in combination (Table 3, rows 7 to 9). Single-residue alanine replacements within the L6-2 region had no effect on processing of Ycf1p (data not shown). Furthermore, replacement of the entire L6-2 region with six alanines (L6-2→A) also had no effect on processing, suggesting that a specific sequence per se is not critical for processing (Fig. 4, lane 7). However, further mutagenesis indicates that the particular sequence content of the L6-2 region does influence processing. Replacement of the L6-2 region with six glutamates (L6-2→E) resulted in inefficient processing of Ycf1p and a lower overall steady-state level of protein, presumably reflecting degradation, compared to the other mutants (Fig. 4, lane 8). We also replaced the L6-2 region with the six amino acids from the neighboring region (L6-2→L6-1), thereby creating a duplication of L6-1. Duplication of the L6-1 region significantly impaired Ycf1p processing (Fig. 4, lane 9). Thus, the ΔL6ins, ΔL6-2, and L6-2→L6-1 mutants, in which proteolytic processing is defective, provide useful tools for determining whether processing and function of Ycf1p are at all correlated (Table 3). The L6-2→L6-1 mutant is particularly advantageous, since it allows us to address the importance of proteolytic processing, without having changed the overall size of L6. Importantly, each Ycf1p mutant expressed from a multicopy plasmid or from the chromosome properly localizes to the vacuolar membrane (data not shown), indicating that any defects in proteolytic processing or activity are not due to mislocalization.
L6ins ycf1 mutants affect substrate specificity when expressed at chromosomal levels.
Previous studies have shown that ycf1Δ cells are sensitive to toxic compounds such as cadmium and arsenite (16, 43, 47) and that Ycf1p directly mediates the transport of cadmium-glutathione complexes (28). We initially examined the ability of the Ycf1p processing mutants to confer resistance to cadmium when the mutant proteins are overexpressed from multicopy 2μ plasmids. Under these conditions, the deletion of the entire L6ins (or portions thereof) did not affect cadmium resistance, regardless of the status of Ycf1p processing (data not shown). Furthermore, moving the processing site to L12 also had no effect on cadmium resistance (data not shown) even though this created a novel functional processing site in Ycf1p (Fig. 3A). These results were not completely surprising considering our earlier observation that Ycf1p expressed from a multicopy 2μ plasmid suppresses the cadmium sensitivity of a pep4Δ mutant (Fig. 1B), suggesting that overexpression of mutant forms of Ycf1p may mask differences that are more subtle.
To circumvent any overexpression artifacts, we assessed the cadmium and arsenite resistance of the Ycf1p mutant proteins expressed from the chromosome. The results from the cadmium spot test show that strains bearing wild-type or GFP-tagged Ycf1p and some of the mutants (Fig. 5, rows 2, 3, and 4 to 6, respectively) confer a wild-type level of resistance to cadmium, while the other mutants (Fig. 5, rows 7 to 10) confer a significantly lower level of cadmium resistance (see also Table 3). Notably, each of these phenotypic groups consists of some mutants that are processed normally and some that are not (Fig. 5, right column, and Table 3), clearly indicating that the degree of cadmium resistance and the processing status are unrelated.
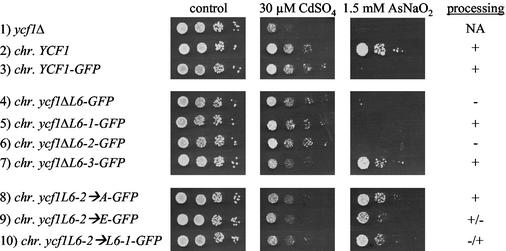
FIG. 5.
Mutations within the L6ins affect substrate specificity when Ycf1p is expressed from the chromosome in the SM1058 strain background. The activity of wild-type Ycf1p (untagged and GFP tagged) and mutant forms of Ycf1p-GFP expressed from the chromosome in the SM1058 strain background is shown. An aliquot (5 μl) of cells at 0.1 OD600, and serial 10-fold dilutions thereof, were spotted onto control plates (no CdSO4 or AsNaO2) and plates containing 30 μM CdSO4 or 1.5 mM AsNaO2 and then incubated at 30°C for 2 to 6 days. The strains used in rows 1 to 10 are SM3851, SM1058, SM4643, SM4644, SM4729, SM4730, SM4731, SM4717, SM4718, and SM4697, respectively. The indicated processing phenotypes were determined by the immunoblot analysis shown in Fig. 4B (see also Table 3). NA, not applicable.
