Abstract
A primer design strategy named CODEHOP (consensus-degenerate hybrid oligonucleotide primer) for amplification of distantly related sequences was used to detect the priming glycosyltransferase (GT) gene in strains of the Lactobacillus casei group. Each hybrid primer consisted of a short 3′ degenerate core based on four highly conserved amino acids and a longer 5′ consensus clamp region based on six sequences of the priming GT gene products from exopolysaccharide (EPS)-producing bacteria. The hybrid primers were used to detect the priming GT gene of 44 commercial isolates and reference strains of Lactobacillus rhamnosus, L. casei, Lactobacillus zeae, and Streptococcus thermophilus. The priming GT gene was detected in the genome of both non-EPS-producing (EPS−) and EPS-producing (EPS+) strains of L. rhamnosus. The sequences of the cloned PCR products were similar to those of the priming GT gene of various gram-negative and gram-positive EPS+ bacteria. Specific primers designed from the L. rhamnosus RW-9595M GT gene were used to sequence the end of the priming GT gene in selected EPS+ strains of L. rhamnosus. Phylogenetic analysis revealed that Lactobacillus spp. form a distinctive group apart from other lactic acid bacteria for which GT genes have been characterized to date. Moreover, the sequences show a divergence existing among strains of L. rhamnosus with respect to the terminal region of the priming GT gene. Thus, the PCR approach with consensus-degenerate hybrid primers designed with CODEHOP is a practical approach for the detection of similar genes containing conserved motifs in different bacterial genomes.
The discovery of microbial compounds that positively modulate the biological response of immune cells and enhance the host's ability to resist microbial infection is a substantial challenge. Microbial polysaccharides have long been believed to have benign biological properties because they are considered to be classic T-cell-independent antigens that do not elicit cell-mediated immune responses. Recently, certain polysaccharides of microbial origin have been shown to act as potent immunomodulators with specific activity for both T cells and antigen-presenting cells, such as monocytes and macrophages (36). The use of immunomodulating agents provides distinct advantages over conventional therapies. For example, the enhancement of the host immune system's innate ability to combat bacterial infection might obviate the problems associated with antibiotic resistance.
Microbial exopolysaccharides (EPS) are either present as capsular polysaccharide associated with the cell surface or secreted as extracellular polysaccharide into the environment of the cell (34). EPS produced by lactic acid bacteria (LAB) have received increasing attention, mainly because of their generally-regarded-as-safe status (34), their rheological properties in food (4), and their potential beneficial properties for health, such as antiulcer activities (21) and immune stimulation (5, 22). The EPS production of Lactobacillus rhamnosus RW-9595M is among the highest measured for EPS-producing (EPS+) strains of LAB (7). The EPS from L. rhamnosus RW-9595M exhibited various abilities to stimulate proinflammatory cytokines in mouse splenocytes as well as in peripheral blood mononuclear cells from several healthy human donors. Under in vitro stimulation with the EPS, these peripheral blood mononuclear cells were able to elicit significant amounts of interleukin-12 (IL-12), a proinflammatory cytokine well known for its gamma interferon (IFN-γ)-inducing capacity. The EPS from L. rhamnosus RW-9595M seems to help a Th1 type of immune response. Th1 cytokines, IL-12, and IFN-γ appear to have prominent roles in cellular immunity that result in resistance to most infectious agents and reduce the manifestations of allergy (14). The potential use of L. rhamnosus and its EPS as probiotic and prebiotic in humans and animals, respectively, stimulates a growing need for fundamental knowledge of EPS biosynthesis, which is presently fairly limited.
Over the past few years, several studies have been initiated to further understand the molecular biology and genetics of EPS biosynthesis by LAB. Recent reports have characterized genes involved in polysaccharide synthesis for Streptococcus thermophilus (3, 9, 10, 11, 31, 32), Lactococcus lactis subsp. cremoris (13, 38, 39), and a few Lactobacillus species (4, 11). These studies revealed that EPS genetic determinants can be located either on a plasmid or on chromosomal DNA and that the organization of the EPS gene clusters appears to be divided into four regions (6, 31, 38). The first region contains regulatory genes, and the second codes for proteins proposed to be involved in determining polymer chain length. The fourth region encompasses genes involved in transport and polymerization. The third region contains genes similar to glycosyltransferases (GTs) specifically required for biosynthesis of the EPS repeating unit. The GTs assemble the EPS repeating unit by sequential transfer of nucleotide sugar residues onto a lipid carrier or onto a growing chain (25, 33, 42, 43). The first step in the assembly of the repeating unit is the transfer of a sugar-1-phosphate to a lipophilic carrier molecule that is anchored in the membrane. This step is achieved by the first GT, referred to here as the priming GT, which, unlike other GTs, recognizes a lipid carrier as well as the sugar residue and does not catalyze a glycosidic linkage. Moreover, the inactivation of the priming GT gene frequently leads to a dramatic decrease or an interruption of EPS production at the cell surface or released into the environment (18, 31, 38). Therefore, the priming GT gene product plays a key role in EPS biosynthesis. In general, most GT genes needed for the synthesis of the repeating unit are often unique or have little similarity to each other (16, 19, 24, 28, 31, 38, 42). However, genes encoding the priming GT found in various gram-positive and gram-negative bacteria are fairly similar, particularly in the carboxy terminus (42), which defines a conserved region present in a group of bacterial sugar transferases (Pfam accession no. PF02397).
