Abstract
Regions of both colicin Ia and diphtheria toxin N-terminal to the channel-forming domains can be translocated across planar phospholipid bilayer membranes. In this article we show that the translocation pathway of diphtheria toxin allows much larger molecules to be translocated than does the translocation pathway of colicin Ia. In particular, the folded A chain of diphtheria toxin is readily translocated by that toxin but is not translocated by colicin Ia. This difference cannot be attributed to specific recognition of the A chain by diphtheria toxin's translocation pathway because the translocation pathway also accommodates folded myoglobin.
INTRODUCTION
A number of toxins (e.g., cholera, ricin, diphtheria, tetanus, anthrax) act by having an enzymatic moiety that, when delivered to the cytosol, ultimately leads to cell death. Gill (1) named these “A-B toxins”: the “A” portion is the active, enzymatic part of the toxin, and the “B” portion is the part of the toxin that binds to the cell membrane and through whose agency the A portion is translocated to the cytosol. In some cases (e.g., ricin and cholera toxin), translocation is achieved through the utilization of the cell's own translocation machinery (2), but in other cases (e.g., diphtheria toxin and anthrax toxin), all of the translocation machinery is built into a part of the B moiety, as manifested by its ability to translocate the A component across a planar lipid bilayer without the aid of any cellular components (3–5).
What is the mechanism of translocation? The B portion of these toxins makes pores (also called channels) in planar lipid bilayers (6,7), and mutants that are defective in pore formation are also defective in intoxicating cells (8,9), making it tempting to suggest that the pore lumen acts as a conduit for the A moiety. Although this has indeed been shown to be the case for anthrax toxin (5), there is no conclusive evidence that the pore is the translocation path for the other channel-forming toxins. (Koriazova and Montal (4) claim to have established this as the translocation mechanism for botulinum neurotoxin. However, a more likely explanation for the blocking of the channel by the A chain, which they claim as evidence for this, is that it results from the A chain blocking the channel from the trans side after it has been translocated across the membrane to that side.) In this article, we demonstrate that the translocation pathway for the A chain of diphtheria toxin is large enough to accommodate the folded chain (although we make no claim that the pathway is through the pore lumen). We do this by comparing the translocation pathway of diphtheria toxin to that of colicin Ia.
Comparison of the channel-forming domains and the channels formed by diphtheria toxin and colicin Ia
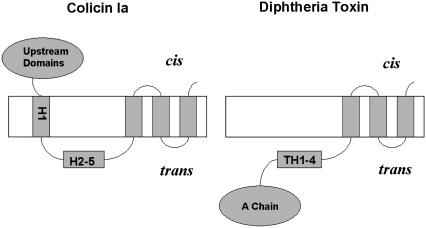
The channel-forming domain of diphtheria toxin (the T domain) and that of colicin Ia are about the same size (∼175 residues). Each consists of a bundle of 10 α-helices, and in both, helices 8 and 9 form a hydrophobic hairpin that is (marginally) long enough to span the bilayer (10,11). There is compelling direct evidence that the T domain channel is monomeric, at least in our planar bilayer system (12), and strong indirect evidence that the colicin Ia channel is as well (13). The topographies of the open state of the two channels have been mapped (14,15), and there are very interesting similarities and differences between them (Fig. 1). In both, the hydrophobic hairpin near the C-terminal end is inserted into the bilayer to form two transmembrane segments (not necessarily α-helices). For the T domain channel, there is one additional transmembrane segment, making a total of three, with everything upstream of it, including the A chain, translocated across the membrane to the trans side. For the colicin Ia channel there are two additional transmembrane segments, making a total of four, with everything upstream of it still on the cis side (the side to which the protein was added). A summary of the relationship between the T domain and the colicin Ia channel-forming domain is shown in Table 1.
FIGURE 1.
Diagrams of the topographies of the colicin Ia and diphtheria toxin channels. The colicin Ia channel has four transmembrane segments, and the upstream N-terminal domains remain on the cis side. The diphtheria toxin channel has three transmembrane segments, and the entire N-terminal region, including the A chain, has been translocated to the trans side. H1, H2-5, and TH1-4 refer to α-helices in the water-soluble forms of colicin Ia and diphtheria toxin, respectively; they are not necessarily α-helices in the channel state of these proteins.
TABLE 1.
Comparison of the channel-forming domains and the channels formed by diphtheria toxin and colicin Ia
| Characteristics | Colicin Ia | Diphtheria toxin |
|---|---|---|
| Number of residues in channel-forming domain | ∼175 | ∼175 |
| Crystallographic structure of water-soluble form | 10 α-helices | 10 α-helices |
| Number of protein subunits forming the channel | 1 | 1 |
| Number of transmembrane segments in the channel | 4 (Whole colicin) 3 (Channel-forming domain) | 3 |
| Two of the transmembrane segments are formed by hydrophobic hairpins | Yes | Yes |
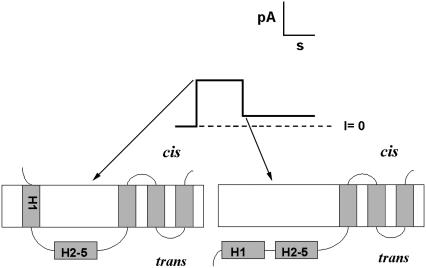
The topography we have just described for the colicin Ia channel, and shown in Fig. 1, is that formed by the whole colicin Ia molecule, which has ∼450 amino acid residues N-terminal to the channel-forming domain. If formed from the channel-forming domain alone, the channel first goes through a four-transmembrane-segment state as shown in Fig. 1 but then ends up in a three-transmembrane-segment state, comparable to that formed by the T domain, with everything upstream of the third transmembrane segment translocated across the membrane to the trans side (16) (Fig. 2). The transition time between the four- and three-transmembrane-segment state (i.e., the dwell time in the four-transmembrane-segment state) is a function of the diameter of the molecular “stopper”, or upstream sequences, attached to the N-terminus of the channel-forming domain: the larger its size, the longer the dwell time (17); if the stopper diameter is too large, the channel remains in the four-transmembrane-segment state because the pathway is not wide enough to accommodate the stopper.