Strikingly different results were observed when arsenite resistance was examined. First, wild-type YCF1-GFP, when chromosomally integrated, was unable to confer resistance to arsenite compared to untagged chromosomal YCF1, although both GFP-tagged and untagged Ycf1p confer cadmium resistance (Fig. 5, compare rows 2 and 3). Thus, the C-terminal cytosolically disposed GFP tag may influence Ycf1p transport activity for some, but not all substrates. Second, the mutants that show reduced cadmium resistance display an unanticipated gain of function for arsenite resistance compared to the Ycf1p-GFP parental construct (Fig. 5, compare rows 7 to 10 with row 3, and Table 3). Together, these findings for cadmium and arsenite resistance of chromosomal ycf1 mutants suggest that proteolytic processing of L6 is not a prerequisite for Ycf1p to function but that the L6 region appears to be involved in substrate specificity, either directly or indirectly.
Strain differences suggest that Ycf1p may interact with a second component to transport certain substrates.
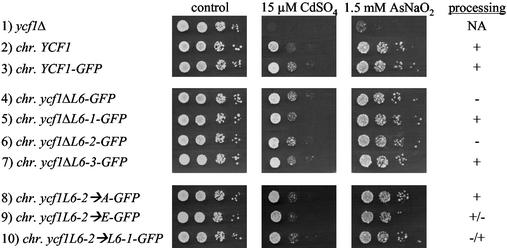
We also examined the cadmium and arsenite resistance of the Ycf1p mutant proteins expressed from the chromosome in a different strain background, BY4741, which is the parental strain for the yeast knockout collection and a derivative of the strain S288C (17) (Fig. 6). Notably, the ycf1 null mutant is sensitive to both compounds (Fig. 6, row 1). However, we were surprised to find that, in contrast to the results presented in Fig. 5 for the SM1058 strain background, neither the GFP tag nor any of the L6 deletion or point mutations affects cadmium or arsenite resistance when expressed chromosomally in the BY4741 strain background. There was no phenotype for any of the L6 mutants, even with higher concentrations of cadmium or arsenite (data not shown). Based on these results, we predict that an additional component(s) that differs between the two strain backgrounds is important for influencing the ability of Ycf1p to confer resistance to certain substrates.
FIG. 6.
Mutations within the L6ins do not affect the substrate specificity when Ycf1p is expressed from the chromosome in the BY4741 strain background. The activity of wild-type Ycf1p (untagged and GFP tagged) and mutant forms of Ycf1p-GFP expressed from the chromosome in the BY4741 strain background is shown. An aliquot (5 μl) of cells at 0.1 OD600, and serial 10-fold dilutions thereof, were spotted onto control plates (no CdSO4 or AsNaO2) and plates containing 15 μM CdSO4 or 1.5 mM AsNaO2 and then incubated at 30°C for 2 to 6 days. The strains used in rows 1 to 10 were SM4719, BY4741, SM4720, SM4721, SM4726, SM4727, SM4728, SM4723, SM4724, and SM4722, respectively. The processing phenotypes were determined by the immunoblot analysis shown in Fig. 4C. NA, not applicable.
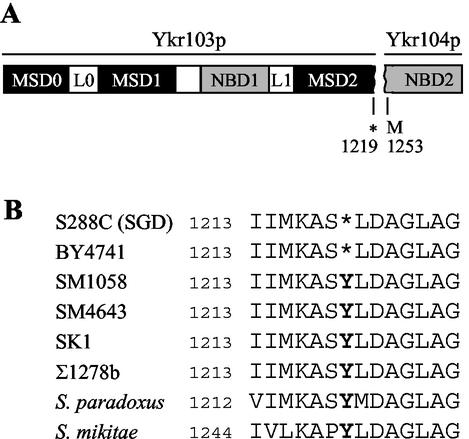
Identification of a new MRP in yeast.