In this study we applied a PCR strategy that uses hybrid oligonucleotide primers containing both a consensus and a degenerate region to detect the priming GT gene in strains of the Lactobacillus casei group. This approach has been used successfully to analyze members of multigene families as well as to isolate gene homologs from different genomes (26). However, to our knowledge this paper is the first to report the use of this strategy to find genes involved in an essential step of bacterial EPS biosynthesis. This method will facilitate the characterization of genetic determinants of EPS production in LAB as well as the screening and selection of EPS+ probiotic bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. The strain XL1-Blue MRA-P2 of Escherichia coli was grown in Luria-Bertani (LB) broth (Bioshop Canada, Burlington, Ontario, Canada) at 37°C with agitation at 200 rpm. Kanamycin was added to the LB broth at a concentration of 50 μg/ml when required for the selection of transformants. Strains of Lactobacillus spp. were grown at 37°C in MRS broth (Difco Laboratories, Detroit, Mich.), and strains of Streptococcus spp. were grown in M17 broth (Difco) at 37°C. All strains were subcultured twice and incubated between 18 and 24 h. Stock cultures were stored at −80°C in brain heart infusion broth (Difco) with 15% (vol/vol) glycerol. The EPS phenotype was evaluated by visual examination as well as by quantitative measurement of purified polymer (7).
TABLE 1.
Source of LAB strains used in this study, presence of the EPS phenotype, and detection of the priming GT gene with hybrid (G-∗-Bact-a-F-36 and G-∗-Bact-a-R-27) and specific primers (G-Lr-Bact-b-F-20 and G-Lr-Bact-b-R-20)
| Strain | Source or referencea | EPS phenotypeb | Detection with:
|
|
|---|---|---|---|---|
| Hybrid primers | Specific primers | |||
| S. thermophilus Sfi6 | Nestlé Culture Collection | + | + | − |
| S. thermophilus NCFB 2393 | NCIMB | + | + | − |
| S. thermophilus TA-040 | Rhône-Poulenc Canada | + | + | − |
| S. thermophilus FYE-41 | Rhône-Poulenc Canada | + | + | − |
| L. casei ATCC 334 | ATCC | − | − | − |
| L. casei ATCC 335 | ATCC | − | + | − |
| L. casei ATCC 4007 | ATCC | − | − | + |
| L. casei ATCC 4646 | ATCC | − | + | + |
| L. casei ATCC 4913 | ATCC | − | − | − |
| L. casei ATCC 4940 | ATCC | − | − | + |
| L. casei ATCC 11578 | ATCC | − | − | + |
| L. casei ATCC 11582 | ATCC | − | − | − |
| L. casei ATCC 11974 | ATCC | − | − | − |
| L. casei ATCC 25180 | ATCC | − | − | − |
| L. casei ATCC 25302 | ATCC | − | + | − |
| L. casei ATCC 25303 | ATCC | − | + | + |
| L. casei ATCC 27092 | ATCC | − | + | − |
| L. casei ATCC 27216 | ATCC | − | + | − |
| L. casei ATCC 29599 | ATCC | − | − | − |
| L. casei ATCC 39392 | ATCC | − | − | − |
| L. casei ATCC 39539 | ATCC | − | + | − |
| L. casei DSM 20207 | DSMZ | − | + | + |
| L. casei DSM 20244 | DSMZ | − | + | − |
| L. casei RW-3703M | D. Roy (FRDC) | + | + | + |
| L. casei type V | Institut Rosell | + | + | + |
| L. zeae ATCC 393 | ATCC | − | − | − |
| L. zeae ATCC 15820 | ATCC | − | − | − |
| L. rhamnosus ATCC 7469 | ATCC | − | + | + |
| L. rhamnosus ATCC 8530 | ATCC | − | + | + |
| L. rhamnosus ATCC 9595 | ATCC | − | + | + |
| L. rhamnosus ATCC 10863 | ATCC | − | + | + |
| L. rhamnosus ATCC 11443 | ATCC | − | + | + |
| L. rhamnosus ATCC 11981 | ATCC | − | + | + |
| L. rhamnosus ATCC 11982 | ATCC | − | + | + |
| L. rhamnosus ATCC 12116 | ATCC | + | + | + |
| L. rhamnosus ATCC 14957 | ATCC | − | + | + |
| L. rhamnosus ATCC 15008 | ATCC | − | + | + |
| L. rhamnosus ATCC 21052 | ATCC | + | + | + |
| L. rhamnosus ATCC 27773 | ATCC | + | + | + |
| L. rhamnosus ATCC 39595 | ATCC | + | + | + |
| L. rhamnosus ATCC 53103 | ATCC | − | + | − |
| L. rhamnosus R | Institut Rosell | + | + | + |
| L. rhamnosus RW-6541M | D. Roy (FRDC) | + | + | + |
| L. rhamnosus RW-9595M | D. Roy (FRDC) | + | + | + |
| E. coli XL1-Blue MRA-P2 | Stratagene | − | − | − |
Institutional names (and locations): Nestlé Culture Collection (Lausanne, Switzerland), NCIMB (National Collections of Industrial and Marine Bacteria, Aberdeen, Scotland, United Kingdom), Rhône-Poulenc Canada (Mississauga, Ontario, Canada), Institut Rosell (Montreal, Quebec, Canada), ATCC (American Type Culture Collection, Manassas, Va.), DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany), FRDC (Food Research and Development Centre, St.-Hyacinthe, Quebec, Canada), and Stratagene (La Jolla, Calif.).