FIGURE 2.
Schematic of a single-channel record obtained from a bilayer treated with the channel-forming portion of colicin Ia. The channel initially opens to the same conductance as that obtained with whole colicin Ia; the topography corresponding to this state has four transmembrane segments, with H1 being one of these segments. (Compare this diagram with that for whole colicin Ia in Fig. 1.) The conductance subsequently drops to a lower conductance; the topography corresponding to this state has three transmembrane segments, with H1 having been translocated to the trans side. H1 and H2-5 refer to α-helices in the water-soluble form of colicin Ia; they are not necessarily α-helices in the channel state of this protein.
What is the relevance of this to the pathway formed by the T domain for the translocation of the A chain of diphtheria toxin? In contrast to colicin Ia, one does not see an intermediate four-transmembrane-segment state for the T domain channel, even with the A chain attached at its N-terminus. Why is the translocation of the A chain so rapid that it does not reveal this state? Is it because its ∼200-residue peptide chain is unfolded, or is the translocation pathway so wide that even the folded chain can go rapidly through it, without lingering in a four-transmembrane-segment state like colicin Ia? We address this question here by looking at A chain translocation, or failure to translocate, when it is attached to the N-terminus of the colicin Ia channel-forming domain. We argue from our results that the translocation pathway created by the T domain is much wider than that created by colicin Ia's channel-forming domain, wide enough to easily accommodate the folded A chain. We further show that the translocation pathway is not specific for the A chain, because the T domain of diphtheria toxin can also translocate folded myoglobin (MG).
MATERIALS AND METHODS
Synthesis of A-Ia
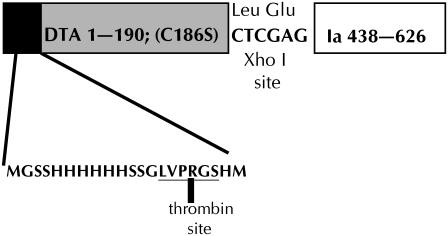
Plasmid pKSJ282 expressing the A chain of diphtheria toxin attached at the N-terminus of the colicin Ia channel-forming domain (A-Ia) was constructed as follows. Plasmid pKSJ155, which expresses a protein called Ia CT-M (16), has residues 438–626 of colicin Ia, as well as the colicin Ia immunity protein, cloned between the Xho I and BamH I sites of pET15-b (Novagen, Madison, WI). This plasmid has an intact Nde I site immediately upstream of the XhoI site, which is downstream from the translation initiation site, histidine-tag (His6-tag), and thrombin recognition sequences of pET15-b. The plasmid was digested first with Nde I (New England Biolabs, Beverly, MA), and the resulting DNA was cleaned up using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA). The DNA was then digested with Xho I (New England Biolabs), treated with calf intestine alkaline phosphatase (New England Biolabs), and gel-purified. A DNA fragment containing the sequence for diphtheria toxin A chain (DTA), but with the cysteine at position 186 mutated to serine, was isolated by PCR from plasmid LFN-DTA (C186→S) (18) containing LFN (the N-terminal portion of anthrax toxin lethal factor) and DTA with C186→S separated by a spacer sequence of 18 residues that includes His6. The forward PCR primer, 5′-GCGCCATATGGGTGCTGACGACGTTGTTGAC-3′, creates an Nde I site before the first codon for DTA. The reverse primer, 5′-GCGCCTCGAGACGGTTACCTGCAGAAGCCT-3′, creates an Xho I site at the end of the DTA gene and omits the translation terminator, so the protein reads through. The resulting purified PCR fragment of ∼580 base pairs was ligated with the digested pKSJ155 to create pKSJ282. The protein expressed from pKSJ282 thus has the initiation and His6-tag and thrombin-recognition sequences from pET15-b, followed by the entire DTA sequence, a two-amino-acid linker encoded by the Xho I site, followed by residues 438–626 of colicin Ia (the channel-forming domain). A schematic representation of the fusion protein (DTA-Ia) is shown in Fig. 3.
FIGURE 3.
Linear sequence of the A-Ia construct used for these experiments. The 190 residues of the A chain of DTA are preceded by the His6-tag and thrombin recognition sequences from pET15-b and connected to the channel-forming domain (residues 438–626) of colicin Ia by a two-amino-acid linker encoded by the XhoI restriction site.
Protein was expressed and purified from pKSJ282 in E. coli BL21(DE3) that was induced for 2 h with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation, resuspended in binding buffer 1 (20 mM sodium phosphate, pH 7.4, 500 mM NaC1, 10 mM imidazole), broken in an Emulsiflex C5 cell disrupter (Avistin, Ottawa, Canada), and then centrifuged at 150,000 × g to remove cellular debris. The clarified extract from 1 liter of culture, in 40 ml binding buffer 1, was loaded onto a 1-ml His Trap HP nickel chelation column (Amersham Biosciences/GE Healthcare, Piscataway, NJ). The column was washed with 25 column volumes of binding buffer 1, then with 10 volumes of the same buffer containing 30 mM imidazole, and purified DTA-Ia (abbreviated A-Ia) was eluted with 250 mM imidazole in binding buffer 1. The yield of purified protein was ∼15 mg of protein; although the stated capacity of this column was ≥40 mg, there was a significant amount of product protein that did not bind to the column.