Several ABC transporters appear to interact with other membrane transporters or channels. Examples include the human MRP-type protein SUR1, which interacts with the KATP channel KIR6.2 (1), and partial molecules of the yeast ABC transporter Ste6p, which complement mutations in full-length Ste6p possibly via hetero-oligomer formation (4, 5, 7). Full-length ABC transporters may also interact as homodimers or heterodimers, as has been proposed for human MRP1 (36) or yeast Ycf1p and Bpt1p (38). Thus, when considering candidate genes that may represent the putative second component responsible for the strain differences described above, we examined the possibility that another ABC transporter may be involved. We focused on YKR103w and YKR104w, reported in the SGD as two adjacent ORFs that code for partial ABC transporters, separated by a nonsense codon, followed by 33 sense codons (Fig. 7A). We hypothesized that the nonsense codon might represent a mutation in S288C (the strain used to sequence the genome and the strain from which BY4741 is derived), whereas in SM1058 (also known as EG123 and whose lineage is distinct from S288C [40]), YKR103/104w may actually code a single “full-length” MRP-type transporter homologous throughout its length to Ycf1p and Bpt1p (6). Expression of the full-length versus truncated transporter, then, might influence the activity of Ycf1p.
FIG. 7.
Identification of a new full-length MRP type transporter in yeast. (A) The partial ABC transporters Ykr103p and Ykr104p are represented schematically, with subdivisions indicating the predicted domains based on sequence alignments with Ycf1p. Ykr103p is comprised of MSD0, L0, MSD1, NBD1, L1, and the majority of MSD2 (with the possible exception of the last transmembrane span according to Kyte and Doolittle hydropathy analysis), and Ykr104p is comprised of NBD2. The abbreviations for the domains are as described in Fig. 2. The numbers shown by the stop codon (*) for Ykr103p and a potential initiating methionine (M) for Ykr104p correspond to the amino acid positions in a hypothetical full-length ABC transporter encoded by the two ORFs YKR103w and YKR104w. (B) The amino acid sequence of the Ykr103p and Ykr104p junction is shown for different S. cerevisiae strains and for the homologues found in different Saccharomyces species, as determined by DNA sequence analysis (BY4741, SM1058, SM4643, SK1, and Σ1278b) or obtained from the SGD website (S. paradoxus and S. mikatae). The sequence deposited in SGD for S288C and our sequence results for BY4741 both contain a stop codon (*) at amino acid 1219, whereas other S. cerevisiae strains, as well as additional “wild” yeast strains, contain a tyrosine (Y) and thus code for a full-length ABC transporter. The region shown for the homologues in S. paradoxus and S. mikatae was chosen based on sequence alignments with YKR103/104w. The GenBank accession numbers for NFT1 in S. cerevisiae strains EG123, Σ1278b, and SK1 are AY230264, AY230265, and AY230266, respectively.
To examine the possibility that YKR103/104w varies between strains, we used PCR amplification of genomic DNA, followed by DNA sequence analysis to confirm that, in the S288C-derived strain BY4741, YKR103/104w are separate ORFs, with a nonsense codon (TAG) at codon 1219 of YKR103w. In contrast, we found that in our laboratory strain SM1058, codon 1219 (TAT) encodes a tyrosine residue. Thus, in SM1058, the combined YKR103/104w ORF encodes a new full-length MRP-type transporter, which we have designated NFT1 (Fig. 7B). In at least some S. cerevisiae strain backgrounds, then, the yeast MRP subfamily consists of six full-length transporters: YCF1, BPT1, YBT1, YOR1, YHL035c, and NFT1.
Interestingly, other S. cerevisiae strains, whose lineages derive independently of S288C (SK1 and Σ1278b), and other species of yeast from the wild (S. paradoxus and S. mikatae), also encode the full-length NFT1 gene (Fig. 7B). The fact that the NFT1 gene product is full-length in SM1058 and truncated in BY4741 may account for the different activities of some of the Ycf1p mutants in these two strain backgrounds; however, proof of this hypothesis awaits further analysis.
DISCUSSION
Identification of a sequence within MSD1 that is necessary and sufficient for Ycf1p processing.
Previous studies by us and others have shown that the MRP-type ABC transporter Ycf1p resides in the vacuolar membrane (28, 31, 47), where it undergoes PEP4-dependent processing. An important goal in the present study was to generate mutations within Ycf1p that block cleavage, permitting us to assess the connection between processing and the transport activity of Ycf1p.