The EPS+ phenotype was attributed by visual examination or by quantification after purification.
DNA purification.
Genomic DNA of bacterial strains was prepared according to Vincent et al. (41) from stationary-phase cultures in MRS, M17, or LB broth. The concentration of the purified DNA was determined by either a fluorometer TKO-100 or DyNA Quant 200 (Hoefer, San Francisco, Calif.) in capillary tubes with the Hoechst 33528 dye.
DNA amplification by PCR with hybrid primers.
Sequences of genes encoding the GT catalyzing the first step in polysaccharide biosynthesis were chosen from the GenBank databases. The Block Maker program located at the web site of the Fred Hutchinson Cancer Research Center in Seattle, Wash. (http://blocks.fhcrc.org/blocks/blockmkr/make_blocks.html), was used to generate blocks of similar amino acids of six selected priming GT proteins: Streptococcus agalactiae (CpsE, accession no. Q04664), Streptococcus thermophilus Sfi6 (EpsE, AAC44012), S. thermophilus NCFB 2393 (CpsE, CAA64433), L. lactis subsp. cremoris NIZO B40 (EpsD, AAC45231), Synechocystis spp. strain PCC6803 (RfbP, BAA18441), and Sinorhizobium meliloti Rm2011 (ExoY, AAA26264). The blocks created by MOTIF from these distantly related sequences were imported into the CODEHOP program (consensus degenerate hybrid oligonucleotide primer) with a direct link at the same site (26).
Each hybrid primer consisted of a short 3′ degenerate core region based on four highly conserved amino acids and a longer 5′ consensus clamp region. The primers were designed by taking into account information on L. rhamnosus codon usage. For the forward (G-*-Bact-a-F-36) and the reverse (G-*-Bact-a-R-27) hybrid primers, the four highly conserved amino acids were DELP and WQVS, respectively, located in the carboxy terminus of the priming GT protein (Table 2).
TABLE 2.
Hybrid and specific oligonucleotide primers used in amplification by PCR of the priming GT of the eps gene cluster in streptococci and lactobacilli speciesa
| Primer | Sequence (5′-3′) | Length (bp) | Annealing temperature (°C) |
|---|---|---|---|
| Hybrid | |||
| G-∗-Bact-a-F-36 | TCATTTTATTCGTAAAACCTCAATTGAYGARYTNCC | 36 | 52 |
| G-∗-Bact-a-R-27 | AATATTATTACGACCTSWNAYYTGCCA | 27 | 52 |
| Specific | |||
| G-Lr-Bact-a-F-26 | ATGAGTTTGGTTGGACCAAGACCTCC | 26 | 58 |
| G-Lr-Bact-b-F-20 | TTGCCAAATATTGGAGGGGT | 20 | 58 |
| G-Lr-Bact-b-R-20 | TTTAATAGGCTCCAGTTGGA | 20 | 58 |
F indicates a forward primer, while R indicates a reverse primer. Bold characters in primers highlight the degenerate region.
The PCR conditions for the hybrid primers were 1× PCR buffer II containing 4 mM MgCl2 (ABI, Perkin-Elmer, Foster City, Calif.), 200 μM deoxynucleoside triphosphate, 50 pmol of each primer, and 2.5 U of AmpliTaq Gold (Perkin-Elmer) in a 50-μl reaction volume. Amplification reactions were performed in a GeneAmp PCR system 2400 (Perkin-Elmer). The PCR program consisted of 45 cycles, after an initial incubation at 94°C for 9 min to activate the AmpliTaq enzyme and to allow complete denaturation of the DNA template. The first 5 cycles consisted of a denaturation step at 94°C for 30 s, an annealing step at 62°C for 30 s, and an elongation step at 72°C for 30 s. The last 40 cycles consisted of a denaturation step at 94°C for 30 s, an annealing step at 52°C for 30 s, an elongation step at 72°C for 30 s, and a final elongation step at 72°C for 10 min. After amplification all PCR products were conserved at 4°C. Agarose gel electrophoresis and staining were carried out with standard procedures (35). Amplified products were purified by using the QIAquick Gel Extraction kit (Qiagen, Chatsworth, Calif.) in accordance with the manufacturer's recommendations.
DNA amplification by PCR with specific primers.