Purified A-Ia was dialyzed into 20 mM Tris-Cl, pH 8.4, 150 mM NaCl before removal of the His6-tag by digesting the protein with 1 unit of thrombin (Novagen) per milligram of purified protein for 5 h at room temperature. The reaction was stopped by addition of phenylmethylsulfonyl fluoride to a concentration of 1 mM. Polyacrylamide gel electrophoresis confirmed that the digestion was complete. The resulting protein thus begins with the glycine after the arginine residue in the thrombin-recognition sequence (see Fig. 3). The protein was stored at −20°C at a concentration of 2 mg/ml. Aliquots were taken and diluted at least 100-fold for the single-channel experiments.
Synthesis of MG-T
The MG-T chimera contained (from N- to C-terminus) a His6-tag (MGSSHHHHHHSSGLVPRGSH) fused to the sequence of sperm whale myoglobin (MG) (19), followed by a linker (SAGNIEG) and then T domain (residues 193–378 of diphtheria toxin). Construction of MG-T was based on three-step PCR (20). The 5′ primer contained an Nde I restriction site and an N-terminal MG sequence. The internal forward primer contained the C-terminal MG sequence, the linker sequence, and the N-terminal T domain sequence. The internal reverse primer was complementary to the internal forward primer. The 3′ primer contained a sequence complementary to the C-terminal of the T domain and a Not I restriction site. The first PCR step used the 5′ primer, internal reverse primer, and template plasmid with the corresponding MG gene. The second PCR step used the internal forward primer, the 3′ primer, and the plasmid template with the T domain gene. These PCR products were purified by agarose electrophoresis, mixed, and then subjected to several PCR cycles. Then the 5′ primer and the 3′ primer were added, and a 40-cycle PCR was performed. The product sequences were purified on agarose gel and inserted into plasmid pET-28a using restriction sites Nde I and Not I. DNA sequencing confirmed that the final construct had the correct sequences.
A pET-28a plasmid containing the MG-T insert was transformed into E. coli strain BL21(DE3). A single colony was picked to inoculate 20 ml LB medium containing 50 μg/ml kanamycin at 37°C. The overnight culture was transferred to 2 L LB medium containing 50 μg/ml kanamycin, grown at 37°C until the OD600 reached 0.6, and then induced with 250 μg/ml IPTG for 3 h. Cells were collected by centrifugation and stored temporarily at −20°C. Cells were suspended in binding buffer 2 (20 mM Tris-HCl, pH 8.0, 150 mM NaCl) containing 100 μg/ml lysozyme. After incubation at room temperature for 30 min, the sample was sonicated on ice using a probe sonicator until the suspension was no longer viscous. The sonicated extract was then centrifuged at 20,000 × g at 4°C for 20 min. The supernatant containing MG-T was purified with TALON Metal Affinity Resin from Clontech (Mountain View, CA). One milliliter of TALON beads was washed twice with >5 volumes binding buffer 2 and then mixed with the supernatant from the cell extract. The mixture was agitated gently for 20 min to allow the His6-tagged protein to bind to the resin. The beads were collected by low-speed centrifugation and transferred to a 10-ml gravity-flow column. The column was washed three times with 5 ml binding buffer 2, and then the protein was eluted with 3 ml elution buffer (binding buffer 2 plus 200 mM imidazole). MG-T was further purified by Source Q anion exchange chromatography. The sample was diluted to 50 ml with 20 mM Tris-HCl, pH 8.0, and then loaded onto the Source Q column. The protein was eluted by a salt gradient (20 mM Tris-HCl, pH 8.0, 0 to 0.5 M NaCl). The purity of the isolated protein (>90%) was confirmed by SDS-PAGE, and protein concentration was determined using the Bradford assay.
Previous studies have shown that myoglobin binds hemin tightly at both low and neutral pH (21). We confirmed that MG-T also binds hemin tightly. Addition of MG-T to hemin at pH 8 resulted in a change in the hemin absorbance spectrum from that of free hemin to that of hemin bound to myoglobin (21). In addition, we confirmed by titration with hemin that MG-T is maximally bound to hemin at both pH 8.0 and 5.3. At both pH values, when hemin was titrated into MG-T, maximal quenching of MG-T tryptophan by bound hemin was observed when at most a modest molar excess of hemin to myoglobin was added (data not shown).
T domain and T domain with the A fragment at its N-terminus (A-T)
The T domain with the second serine after the (His)6 in the N-terminal His6-tag mutated to a cysteine and then biotinylated was the same sample as that used in the experiments described by Senzel et al. (22). The A-T with an N-terminal His6-tag was the same as that used in the experiments of Oh et al. (3). In a separate construct, biotin was attached to a genetically engineered biotin-accepting site (23) at the N-terminus of the A chain of A-T.
Bilayer experiments
Planar phospholipid bilayer membranes (∼50 μm in diameter) were formed at room temperature by a modification of the folded film technique (24) as described by Huynh et al. (25). The lipid used was asolectin (lecithin type II S; Sigma Chemical Company, St. Louis, MO) from which neutral lipids were removed by the method of Kagawa and Racker (26). After the membrane formed, channel-forming proteins to be tested were added to the cis solution, and single-channel behavior was monitored. All of the A-Ia experiments were done in symmetric salt solutions containing 1 M KC1, 5 mM CaCl2, 1 mM EDTA, and either 20 mM MES (pH 6.2), 20 mM malic acid (pH 4.85 or 4.5), or 25 mM succinic acid (pH 4.0). The pH 6.2 solution also contained urea at molar concentrations of 0, 2.13, 2.83, 4.25, 6.38, or 8.5. The A-T experiments were done with 1 M KC1, 5 mM CaC12, 1 mM EDTA on both sides of the membrane and 20 mM MES, pH 6.2 in the cis solution (the side to which A-T was added) and 20 mM HEPES, pH 7.2 in the trans solution. MG-T experiments were done with 1 M KCl, 2 mM CaCl2, 1 mM EDTA on both sides of the membrane and 30 mM MES, pH 5.3, on the cis side and 50 mM HEPES, pH 7.2, on the trans side. Experiments were done under voltage-clamp conditions; the voltages are those of the cis solution with respect to the trans solution, whose potential was held at virtual ground. Currents were filtered at 100 Hz by the chart recorder.