The estimated size of the Ycf1p cleavage products, together with sequence alignment between Ycf1p and other members of the yeast MRP subfamily, revealed a 17-amino-acid insertion, designated L6ins, that is unique to Ycf1p and is in the vicinity of the cleavage site (Fig. 2B). L6ins lies within a predicted lumenal loop (L6) of Ycf1p. Deletion of L6ins resulted in accumulation of full-length Ycf1p and, strikingly, its transfer to L12 of Ycf1p or L6 of another MRP protein, Bpt1p, resulted in novel cleavage (Fig. 3). These findings indicate that the L6ins is necessary and sufficient to direct proteolytic processing, at least when present in the lumenal loop of a vacuolar MRP-type membrane protein, where it is accessible to vacuolar proteases. Cleavage could occur within the L6ins or somewhere in its vicinity. For instance, the addition of the L6ins to L12 of Ycf1p and L6 of Bpt1p could expose a nearby protease-sensitive site that is normally occluded in these loops.
Interestingly, a previous study showed that YCF1, when heterologously expressed in Sf21 insect cells, also undergoes proteolytic processing to yield approximately the same size C-terminal product as that observed in its native yeast cell environment (35). Since Ycf1p localizes to the plasma membrane in Sf21 cells, rather than to the lysosome, this cleavage may not be truly analogous to that which occurs in yeast cells. Nevertheless, it is possible that cleavage occurs within or near the L6ins even in insect cells, indicating that posttranslational proteolytic processing may be inherent to the biology of Ycf1p. Notably, human MRP1, a close homologue of Ycf1p but missing the L6ins sequence, is not proteolytically processed either in mammalian or Sf21 insect cells (20, 35).
Proteolytic processing is unrelated to the function of Ycf1p.
Since Ycf1p is not processed in a pep4Δ mutant, and because the pep4Δ mutant is sensitive to cadmium, it seemed possible that the processing site mutants might also be sensitive to cadmium. However, mutations in L6ins of Ycf1p (discussed below) showed that the ability of Ycf1p to confer resistance to cadmium is completely unrelated to its processing phenotype. Thus, the sensitivity of the pep4Δ mutant to cadmium must not due to lack of Ycf1p processing but instead may be caused by one of the many pleiotropic phenotypes associated with the pep4Δ mutant (21, 45); such pleiotropic effects could indirectly alter Ycf1p function or cellular cadmium levels.
A subset of mutations in L6ins resulted in Ycf1p proteins that cannot be processed but retain the ability to confer the wild-type level of cadmium resistance (Fig. 5 and Table 3). Thus, proteolytic processing of Ycf1p is clearly not required to activate Ycf1p. Proteolytic processing does not appear to “deactivate” Ycf1p either, since Ycf1pΔL6ins (unprocessed) does not confer a greater resistance to cadmium than wild-type Ycf1p (processed). Thus, processing appears to represent a “neutral” event in terms of Ycf1p activity. Full functionality of split molecules is not unprecedented for ABC transporters, and indeed the natural coexpression of independently encoded ABC modules in bacteria and humans (19, 52), or the artificial coexpression of half, quarter-three-quarter, and other combinations of partial molecules for a variety of eukaryotic ABC proteins (i.e., Ste6p, MRP1, and MRP2) also promotes normal function (2, 3, 7, 14). However, Ycf1p is the only full-length ABC transporter known to undergo processing as a normal part of its life cycle in vivo. Although our studies have not uncovered a function for Ycf1p processing, it is possible that a role may be revealed under as-yet-unexamined physiological condition(s).
Mutations within L6ins affect substrate specificity.
Resistance to both cadmium and arsenite is Ycf1p dependent, as evidenced by the sensitivity of a ycf1Δ strain to both compounds (Fig. 5) (16, 43, 47). Surprisingly, we found that certain L6ins mutations in Ycf1p can differentially affect cellular resistance to these compounds, at least in our laboratory strain background SM1058. One group confers resistance to cadmium but not arsenite, while another group confers resistance to arsenite and reduced resistance to cadmium (Fig. 5 and Table 3). Furthermore, each group contains some ycf1 mutants that are proteolytically processed and some that are not. One explanation for the two distinct classes is that mutant Ycf1p can exist in two conformations (“A” and “B”) in which different domains or distinct regions within the active site of Ycf1p interact to bind, recognize, or transport various substrates. According to this scenario, the first group of mutant proteins (Ycf1p-GFP, ΔL6ins, ΔL6-1, and ΔL6-2) confer cadmium resistance but not arsenite resistance because they assume conformation “A.” The other mutant proteins (ΔL6-3, L6-2→A, E, or L6-1) assume conformation “B,” conferring arsenite resistance and reduced cadmium resistance.