A set of specific primers for L. rhamnosus, G-Lr-Bact-a-F-20 and G-Lr-Bact-b-R-20, were designed from the sequence for L. rhamnosus strain RW-6541M (GenBank accession no. AF384150) in order to amplify the entire putative undecaprenyl-phosphate glycosyl-1-phosphate transferase (or priming GT) gene in strains of the L. casei group (Table 2). In addition, primer G-Lr-Bact-a-F-26 was designed to amplify the 276-bp 3′ region of the gene when used with primer G-Lr-Bact-b-R-20 (Table 2).
PCR conditions were 1× PCR buffer containing 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate, 20 pmol of each primer, and 2 U of Taq polymerase (Pharmacia, Montreal, Quebec, Canada) in a 50-μl reaction volume overlaid with mineral oil. Reactions were performed in a Perkin-Elmer GeneAmp 9600 PCR system. The PCR program consisted of 35 cycles, after an initial incubation at 94°C for 9 min. The cycles consisted of a denaturation step at 94°C for 30 s, an annealing step at 58°C for 30 s, an elongation at 72°C for 30 s, and a final elongation step at 72°C for 10 min. After amplification, all PCR products were conserved at 4°C. Amplified products were visualized by agarose gel electrophoresis, and fragments were purified by using the QIAquick gel extraction kit (Qiagen) in accordance with the manufacturer's recommendations.
DNA sequencing and analysis.
Purified PCR products were cloned in the vector pCR 2.1-TOPO by using the TOPO TA Cloning kit (Invitrogen, Carlsbad, Calif.) with chemically competent cells in accordance with the manufacturer's recommendations. Plasmids from transformants purified with the QIAprep Spin Miniprep kit (Qiagen) were used for automated sequencing. The sequences on both strands from two clones were then determined with M13 forward and reverse primers by the DNA sequencing service of the Life and Health Sciences Pavilion of Laval University (Quebec, Canada).
Nucleotide and amino acid sequence analyses were performed with either the OMIGA version 2.0 software (Oxford Molecular, Madison, Wis.) or programs accessible on the internet. Translation of nucleotide sequences was performed by using the OMIGA software or the ExPASy translate routine at the ExPASy Molecular Biology Server of the Swiss Institute of Bioinformatics (http://ca.expasy.org/). Similarity searches were performed with the Advanced BLAST algorithm (1, 2) available at the National Center for Biotechnology Information site (http://www.ncbi.nlm.nih.gov/) and the FASTA algorithm version 3.0 (23) from the European Bioinformatics Institute (http://www.ebi.ac.uk/fasta33/index.html). Sequence alignments were conducted with the ClustalW (35) algorithm as implemented in the OMIGA software or at the ClustalW server at the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/index.html).
Phylogenetic analysis.
Multiple sequences from the conserved C-terminal region of the priming GT gene of various EPS+ strains of gram-negative and gram-positive bacteria were aligned and boxed by using the ClustalW program (35) and the Boxshade program, respectively, from the European Molecular Biology network (http://www.ch.embnet.org/software/BOX_form.html). The phylogenetic tree was created by the neighbor-joining method of Saitou and Nei (27) and the p-distance measure and pairwise deletion as implemented in the MEGA program (17). The bootstrap method (8) was employed to determine the statistical confidence of the phylogenetic relationships. A total of 500 bootstrap trees were generated for each data set.
Nucleotide sequence accession numbers.
The sequences obtained in this study are available under GenBank accession nos. AF323521 to AF323529.
RESULTS
Detection of priming GT genes with hybrid primers.
Hybrid primers for the priming GT gene were designed with the CODEHOP program (26) by using sequences of genes encoding the GT catalyzing the first step in polysaccharide synthesis from six EPS+ strains of gram-negative and gram-positive bacteria, targeting the conserved C-terminal portion (15, 42). The detection of the priming GT gene was conducted with DNA isolated from 40 commercial isolates and reference strains of L. rhamnosus, L. casei, and L. zeae (Table 1). Two strains, S. thermophilus Sfi6 (31) and S. thermophilus NCFB 2393 (10), were used as positive controls, since their sequences were used to design the hybrid primers for the priming GT gene. Two other commercial isolates of S. thermophilus were also tested (Table 1). A PCR product of the expected size of 189 bp was amplified for all 4 strains of S. thermophilus, 11 strains of L. casei, and 17 strains of L. rhamnosus, but not for 10 strains of L. casei and 2 strains of L. zeae (Table 1). The PCR products obtained for streptococci-positive controls corresponded to the expected size according to the position of the primers in the holotype genes.
Detection of putative priming GT genes with specific primers.
In order to validate the hybrid primer strategy, specific primers were designed from the RW-6541M GT gene (GenBank accession no. AF384150; Table 2). By using the specific primers G-Lr-Bact-a-F-26 and G-Lr-Bact-b-R-20, the 276-bp PCR products obtained from eight strains of L. rhamnosus and one strain of L. casei were sequenced and revealed to be identical to those obtained with the hybrid primers over the region covered by both PCR approaches (GenBank accession nos. AF323521 to AF323529). In addition, to detect the presence of the putative undecaprenyl-phosphate glycosyl-1-phosphate transferase gene in strains of the L. casei group, specific primers (G-Lr-Bact-b-F-20 and G-Lr-Bact-b-R-20) were also designed from the RW-6541M GT gene (AF384150; Table 2). A PCR product of 694 bp was amplified for all strains of L. rhamnosus that were positive with the hybrid primers, except for strain ATCC 53103. In addition, eight strains of L. casei exhibited a PCR product, while no strain of S. thermophilus or L. zeae gave positive results with these specific primers (Table 1).