Tryptophan fluorescence measurements
The tryptophan fluorescence measurements on the urea-dependent unfolding of diphtheria toxin A chain were made on a Shimadzu RF-5301PC spectrofluorophotometer using a 1-cm pathlength quartz cuvette. A chain with an N-terminal 19-residue His6-tag and a C186A substitution to avoid disulfide bond formation was prepared as described previously (27). Emission spectra were recorded from 310 to 500 nm using a 290 nm excitation. Both the excitation and emission slit widths were 3 nm, and all spectra were autozeroed at 500 nm. The protein concentration was 27 μg/ml in 1 M KCl, 20 mM MES, 5 mM CaCl2, 1 mM EDTA, pH 6.2 and 0–8.5 M urea, with a sample volume of 1 ml.
RESULTS
Colicin Ia experiments
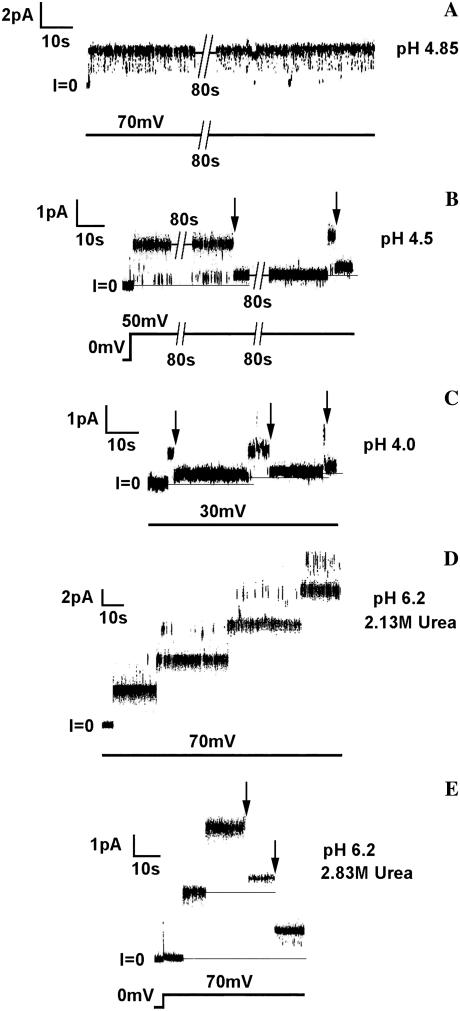
At pH 4.85 or higher, the conductance of the channel formed by the channel-forming part of colicin Ia with diphtheria toxin's A chain attached to its N terminus (A-Ia) was the same as that of the four-transmembrane-segment state of colicin Ia; transitions to the smaller conductance exhibited by the three-transmembrane-segment state rarely occurred (Fig. 4 A). In contrast, at pH 4.5, these transitions readily occurred (Fig. 4 B), and at pH 4.0, they were so rapid as to be sometimes almost unresolvable at our 100-Hz filtering (Fig. 4 C). (The record shown in Fig. 4 C was selected to illustrate representative resolvable transitions of varying durations.) The effect of pH on transitions is even more dramatic than it at first appears in Fig. 4. As was previously shown, the dwell time in the four-transmembrane-segment state becomes shorter as the voltage is increased (16). Thus, the rare transitions occurring at +70 mV at pH 4.85 (none is seen in Fig. 4 A) are in marked contrast to the readily occurring transitions at + 50 mV at pH 4.5 (Fig. 4 B) and are in even more striking contrast to the rapid transitions seen at + 30 mV at pH 4.0 (Fig. 4 C). In fact, at voltages above + 30 mV at pH 4.0, most of the transitions were too rapid to be resolved at 100-Hz filtering.
FIGURE 4.
Behavior of single channels formed by A-Ia under various conditions of pH and urea concentrations. In all experiments, the membrane separated symmetric solutions containing 1 M KC1, 5 mM CaC12, and 1 mM EDTA. In addition, the solutions contained (A) 20 mM potassium malate, pH 4.85; (B) 20 mM potassium malate, pH 4.5; (C) 25 mM potassium succinate, pH 4.0; (D) 20 mM MES, 2.13 M urea, pH 6.2; and (E) 20 mM MES, 2.83 M urea, pH 6.2. After the membrane formed, A-Ia was added to the cis solution at a concentration of: (A) 1 ng/ml; (B) 12 ng/ml; (C) 11 ng/ml; (D) 5 ng/ml; and (E) 7 ng/m1. The arrows mark the transitions from the four-transmembrane-segment state to the three-transmembrane-segment state (see Fig. 2). Note that although the dwell time in the four-transmembrane-segment state is voltage dependent, becoming shorter as the voltage is increased (16), we see in these representative records that there is no transition at +70 mV at pH 4.85 (A), but there are rapid transitions at only +30 mV at pH 4.0 (C). That is, the lower the pH, the more readily is the A chain translocated. Similarly, we see that at +70 mV and pH 6.2, there are no transitions observed in 2.13 M urea, even with four open channels (D), whereas each of the two channels that are open in 2.83 M urea undergoes transitions (E). (The rapid upward flickerings seen in the records, particularly at +70 mV, are to higher-conductance states and are typically seen with colicin Ia channels.) The records were filtered at 100 Hz by the chart recorder.