It is somewhat surprising that substrate specificity can be affected by alterations in a region of the Ycf1p transporter on the side of the membrane where the substrate is released, as opposed to the side where the substrate encounters the transporter. Perhaps the L6ins mutations in Ycf1p selectively affect substrate release rather than substrate binding. Alternatively, the differential transport of cadmium and arsenite observed for some of the ycf1 lumenal loop mutations may relate to the mode of substrate transport, i.e., cadmium is transported by Ycf1p as a complex with glutathione (28) while, at least for MRP1, arsenite may be cotransported with glutathione (37). Another recent study also identified mutations that altered the substrate specificity of Ycf1p; in this case the mutations mapped to cytosolic and membrane-spanning regions of Ycf1p (12). Similarly, for human MRP1, residues within several membrane spans appear to affect substrate specificity (18, 25, 42, 53-55). Taken together, the studies on yeast and human MRP transporters provide evidence that the recognition, binding, and/or transport of particular substrates can involve regions on both sides of the membrane, as well as regions spanning the membrane.
Strain variation in the phenotype of ycf1 mutants.
In the course of our Ycf1p studies, we used two differing strain backgrounds: (i) SM1058, also designated EG123, which is our standard laboratory strain and has a poorly defined lineage (40), and (ii) BY4741, an S288C derivative that is the parental strain for the yeast deletion collection (17). Quite surprisingly, we found that none of the L6ins Ycf1p mutants used in the present study affected cellular cadmium or arsenite resistance in BY4741 (Fig. 6), in contrast to the results in the SM1058 strain background discussed above. Although the L6ins mutant phenotypes were indistinguishable from wild-type in BY4741, the ycf1Δ mutation in this background remained sensitive to these compounds, indicating that resistance is still Ycf1p dependent.
These results suggest that another cellular component(s), which may be directly or indirectly involved in YCF1-mediated resistance to toxic compounds such as cadmium or arsenite, may be different in the two strains. There are several possibilities for such a component. It could act (i) directly on Ycf1p, for instance, to form a heterodimer; (ii) indirectly, perhaps altering general properties of how Ycf1p functions; or (iii) at a step different from Ycf1p, for instance, to influence the intracellular level of toxic compounds or their targets. Phenotypic variation among yeast strains is not uncommon and has been used as a starting point to identify novel components for a number of cellular processes, including pseudohyphal growth, flocculation, and cell polarity (23, 26). Clearly, an important next step will be to determine whether the ycf1 “modifier” segregates as a single, or multiple, gene trait.
Discovery of a new ABC transporter in yeast, Nft1p.
One possible cellular component that we considered might contribute to the strain differences discussed above could be another MRP protein. We investigated two uncharacterized ORFs, YKR103w and YKR104w. We had previously observed in an analysis of yeast ABC proteins that these adjacent sequences in the yeast genome appear to encode two pieces of a single full-length MRP protein (6, 44). We discovered that in the strain SM1058, YKR103/104w is one continuous ORF rather than two ORFs separated by a nonsense codon, as in the strains BY4741 and S288C (Fig. 7). Because YKR103/104w actually represents a single gene encoding a new full-length MRP-type transporter, we have named this gene NFT1. Interestingly, possession of a full-length NFT1 gene appears to be the “wild-type” situation for yeast, since other S. cerevisiae laboratory strains (Σ1278b and SK1) that differ in lineage from S288C also possess the full-length NFT1 gene, as do other Saccharomyces species (S. paradoxus and S. mikitae). Differences between S288C and other S. cerevisiae strains, as well as other Saccharomyces species, have been reported for a variety of genes, such as the aquaporin genes (9). It is possible that the manner in which S288C was cultivated in the laboratory led to an unintentional selection against the presence of full-length NFT1.