Sequence analysis of putative priming GT genes.
Strong similarity was found with genes that have been postulated or established to encode GTs catalyzing the first step of polysaccharide biosynthesis in various EPS+ strains of gram-positive and gram-negative bacteria. For S. thermophilus, the priming GT sequences determined in this study for strains Sfi6 and FYE-41 are 100% identical to the priming GT (epsE) sequence isolated and characterized by Stingele et al. (31). This confirms that the hybrid primers detected the sequence of the priming GT gene in the positive control strain Sfi6 and reveals that the sequence from strain FYE-41 is identical to that from strain Sfi6. The predicted amino acid sequences from strain TA-040 of S. thermophilus only show 50% identity with those of S. thermophilus Sfi6 (Table 3). However, TA-040 shows greater identity (90 to 95%) with strains of Streptococcus pneumoniae type 19F, S. agalactiae type Ia, and Streptococcus suis type 2 (Table 3).
TABLE 3.
Comparison of predicted amino acid sequences of priming-like GT genes of streptococci (for 43 residues) and lactobacilli (for 91 residues) with selected database entriesa
| Strain or group | % Identity/% similarity of predicted sequences
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Efa2957 CpsD | Lgas 0005 | Lheeps1 Eps1 | LbuLfi5 EpsE | Ooen 0781 | ScoA3 SCF62.07 | Blo0237 CpsD | Ssu2E Cps2E | Saglae CpsIaE | Spn19f CpsI9fE | Spn14E Cps14E | SthNCFB CpsE | SthSfi6 EpsE | Sen WbaP | LerB40 EpsD | Xca GumD | LrhX202 Cap5M | Sau5M Cap5M | Sme ExoY | LcrB35 EpsD | |
| S. thermophilus | ||||||||||||||||||||
| TA-040 | 59/67 | 63/70 | 62/67 | 60/72 | 65/72 | 58/67 | 57/66 | 90/95 | 90/93 | 88/90 | 90/95 | 95/97 | 50/59 | 58/67 | 57/64 | 46/53 | 46/62 | 45/63 | 44/60 | 40/57 |
| FYE-41 | 52/64 | 52/61 | 38/57 | 52/66 | 52/61 | 54/64 | 45/59 | 50/64 | 52/60 | 53/60 | 52/64 | 50/57 | 100/100 | 54/66 | 53/62 | 41/54 | 33/41 | 43/63 | 46/66 | 46/67 |
| L. rhamnosus | ||||||||||||||||||||
| Group 1 | 58/76 | 67/79 | 69/73 | 62/77 | 55/71 | 56/71 | 51/65 | 54/73 | 53/72 | 52/72 | 51/71 | 52/72 | 46/64 | 53/67 | 36/57 | 35/53 | 34/53 | 36/59 | 45/62 | 33/58 |
| Group 2 | 57/77 | 65/77 | 67/82 | 61/76 | 54/72 | 55/72 | 52/67 | 53/72 | 53/72 | 52/73 | 51/71 | 52/73 | 46/65 | 52/65 | 35/56 | 34/53 | 33/52 | 36/59 | 44/62 | 35/54 |
| L. casei Type V | 62/78 | 69/78 | 68/82 | 62/75 | 59/71 | 54/63 | 48/62 | 53/73 | 56/67 | 49/70 | 49/69 | 51/68 | 47/66 | 49/63 | 38/58 | 34/53 | 35/53 | 36/57 | 43/61 | 32/53 |
Data are presented as % identity/% similarity. Group 1 represents the identical sequence of strains ATCC 12116, ATCC 27773, ATCC 9595, ATCC 7469, RW-9595M, and R. Group 2 consists of strains ATCC 21052 and RW-6541M. Abbreviations: Efa2957, E. faecium (ZP_00038042.1); Lgas005, L. gasseri (ZP_00045843.1); Lheeps1, L. helveticus (CAC07462.1); LbuLfi5, L. delbrueckii subsp. bulgaricus Lfi5 (AAG44709.1); Ooen, O. oeni MCW (ZP_00069765.1); ScoA3, S. coelicolor A3(2) (NP_624703.1); Blo0237, B. longum NCC 2705 (NP_695447.1); Ssu2E, S. suis 2E (AAD24451.1), Saglae, S. agalactiae Iae (BAA82279.1); Spn19f, S. pneumoniae 19f (AAC44962.1); Spn14E, S. pneumoniae 14 (CAA59777.1); SthNCFB, S. thermophilus NCFB 2393 (JC5726); SthSfi6, S. thermophilus Sfi6 (AAC44012.1); Sen, Salmonella enterica (CAA43081.1); LcrB40, L. lactis subsp. cremoris NIZO B40 (NP_053030.1); Xca, X. campetris (CAA49577.1); LrhX202, L. rhamnosus X202 (AAG01983.1); Sau5M, S. aureus Cap5M (NP_370685.1); Smo, S. meliloti (AAA26264.1); LcrB35, L. lactis subsp. cremoris NIZO B35 (AAD22526.1).