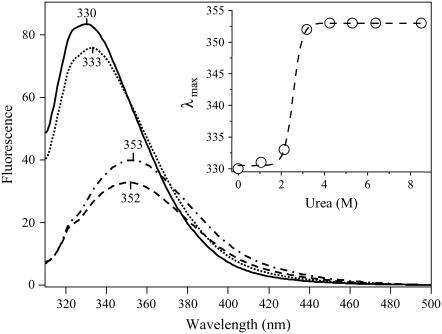
These results are explicable in terms of the known unfolding of the A chain at low pH (28): the translocation pathway created by the colicin Ia channel is large enough to allow passage through it of the unfolded A chain but not large enough to accommodate the folded chain. As an independent indication that the state of A chain folding is relevant to its translocation through the colicin Ia translocation pathway, we looked at the effect of a denaturant, urea, on translocation. At pH 6.2, A chain translocation did not occur in the absence of urea and only rarely (not seen in the figure) in 2.13 M urea (Fig. 4 D) but did occur frequently at urea concentrations of 2.83 M and above (Fig. 4 E). This correlates well with the observation that at pH 6.2, the free A chain is folded at urea concentrations of 0 to 2.13 M and completely unfolded at urea concentrations of 3.18 M and above, as measured by the wavelength shift of tryptophan fluorescence (Fig. 5).
FIGURE 5.
Urea titration of DTA (C186A mutant) monitored by tryptophan fluorescence. The exposure of DTA to increasing amounts of urea from 0 M to 8.5 M results in both a decrease in the fluorescence intensity and a red-shift of the emission maximum of the tryptophan fluorescence spectrum; both effects are consistent with the exposure of tryptophan residues to aqueous solvent because of the unfolding of DTA. Shown are illustrative spectra of DTA in 0 M (solid), 2.13 M (dots), 3.18 M (dash), and 8.50 M (dot-dash) urea. (Inset) Plot of the emission maximum as a function of urea concentration (open circles). The data are described (R2 = 0.999) by a sigmoidal function (dashed line) indicative of a two-state unfolding process. The midpoint of the transition is 2.6 M.
Diphtheria toxin experiments
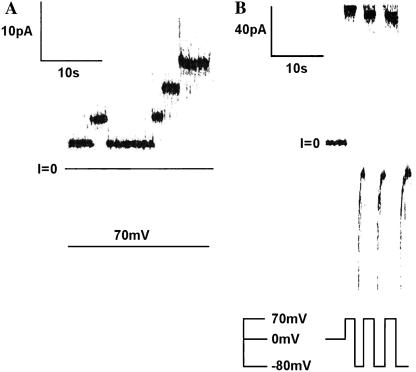
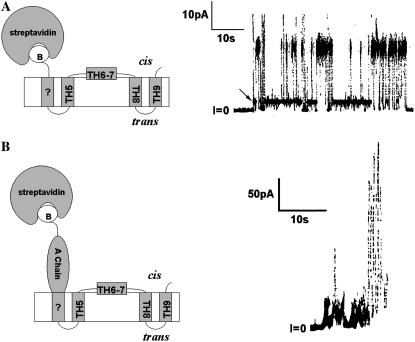
In contrast to the results just described for experiments with A-Ia, we have never seen transitions with A-T (the A chain attached to the N-terminus of the T domain) from a higher conductance state to the three-transmembrane-segment state, even at pH 6.2, where the A chain is folded; the channels always initially open to the three-transmembrane-segment state (Fig. 6 A). (The rapid channel closures at negative voltages (Fig. 6 B) are a consequence of the histidine tag at the N terminus of the A chain, which blocks the channels from the trans side and thus demonstrates that the A chain has been translocated to that side (3).) A possible explanation for this observation is that the T domain actually does initially form a four-transmembrane-segment channel before going to the three-transmembrane-segment channel, but that these two channel structures, unlike those formed by colicin Ia, have the same single-channel conductance. This explanation, however, is not tenable. If the large-diameter, tightly folded streptavidin, instead of the A chain, is attached to the N-terminus of the T domain, which forces the N-terminus to remain on the cis side, then one indeed sees a larger-conductance channel (Fig. 7 A), which presumably has four transmembrane segments. We have no direct evidence that this large-conductance channel has four transmembrane segments and make this presumption purely by analogy to our previous results with colicin Ia (17). Its transmembrane topography, however, is not relevant to the main conclusion that can be drawn from the experiment. Namely, if there were an observable intermediate channel state as the A chain translocates to the trans side, the conductance of that intermediate channel would not be the same as that of the final, three-transmembrane-segment channel. Interestingly, if streptavidin is attached to the N-terminus of A-T, an ill-defined, “junky” conductance results (Fig. 7 B). It is as if A chain insertion into the translocation pathway, without being able to fully translocate because it is trapped by streptavidin at its N-terminus, creates an unstable structure, or structures, instead of a stable channel state.
FIGURE 6.