The observed strain differences in cadmium and arsenite resistance of certain ycf1 L6 mutations expressed in SM1058 versus BY4741 could potentially be attributed to one strain background expressing the full-length MRP transporter (Nft1p) and the other expressing one or both of the partial molecules (Ykr103p and Ykr104p). At least two models can be envisioned to explain how NFT1 or YKR103/104w might account for the strain differences we observed with certain Ycf1p mutant proteins. First, the expression of Ykr103p and/or Ykr104p in the BY4741 strain background could positively influence the activity of the Ycf1p mutants, perhaps by forming a heterodimer with Ycf1p. Such partial-molecule complementation has been observed for mutations in another ABC transporter, Ste6p (4, 7). Another possibility is that the full-length transporter, Nft1p, could negatively influence the activity of Ycf1p in the SM1058 strain background, thereby revealing the mutant phenotypes. Although it is tempting to speculate that Nft1p is involved in Ycf1p-mediated cadmium and arsenite resistance, it is indeed possible that neither Nft1p nor Ykr103p and/or Ykr104p are affecting the activity of Ycf1p and that the strain differences are due to another cellular component(s). Elucidation of the function of Nft1p is currently under investigation.
Acknowledgments
We thank R. Rao for helpful comments on the manuscript and J. Boeke and M. Rose for strains.
This work was supported by grants DK58029 and GM51508 from the National Institutes of Health to S.M.
REFERENCES
- 1.Aguilar-Bryan, L., J. Bryan, and M. Nakazaki. 2001. Of mice and men: K(ATP) channels and insulin secretion. Rec. Prog. Horm. Res. 56:47-68. [DOI] [PubMed] [Google Scholar]
- 2.Bakos, E., R. Evers, G. Calenda, G. E. Tusnady, G. Szakacs, A. Varadi, and B. Sarkadi. 2000. Characterization of the amino-terminal regions in the human multidrug resistance protein (MRP1). J. Cell Sci. 113:4451-4461. [DOI] [PubMed] [Google Scholar]
- 3.Bakos, E., R. Evers, G. Szakacs, G. E. Tusnady, E. Welker, K. Szabo, M. de Haas, L. van Deemter, P. Borst, A. Varadi, and B. Sarkadi. 1998. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J. Biol. Chem. 273:32167-32175. [DOI] [PubMed] [Google Scholar]
- 4.Berkower, C., and S. Michaelis. 1993. Effects of nucleotide binding fold mutations on STE6, a yeast ABC protein, p. 130-146. In L. Reuss, J. M. Russell, and M. L. Jennings (ed.), Molecular biology and function of carrier proteins. Rockefeller University Press, New York, N.Y. [PubMed]
- 5.Berkower, C., and S. Michaelis. 1991. Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP-binding cassette (ABC) protein superfamily. EMBO J. 10:3777-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkower, C., and S. Michaelis. 1995. Sequence comparison of yeast ATP-binding cassette (ABC) proteins, vol. LX. Cold Spring Harbor Press, Cold Spring Harbor, N.Y. [DOI] [PubMed]
- 7.Berkower, C., D. Taglicht, and S. Michaelis. 1996. Functional and physical interactions between partial molecules of STE6, a yeast ATP-binding-cassette transport protein. J. Biol. Chem. 271:22983-22989. [DOI] [PubMed] [Google Scholar]
- 7a.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 8.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 9.Carbrey, J. M., M. Bonhivers, J. D. Boeke, and P. Agre. 2001. Aquaporins in Saccharomyces: characterization of a second functional water channel protein. Proc. Natl. Acad. Sci. USA 98:1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, P., S. Sapperstein, J. D. Choi, and S. Michaelis. 1997. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J. Cell Biol. 136:251-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elble, R. 1992. A simple and efficient procedure for transformation of yeasts. BioTechniques 13:18-20. [PubMed] [Google Scholar]