Among L. rhamnosus strains, two groups can be distinguished by their identical nucleotide sequences. Group 1 consists of strains ATCC 12116, ATCC 27773, ATCC 9595, ATCC 7469, RW-9595M, and R, while group 2 consists of strains ATCC 21052 and RW-6451M. The 3′ region of the priming GT gene of these two groups differs at only four nucleotide positions. The predicted amino acid sequences are 97% identical. Two amino acid changes are conservative, thus giving 100% similarity between the two groups. The predicted priming GT sequence from L. casei Type V is 79% identical to the consensus sequence of group 1 and is 76% identical to that of group 2 L. rhamnosus strains.
In comparing the putative priming GT sequences from group 1 only with those from the databases, an identity of 69% was found with the Eps1 gene product of Lactobacillus helveticus LH59, of 62% with the EpsE protein of Lactobacillus delbrueckii subsp. bulgaricus Lfi5, and of 56% with SCF62.07 of Streptomyces coelicolor A3 (Table 3). Moreover, an identity of 50% or higher was observed with the priming GT sequences of many streptococci (Table 3). In contrast, the highly similar sequences from all strains of L. rhamnosus characterized in this study showed only 34% identity with the EpsC gene product of L. rhamnosus X202. Comparable low identity was also found with priming GT sequences from L. lactis subsp. cremoris (strains B40 and B35), Staphylococcus aureus (Cap5M), and Xanthomonas campestris (GumD).
Two blocks of high similarity in the C terminus of the priming GT gene product are conserved in L. rhamnosus and L. casei sequences (Fig. 1). Moreover, the search in the Conserved Domain Database with the CD Search program at the National Center for Biotechnology Information web site (2) revealed the presence of a partial bacterial sugar transferase domain. This Pfam family contains a conserved region from 52 bacterial sugar transferases involved in different biosynthetic pathways (Pfam accession no. PF02397).
FIG. 1.
Alignment of the complete C-terminal region of the priming GT gene products. Each superscript number corresponds to sequences found in the tree (see Fig. 2). The boxed sequence identifies amino acids that are identical (black shading) or functionally similar (gray shading). Sequences under the labels B and C correspond to the conserved blocks previously described (40, 42). The asterisks indicate the glutamate (E) and the aspartate (D) residues proposed to be the catalytic residues of the GT. Sequences with superscripts 1 to 9 are from this study, and the remainder are from the GenPept database. Lrh121161, L. rhamnosus ATCC 12116 (accession no. AAG38614.1); Lrh95952, L. rhamnosus ATCC 9595 (AAG38620.1); LrhR3, L. rhamnosus R (AAG38618.1); Lrh277734, L. rhamnosus ATCC 27773 (AAG38615.1); Lrh74695, L. rhamnosus ATCC 7469 (AAG38621.1); Lrh9595M6, L. rhamnosus RW-9595M (AAG38619.1); Lrh210527, L. rhamnosus ATCC 21052 (AAG38616.1); LrhRW6541M8, L. rhamnosus RW-6541M (AAG38617.1); LcaTypeV9, L. casei Type V (AAG38622.1); Lgas000510, L. gasseri (ZP_00045843.1); Efae295711, E. faecium (ZP_00038042.1); Lheeps112, L. helveticus (CAC07462.1); LbuLfi513, L. delbrueckii subsp. bulgaricus Lfi5 (AAG44709.1); Ooen078114, O. oeni MCW (ZP_00069765.1); Sag15, S. agalactiae CpsD (BAA33745); Spn19f16, S. pneumoniae Cps19fE (AAC44962.1); SthTA04017, S. thermophilus TA-040 (AAG38623.1); SthSfi3918, S. thermophilus Sfi39 (AAK61899.1); SthNCFB19, S. thermophilus NCFB 2393 (JC5726); Blo023720, B. longum NCC 2705 (NP_695447.1); BloRfbP21, B. longum NCC 2705 (NP_695455.1); Xcam22, X. campestris GumD (CAA49577.1); LrhX20223, L. rhamnosus X202 EpsC (AAG01983.1); Efae131424, E. faecium (ZP_00036441.1); LcrB89125, L. lactis subsp. cremoris NIZO B891 (AAD22533.1); LcrB4026, L. lactis subsp. cremoris NIZO B40 (NP_053030.1); LcrB3527, L. lactis subsp. cremoris NIZO B35 (AAD22526.1); SthFYE4128, S. thermophilus FYE-41 (AAG38624.1); SthCNRZ29, S. thermophilus CNRZ 368 (CAB52240.1); SthMR-1C30, S. thermophilus MR-1C (AAC31163.1); SthSfi631, S. thermophilus Sfi6 (AAC44012.1).
Phylogenetic analysis.