Demonstration that channels formed by A-T do not initially open to a resolvable higher-conductance state (characterized by four transmembrane segments) before the translocation of the A chain, as does A-Ia, but rather open directly to the conductance characterized by three transmembrane segments, with the A chain already translocated to the trans side. (A) Single-channel record. Before the start of the record, A-T (with a His6-tag attached to the N-terminus) was added to the cis solution to a concentration of 0.64 μg/ml; one channel had already inserted before the voltage was stepped to +70 mV. Note that in contrast to the records shown in Fig. 4 for A-Ia, the channels do not initially open to a higher conductance before attaining their final value. The membrane separated symmetric solutions of 1 M KC1, 5 mM CaC12, 1 mM EDTA, 20 mM MES, pH 6.2. The record was filtered at 100 Hz by the chart recorder. (B) Macroscopic record demonstrating that the A fragment has been translocated to the trans side. Before the start of the record, A-T (with a His6-tag attached to the N-terminus) was added to the cis solution to a concentration of 10.7 μg/ml, and numerous channels had been inserted. The record shows rapid closure when the voltage was switched from +70 to −80 mV and rapid reopening when the voltage was switched back from −80 to +70 mV; this behavior is characteristic of that obtained with a His6-tag attached to an A chain that has been translocated to the trans side (3). The solutions on both the cis and trans sides of the membrane contained 1 M KC1, 5 mM CaC12, and 1 mM EDTA. During the insertion of the channels before the start of the record, the cis solution also contained 20 mM MES, pH 6.2, and the trans solution contained 5 mM HEPES, pH 7.2. The trans solution was then lowered to pH 5.6 with 5 mM potassium succinate, and the above record was obtained. (If A-T channels have inserted at cis pH 6.2, they do not show His6-tag gating if the trans pH is 7.2, or even 6.2. We hypothesize that this is because the A chain has been translocated in its folded state and in this state sterically inhibits the N-terminal His6-tag from interacting with the channel. By our lowering the trans pH to 5.6 after the A chain has been translocated, the A chain has unfolded sufficiently, we believe, to allow the N-terminal His6-tag to interact with the channel.) The record was filtered at 100 Hz by the chart recorder.
FIGURE 7.
Channel behavior when the N-terminus of either the T domain (A) or A-T (B) is forced to remain on the cis side. A cartoon for each of the resulting states is shown on the left side of the panel. (A) Channel formed by the T domain with streptavidin bound to the biotinylated cysteine in the N-terminal His6-tag. The cis solution contained 0.3 μg/ml of T domain whose N-terminal His6-tag was biotinylated and had been preincubated with streptavidin. Note the large size of this channel. (The arrow marks a substate having the same conductance under the pH conditions of the experiment as that of a channel without streptavidin attached. We speculate that the switches to a smaller conductance level seen in this trace result from the lateral movement within the bilayer of the fourth transmembrane segment away from the others, thereby generating a “normal” three-transmembrane-segment channel.) The topography of TH5 to TH9 in the cartoon is that of the “normal” channel (15); in the “normal” channel, everything N-terminal to TH5 resides on the trans side (Fig. 1). The “B” in the cartoon stands for biotin. The solutions on both the cis and trans sides of the membrane contained 1 M KC1, 2 mM CaC12, and 1 mM EDTA; the cis solution also contained 30 mM MES, pH 5.3, and the trans solution contained 50 mM HEPES, pH 7.2. The applied voltage is +60 mV. (From Gordon (33).) (B) Activity generated by A-T with streptavidin bound to a biotin at the N-terminus of the A chain. The cis solution contained 35 ng/ml A-T biotinylated at its N-terminus and 20 μg/ml streptavidin that was present before the addition of A-T. Note the erratic current fluctuations and lack of a clear channel conductance state. The membrane separated symmetric solutions of 1 M KC1, 5 mM CaC12, 1 mM EDTA, 20 mM potassium malate, pH 4.85. The applied voltage is +60 mV.
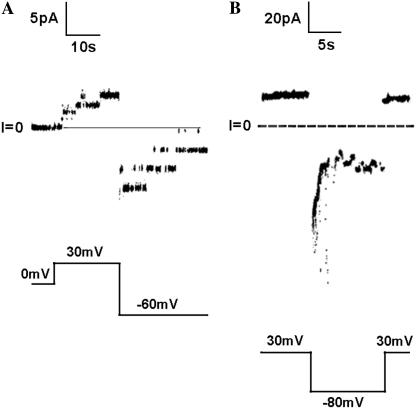
It is possible that the rapid translocation of the folded A chain by the T domain of diphtheria toxin (Fig. 6), in contrast to its inability to be translocated by the channel-forming domain of colicin Ia (Fig. 4, A and D), is a consequence of the T domain being optimized to specifically aid in the unfolding of the A chain; i.e., the T domain translocation pathway cannot, in general, accommodate much larger molecules than can the colicin Ia translocation pathway. This is unlikely, however, a priori, on two counts: first, in the crystal structure of diphtheria toxin, the A chain is folded (10), and second, in the whole toxin, the A chain retains its enzymatic activity (29), demonstrating that the T domain has not promoted its unfolding in free solution. This still leaves the possibility that when the T domain associates with a lipid bilayer, it specifically aids the unfolding of the A chain, and it is only the unfolded chain that can be translocated. This, too, is unlikely, given that the T domain's translocation pathway readily lets through myoglobin (Fig. 8, A and B) attached (by gene fusion) at the N-terminus of the T domain. The minimal cross-sectional diameter of 47 Å for myoglobin (30) is essentially the same as the 44 Å minimal cross-sectional diameter of the folded A chain (31). (The minimal cross-sectional diameter is defined as the diameter of the smallest circle that can be drawn around the projected image of the space-filling model of the molecule and was determined as described by Kienker et al. (17).)
FIGURE 8.