- 12.Falcon-Perez, J. M., M. Martinez-Burgos, J. Molano, M. J. Mazon, and P. Eraso. 2001. Domain interactions in the yeast ATP-binding cassette transporter Ycf1p: intragenic suppressor analysis of mutations in the nucleotide-binding domains. J. Bacteriol. 183:4761-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falcon-Perez, J. M., M. J. Mazon, J. Molano, and P. Eraso. 1999. Functional domain analysis of the yeast ABC transporter Ycf1p by site-directed mutagenesis. J. Biol. Chem. 274:23584-23590. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez, S. B., Z. Hollo, A. Kern, E. Bakos, P. A. Fischer, P. Borst, and R. Evers. 2002. Role of the N-terminal transmembrane region of the multidrug resistance protein MRP2 in routing to the apical membrane in MDCKII cells. J. Biol. Chem. 277:31048-31055. [DOI] [PubMed] [Google Scholar]
- 15.Fujimura-Kamada, K., F. J. Nouvet, and S. Michaelis. 1997. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J. Cell Biol. 136:271-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh, M., J. Shen, and B. P. Rosen. 1999. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:5001-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 18.Haimeur, A., R. G. Deeley, and S. P. Cole. 2002. Charged amino acids in the sixth transmembrane helix of multidrug resistance protein 1 (MRP1/ABCC1) are critical determinants of transport activity. J. Biol. Chem. 277:41326-41333. [DOI] [PubMed] [Google Scholar]
- 19.Higgins, C. F. 2001. ABC transporters: physiology, structure, and mechanism: an overview. Res. Microbiol. 152:205-210. [DOI] [PubMed] [Google Scholar]
- 20.Hipfner, D. R., S. D. Gauldie, R. G. Deeley, and S. P. Cole. 1994. Detection of the Mr 190,000 multidrug resistance protein, MRP, with monoclonal antibodies. Cancer Res. 54:5788-5792. [PubMed] [Google Scholar]
- 21.Jones, E. W. 1991. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 266:7963-7966. [PubMed] [Google Scholar]
- 22.Jones, E. W., G. C. Webb, and M. A. Hiller. 1997. Biogenesis and function of the yeast vacuole, p. 363-470. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Jorgensen, P., B. Nelson, M. D. Robinson, Y. Chen, B. Andrews, M. Tyers, and C. Boone. 2002. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics 162:1091-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a cold spring harbor course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Koike, K., C. J. Oleschuk, A. Haimeur, S. L. Olsen, R. G. Deeley, and S. P. Cole. 2002. Multiple membrane-associated tryptophan residues contribute to the transport activity and substrate specificity of the human multidrug resistance protein, MRP1. J. Biol. Chem. 277:49495-49503. [DOI] [PubMed] [Google Scholar]
- 26.Kron, S. J. 1997. Filamentous growth in budding yeast. Trends Microbiol. 5:450-454. [DOI] [PubMed] [Google Scholar]
- 27.Levitan, D., J. Lee, L. Song, R. Manning, G. Wong, E. Parker, and L. Zhang. 2001. PS1 N- and C-terminal fragments form a complex that functions in APP processing and Notch signaling. Proc. Natl. Acad. Sci. USA 98:12186-12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Z., M. Szczypka, Y. Lu, D. Thiele, and P. Rea. 1996. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J. Biol. Chem. 271:6509-6517. [DOI] [PubMed] [Google Scholar]
- 29.Li, Z. S., Y. P. Lu, R. G. Zhen, M. Szczypka, D. J. Thiele, and P. A. Rea. 1997. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. USA 94:42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason, D. L. 2002. Functional analysis of MRP transporters in Saccharomyces cerevisiae. Ph.D. thesis, The Johns Hopkins University School of Medicine, Baltimore, Md.