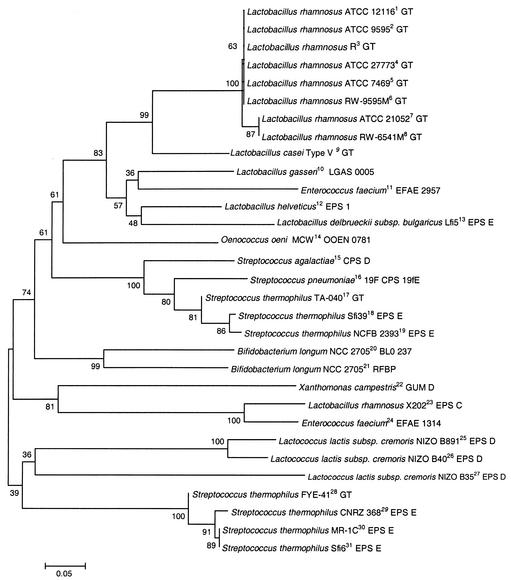
The alignment of the predicted amino acid sequence of the C-terminal region of the priming GT gene products was used to generate a phylogenetic tree (Fig. 2). Lactobacilli sequences fall into two groups (Fig. 2). The first group, including all but one Lactobacillus strain, also includes a sequence from Oenococcus oeni and one from Enterococcus faecium. L. rhamnosus X202 is the only Lactobacillus strain clustered with X. campestris, along with a second predicted GT gene product from the genome of E. faecium (Efae1314). S. thermophilus priming GT sequences also fall into two major groups. Predicted gene products from three strains were grouped with pathogenic streptococci, such as S. agalactiae and S. pneumoniae. Four other S. thermophilus priming GT gene products were grouped with sequences from L. lactis. The Bifidobacterium longum genome sequence carries two potential priming GT genes forming an intermediate group.
FIG. 2.
Phylogenetic tree of the C terminus of the priming GTs of various gram-negative and gram-positive bacteria, generated by the MEGA program. The number associated with the branches refer to bootstrap values (confidence limits) resulting from 500 replicate resamplings. The scale represents the number of amino acid substitutions per site. Sequences with superscripts 1 to 9 are from this study, and the remainder are from the GenPept database (see the legend of Fig. 1 for GenBank accession numbers).
DISCUSSION
The addition of a monosaccharide to a lipid carrier by the priming GT is thought to be a key step in EPS biosynthesis. Previous studies have indicated that inactivation of the priming GT gene alters or interrupts the production of EPS, suggesting the important role of this enzyme in EPS biosynthesis (18, 31, 38). However, detection of this gene is difficult considering that little similarity exists between genes encoding similar functions from distantly related species. Two strategies can be applied to identify distantly related sequences by PCR, by using either consensus or degenerate primers. However, single consensus primers targeting highly conserved regions do not account well for codon usage and are thus most useful for more closely related sequences (26). Degenerate primers were used successfully to identify the priming GT gene, aceA, which is likely to encode the phosphate-prenyl glucose-1-phosphate transferase catalyzing the first step in acetan biosynthesis in Acetobacter xylinum (11). Degenerate primers have the disadvantage that, in order to detect divergent genes, increased degeneracy leads to decreasing the quantity of each primer in the population and to a corresponding lower proportion of the proper primer. Thus, artifactual amplification results from nonspecific priming and low-stringency annealing conditions. The use of consensus-degenerate hybrid primers conceived with the CODEHOP program was designed to overcome these drawbacks (26). Degeneracy is reduced while a conserved region is provided in order to increase events of specific amplification.
In the present study the PCR strategy with hybrid primers was based on four highly conserved amino acids and allowed the detection of the priming GT of 17 strains of L. rhamnosus as well as of other LAB strains, such as L. casei and S. thermophilus. Sequence analysis revealed similarities to the GTs catalyzing the transfer of the first monosaccharide of the EPS repeating unit to a lipid carrier molecule of various EPS+ strains.
The hybrid primers amplified a sequence of 189 bp from strains of the L. casei group. Among these strains, only L. rhamnosus RW-9595M, RW-6541M, ATCC 12116, ATCC 27773, ATCC 21052, ATCC 39595, and R and L. casei Type V and RW-3703M were able to produce a measurable amount of EPS. These results are in agreement with those of Bourgoin et al. (3), who found that the eps locus was detected by hybridization in all S. thermophilus strains, even if only 3 of the 16 strains tested were found to be ropy in milk. The authors proposed that undetectable EPS production or EPS production under other conditions cannot be excluded in strains found to be nonropy (3). Further experimentation by reverse transcription-PCR needs to be carried out in order to clarify the expression of these genes in relation to the level of EPS production.
The specific primers were designed from an L. rhamnosus sequence so that the absence of an amplicon for S. thermophilus was expected. The absence of an amplicon for 13 out of 21 L. casei strains examined indicates divergence from the L. rhamnosus sequence and thus is a good potential source of different priming GT enzymes in lactobacilli of this group.