Demonstration that myoglobin (MG) attached to the N-terminus of the T domain (MG-T) is readily translocated across the membrane in association with channel formation. (A) Single-channel record. Before the start of the record, ∼25 μM MG-T with a His6-tag attached to the N-terminus of myoglobin was incubated (at pH 8.0) for 5 min at room temp with 500 μM hemin (added from a solution of 5 mM hemin in DMSO) to ensure the folded state of the protein (21) and was then added, after a 10-fold dilution, to the cis solution to a concentration of ∼0.15 nM. (The hemin concentration was therefore 3 nM.) The record begins 1 min after the addition. Single channels are seen to open at +30 mV. The channels open to a conductance (∼42 pS) comparable to that of T domain channels without MG attached to the N-terminus (25) and do not initially open to a higher conductance before attaining this value. When the voltage is switched to −60 mV, the channels close, consistent with the N-terminal His6-tag having been translocated across the membrane to the trans side (3). (B) Macroscopic record: the same experiment ∼9 min later, after enough channels have inserted into the membrane to give a macroscopic response. We see that the channels rapidly close when the voltage is switched from +30 mV to −80 mV and rapidly reopen when the voltage is switched back to +30 mV. This is the same behavior as seen with a His6-tag attached to an A chain that has been translocated to the trans side ((3); Fig. 6) and therefore confirms that the myoglobin has been translocated across the membrane. Solutions on both sides of the membrane contained 1 M KCl, 2 mM CaCl2, 1 mM EDTA; the cis solution contained 30 mM MES, pH 5.3; the trans solution contained 50 mM HEPES, pH 7.2. Records were filtered at 100 Hz by the chart recorder.
DISCUSSION
Both the channel-forming domain of colicin Ia (the ∼175-residue C-terminal end) and that of diphtheria toxin (its ∼175-residue T domain) form channels having three transmembrane segments (15,16), but whereas colicin Ia reaches this state by first passing through an observable channel state with four transmembrane segments ((16); Fig. 2), no such four-transmembrane-segment channel state is normally observed for diphtheria toxin. A possible explanation for this difference in behavior is that the translocation pathway created by the T domain is much wider than that created by colicin Ia, and hence, residues N-terminal to it, including the ∼200-residue A chain, do not get “hung up” in it but, rather, can rapidly pass through. The results presented in this article strongly support this explanation.
Although the translocation pathway of colicin Ia lets through the unfolded A chain, it does not let through the folded chain (Fig. 4). The rare translocation events that we have seen of the A chain through colicin Ia's translocation pathway at pH 4.85 and at 2.13 M urea (pH 6.2) are probably attributable to occasional transient unfoldings of the A chain under these conditions. In contrast, the T domain's translocation pathway readily lets through the folded A chain (Fig. 6). Before this work, it was not clear whether the A chain was folded or unfolded under the pH conditions in which we observed its translocation by diphtheria toxin (3). By using the colicin Ia translocation pathway as a measure of A chain folding in a membrane system, we have shown that, above pH 4.85, the A chain is folded (Fig. 4). The large size of the T domain's translocation pathway is consistent with its readily letting through myoglobin (minimal cross-sectional diameter 47 Å (30)) attached to the N-terminus of the T domain (Fig. 8) and is sufficient to account for its letting through the folded A chain (minimal cross-sectional diameter 44 Å (31)) without invoking any special molecular recognition of that entity. On the other hand, there is a limit to the size of a folded protein that can be translocated through the T domain's pathway. In particular, streptavidin (diameter 61 Å (32)) does not go through (Fig. 7 A).
The relation of the protein translocation pathways for diphtheria toxin and colicin Ia to their respective channels is unclear. There is no evidence at present that the channels themselves serve as the protein conduit. The size of the colicin Ia pathway appears to be voltage dependent, suggesting lipid involvement in its structure (17). Besides the differences in the sizes of their protein translocation pathways, there may be one other significant difference between the pathways for colicin Ia and diphtheria toxin. Namely, negative voltages can readily drive “stoppers” (covalently folded proteins and peptides) attached to the N-terminus of the colicin Ia channel-forming domain back from the trans side (to which side they were translocated) to the cis side (17); we have no indication that this is equally true for diphtheria toxin. That is, once the A chain has been translocated, it does not readily return to the cis side in response to large negative voltages.
Acknowledgments
We thank Dr. Stephen Juris for his generous gift of LFN-DTA (C186S) DNA, Dr. Paul Kienker for determining the minimal diameters of myoglobin and DTA, and Drs. Myles Akabas, Paul Kienker, and Stephen Slatin for their comments on the manuscript.
This work was supported by National Institutes of Health grant GM 29210 (A.F.), National Institutes of Health grant GM 31986 (E.L.), Medical Scientist Training Program grant GM07288 and Molecular Biophysics Training Grant (GM08572) at the Albert Einstein College of Medicine (B.S.S.-J.).
References
- 1.Gill, D. M. 1978. Seven toxic peptides that cross cell membranes. In Bacterial Toxins and Cell Membranes. J. Jaljaszewicz and T. Wadstrom, editors. Academic Press, London. 291–322.
- 2.Hazes, B., and R. J. Read. 1997. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry. 36:11051–11054. [DOI] [PubMed] [Google Scholar]
- 3.Oh, K. J., L. Senzel, R. J. Collier, and A. Finkelstein. 1999. Translocation of the catalytic domain of diphtheria toxin across planar phospholipid bilayers by its own T domain. Proc. Natl. Acad. Sci. USA. 96:8467–8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koriazova, L. K., and M. Montal. 2003. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat. Struct. Biol. 10:13–18. [DOI] [PubMed] [Google Scholar]
- 5.Zhang, S., E. Udho, Z. Wu, R. J. Collier, and A. Finkelstein. 2004. Protein translocation through anthrax toxin channels formed in planar lipid bilayers. Biophys. J. 87:3842–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoch, D. H., M. Romero-Mira, B. E. Ehrlich, A. Finkelstein, B. R. DasGupta, and L. L. Simpson. 1985. Channels formed by botulinum, tetanus, and diphtheria toxins in planar lipid bilayers: relevance to translocation of proteins across membranes. Proc. Natl. Acad. Sci. USA. 82:1692–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaustein, R. O., T. M. Koehler, R. J. Collier, and A. Finkelstein. 1989. Anthrax toxin: channel-forming activity of protective antigen in planar phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 86:2209–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silverman, J. A., J. A. Mindell, A. Finkelstein, W. H. Shen, and R. J. Collier. 1994. Mutational analysis of the helical hairpin region of diphtheria toxin transmembrane domain. J. Biol. Chem. 269:22524–22532. [PubMed] [Google Scholar]
- 9.Sellman, B. R., S. Nassi, and R. J. Collier. 2001. Point mutations in anthrax protective antigen that block translocation. J. Biol. Chem. 276:8371–8376. [DOI] [PubMed] [Google Scholar]
- 10.Choe, S., M. J. Bennett, G. Fujii, P. M. Curmi, K. A. Kantardjieff, R. J. Collier, and D. Eisenberg. 1992. The crystal structure of diphtheria toxin. Nature. 357:216–222. [DOI] [PubMed] [Google Scholar]
- 11.Wiener, M., D. Freymann, P. Ghosh, and R. M. Stroud. 1997. Crystal structure of colicin Ia. Nature. 385:461–464. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, M., and A. Finkelstein. 2001. The number of subunits comprising the channel formed by the T domain of diphtheria toxin. J. Gen. Physiol. 118:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slatin, S. L., and P. K. Kienker. 2003. Colicin channels and protein translocation. Parallels with diphtheria toxin.In Pore-Forming Peptides and Protein Toxins. G. Menestrina, M. Dalla Serra, and P. Lazarovici, editors. Taylor and Francis, London. 102–131.