- 31.Mason, D. L., and S. Michaelis. 2002. Requirement of the N-terminal extension for vacuolar trafficking and transport activity of yeast Ycf1p, an ATP-binding cassette transporter. Mol. Biol. Cell 13:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaelis, S., and I. Herskowitz. 1988. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol. Cell. Biol. 8:1309-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oldenburg, K. R., K. T. Vo, S. Michaelis, and C. Paddon. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratovitski, T., H. H. Slunt, G. Thinakaran, D. L. Price, S. S. Sisodia, and D. R. Borchelt. 1997. Endoproteolytic processing and stabilization of wild-type and mutant presenilin. J. Biol. Chem. 272:24536-24541. [DOI] [PubMed] [Google Scholar]
- 35.Ren, X., T. Furukawa, Z. Chen, H. Okumura, S. Aoki, T. Sumizawa, A. Tani, M. Komatsu, X. Mei, and S. Akiyama. 2000. Functional comparison between YCF1 and MRP1 expressed in Sf21 insect cells. Biochem. Biophys. Res. Commun. 270:608-615. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg, M. F., Q. Mao, A. Holzenburg, R. C. Ford, R. G. Deeley, and S. P. Cole. 2001. The structure of the multidrug resistance protein 1 (MRP1/ABCC1). crystallization and single-particle analysis. J. Biol. Chem. 276:16076-16082. [DOI] [PubMed] [Google Scholar]
- 37.Salerno, M., M. Petroutsa, and A. Garnier-Suillerot. 2002. The MRP1-mediated effluxes of arsenic and antimony do not require arsenic-glutathione and antimony-glutathione complex formation. J. Bioenerg. Biomembr. 34:135-145. [DOI] [PubMed] [Google Scholar]
- 38.Sharma, K. G., D. L. Mason, G. Liu, P. A. Rea, A. K. Bachhawat, and S. Michaelis. 2002. Localization, regulation, and substrate transport properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryot. Cell 1:391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siliciano, P., and K. Tatchell. 1984. Transcription and regulatory signals at the mating type locus in yeast. Cell 37:969-978. [DOI] [PubMed] [Google Scholar]
- 41.Sprague, G. F., and J. W. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 657-744. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 42.Stride, B. D., S. P. Cole, and R. G. Deeley. 1999. Localization of a substrate specificity domain in the multidrug resistance protein. J. Biol. Chem. 274:22877-22883. [DOI] [PubMed] [Google Scholar]
- 43.Szczypka, M. S., J. A. Wemmie, W. S. Moye-Rowley, and D. J. Thiele. 1994. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 269:22853-22857. [PubMed] [Google Scholar]
- 44.Taglicht, D., and S. Michaelis. 1998. A complete catalogue of Saccharomyces cerevisiae ABC proteins and their relevance to human health and disease, p. 130-162. In S. V. Ambudkar and M. M. Gottesman (ed.), Methods in enzymology. Academic Press, Inc., New York, N.Y. [DOI] [PubMed]
- 45.Teichert, U., B. Mechler, H. Muller, and D. H. Wolf. 1989. Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J. Biol. Chem. 264:16037-16045. [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wemmie, J., and W. Moye-Rowley. 1997. Mutational analysis of the Saccharomyces cerevisiae ATP-binding cassette transporter protein Ycf1p. Mol. Microbiol. 25:683-694. [DOI] [PubMed] [Google Scholar]
- 48.Wemmie, J., M. Szczypka, D. Thiele, and W. Moye-Rowley. 1994. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 269:32592-32597. [PubMed] [Google Scholar]
- 49.Wemmie, J. A., S. M. Steggerda, and W. S. Moye-Rowley. 1997. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J. Biol. Chem. 272:7908-7914. [DOI] [PubMed] [Google Scholar]
- 50.Wolfe, M. S., and C. Haass. 2001. The role of presenilins in gamma-secretase activity. J. Biol. Chem. 276:5413-5416. [DOI] [PubMed] [Google Scholar]
- 51.Wolfe, M. S., and D. J. Selkoe. 2002. Intramembrane proteases: mixing oil and water. Science 296:2156-2157. [DOI] [PubMed] [Google Scholar]
- 52.Yu, L., R. E. Hammer, J. Li-Hawkins, K. Von Bergmann, D. Lutjohann, J. C. Cohen, and H. H. Hobbs. 2002. Disruption of Abcg5 and Abcg8 in mice reveals their crucial role in biliary cholesterol secretion. Proc. Natl. Acad. Sci. USA 99:16237-16242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, D. W., S. P. Cole, and R. G. Deeley. 2002. Determinants of the substrate specificity of multidrug resistance protein 1: role of amino acid residues with hydrogen bonding potential in predicted transmembrane helix 17. J. Biol. Chem. 277:20934-20941. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, D. W., S. P. Cole, and R. G. Deeley. 2001. Identification of a nonconserved amino acid residue in multidrug resistance protein 1 important for determining substrate specificity: evidence for functional interaction between transmembrane helices 14 and 17. J. Biol. Chem. 276:34966-34974. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, D. W., S. P. Cole, and R. G. Deeley. 2001. Identification of an amino acid residue in multidrug resistance protein 1 critical for conferring resistance to anthracyclines. J. Biol. Chem. 276:13231-13239. [DOI] [PubMed] [Google Scholar]