- 14.Qiu, X. Q., K. S. Jakes, P. K. Kienker, A. Finkelstein, and S. L. Slatin. 1996. Major transmembrane movement associated with colicin Ia channel gating. J. Gen. Physiol. 107:313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senzel, L., M. Gordon, R. O. Blaustein, K. J. Oh, R. J. Collier, and A. Finkelstein. 2000. Topography of diphtheria toxin's T domain in the open channel state. J. Gen. Physiol. 115:421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kienker, P. K., K. S. Jakes, and A. Finkelstein. 2000. Protein translocation across planar bilayers by the colicin Ia channel-forming domain: where will it end? J. Gen. Physiol. 116:587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kienker, P. K., K. S. Jakes, R. O. Blaustein, C. Miller, and A. Finkelstein. 2003. Sizing the protein translocation pathway of colicin Ia channels. J. Gen. Physiol. 122:161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne, J. C., S. R. Blanke, P. C. Hanna, and R. J. Collier. 1995. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol. Microbiol. 15:661–666. [DOI] [PubMed] [Google Scholar]
- 19.Springer, B. A., and S. G. Sligar. 1987. High-level expression of sperm whale myoglobin in Escherichia coli. Proc. Natl. Acad. Sci. USA. 84:8961–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao, G., and E. London. 2005. Behavior of diphtheria toxin T domain containing substitutions that block normal membrane insertion at Pro345 and Leu307: control of deep membrane insertion and coupling between deep insertion of hydrophobic subdomains. Biochemistry. 44:4488–4498. [DOI] [PubMed] [Google Scholar]
- 21.Hargrove, M. S., and J. S. Olson. 1996. The stability of holomyoglobin is determined by heme affinity. Biochemistry. 35:11310–11318. [DOI] [PubMed] [Google Scholar]
- 22.Senzel, L., P. D. Huynh, K. S. Jakes, R. J. Collier, and A. Finkelstein. 1998. The diphtheria toxin channel-forming T domain translocates its own NH2-terminal region across planar bilayers. J. Gen. Physiol. 112:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schatz, P. J. 1993. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N. Y.). 11:1138–1143. [DOI] [PubMed] [Google Scholar]
- 24.Montal, M. 1974. Formation of bimolecular membranes from lipid monolayers. Methods Enzymol. 32:545–554. [DOI] [PubMed] [Google Scholar]
- 25.Huynh, P. D., C. Cui, H. Zhan, K. J. Oh, R. J. Collier, and A. Finkelstein. 1997. Probing the structure of the diphtheria toxin channel. Reactivity in planar lipid bilayer membranes of cysteine-substituted mutant channels with methanethiosulfonate derivatives. J. Gen. Physiol. 110:229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagawa, Y., and E. Racker. 1971. Partial resolution of the enzymes catalyzing oxidative phosphorylation. XXV. Reconstitution of vesicles catalyzing 32Pi triphosphate exchange. J. Biol. Chem. 246:5477–5487. [Google Scholar]
- 27.Hayashibara, M., and E. London. 2005. Topography of diphtheria toxin A chain inserted into lipid vesicles. Biochemistry. 44:2183–2196. [DOI] [PubMed] [Google Scholar]
- 28.Zhao, J. M., and E. London. 1988. Conformation and model membrane interactions of diphtheria toxin fragment A. J. Biol. Chem. 263:15369–15377. [PubMed] [Google Scholar]
- 29.Lory, S., S. F. Carroll, P. D. Bernard, and R. J. Collier. 1980. Ligand interactions of diphtheria toxin. I. Binding and hydrolysis of NAD. J. Biol. Chem. 255:12011–12015. [PubMed] [Google Scholar]
- 30.Urayama, P., G. N. Phillips, Jr., and S. M. Gruner. 2002. Probing substates in sperm whale myoglobin using high-pressure crystallography. Structure. 10:51–60. [DOI] [PubMed] [Google Scholar]
- 31.Bennett, M. J., S. Choe, and D. Eisenberg. 1994. Domain swapping: entangling alliances between proteins. Proc. Natl. Acad. Sci. USA. 91:3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freitag, S., T. Le, I. L. Klumb, P. S. Stayton, and R. E. Stenkamp. 1997. Structural studies of the streptavidin binding loop. Protein Sci. 6:1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon, M. 2002. The Topology and Molecularity of the Channel Formed by Diphtheria Toxin's T Domain or Fear and Loathing in da Bronx. PhD thesis. Albert Einstein College of Medicine, Bronx, NY.