The sequences obtained for the 3′ region of the priming GT gene of eight strains of L. rhamnosus amplified with the specific primers are identical, with the exception of L. rhamnosus strain ATCC 21052 and RW-6541 M, which showed only four different nucleotides. Such high similarity should be correlated with an identical sugar specificity for the enzyme. Van Calsteren et al. (37) have determined the sugar composition and the structure of the EPS repeating unit of L. rhamnosus strains RW-9595M, ATCC 12116, ATCC 27773, ATCC 21052, RW-6541M, and R. For all strains, the EPS was composed of rhamnose, glucose, galactose, and pyruvate in a proportion of 4:2:1:1. They all possess an identical sugar composition and an identical EPS repeating unit. The sequence identity observed for the C terminus of the priming GT gene product in L. rhamnosus thus likely reflects the identity found for the first sugar in the EPS repeating unit structure. However, even though they show identical traits, their respective EPS production varies greatly among strains. For example, strain RW-9595M produced 1,275 mg/liter in basal minimal medium (BMM) while strain R produced 600 mg/liter (7).
In contrast to the high homology among L. rhamnosus strains, the priming GT sequence obtained for L. casei Type V showed a higher divergence from L. rhamnosus. However, L. casei Type V is more similar to L. rhamnosus than to the strains L. helveticus LH59 or L. delbrueckii subsp. bulgaricus Lfi5 characterized by Germond et al. (9). This similarity corresponds with the close relationship between L. casei and L. rhamnosus as determined by 16S ribosomal DNA sequence homology (30).
Phylogenetic analysis of the conserved C-terminal region of the priming GT gene product among EPS+ strains of gram-negative and gram-positive bacteria revealed that all lactobacilli except one are grouped and form a distinctive branch from the other LAB strains. However, the exception is the EpsC protein of L. rhamnosus X202, which appears closer to the GumD protein of X. campestris and to a second GT from E. faecium than to the sequences of other lactobacilli. The streptococcal sequences also form two groups, one group of which includes the pathogenic streptococci. Divergence within the same species has been observed in S. pneumoniae, where two classes of cpsC, cpsD, and cpsE genes were found among capsule loci belonging to several serotypes (20). Moreover, from 19 serotypes of pneumococci expressing glucosyltransferase activity, only 7 hybridized strongly with the Cps14E of S. pneumoniae serotype 14 (16). Thus, pneumococci possess glucosyltransferase genes distinct from the cps14E but encoding enzymes with similar activity (16). Furthermore, based on the genetic and biochemical diversity of the putative GT genes and the EPS, 16 different ropy L. lactis strains could be classified into four groups (40). A similar polymorphism has been observed in the central region of the guaA gene encoding a GMP synthetase in several closely related L. rhamnosus strains, such as L. rhamnosus X202 and C83 (12). The variability is also observed in other species closely related to L. rhamnosus, such as L. casei. In addition, more than one potential priming GT may be present in a genome, as revealed for E. faecium and B. longum. In each case, one gene may be associated with other genes involved in the production of EPS, while the other may be found with genes involved in cell wall biosynthesis.
The partial block B and the complete block C found in the priming GT of L. rhamnosus and L. casei were proposed by Wang et al. (42) to interact with the lipid carrier and to be responsible for sugar specificity, respectively. As reported by Stingele et al. (32), no sugar specificity motifs could be detected for galactosyltransferases or glucosyltransferases among EPS+ species of gram-negative or gram-positive bacteria. However, van Kranenburg et al. (40) noted a conserved tyrosine (Y) residue (position 26) in block C of priming galactosyltransferase sequences that was absent in glucosyltransferases. This particular tyrosine is replaced by phenylalanine (F) in the L. rhamnosus priming GT sequence, which places it with the enzymes for which a glucose specificity has been determined experimentally. Hydrophobic cluster analysis of various β-glycosyltransferases has shown two aspartic acid residues with a spacing of approximately 50 amino acids to be conserved, and these are predicted to be the catalytic residues for enzymes catalyzing a glycosidic linkage between two sugars (29). A glutamate residue (E) and an aspartate residue (D), separated by 50 amino acids and conserved among gram-positive GTs, have been proposed as two possible candidates for the catalytic residues in the priming GT of L. lactis (40). These two residues, also spaced by 50 amino acids, are present in the GT gene product sequence from all strains of lactobacilli identified in this study. Priming GTs link a monosaccharide with an isoprenoid carrier instead of to another sugar, so the catalytic site must take into account this difference in target. Thus, it is not surprising that these proteins form a separate family and are not grouped with other glycosyltransferase families of the Pfam and CAZy databases (carbohydrate active enzyme server at http://afmb.cnrs-mrs.fr/CAZY/index.html).
The PCR approach with consensus-degenerate hybrid primers led to the detection and the identification of the putative priming GT gene not only in EPS+ and EPS− L. rhamnosus but also in other EPS+ LAB strains. The hybrid strategy overcomes the unsuccessful PCR amplification of unknown sequences that are too divergent from known sequences to be readily isolated by standard methods. The reliability of the consensus-degenerate PCR strategy in detecting putative priming GT genes was confirmed by obtaining identical sequences by using specific primers. These results suggest that the hybrid primers would therefore be a useful tool for the isolation of the priming GT gene in uncharacterized species of LAB. The detection of new priming GTs will help diversify the GT genes available for the genetic engineering of LAB in order to produce novel polysaccharides. Further work is in progress on the identification and characterization of glycosyltransferases in the EPS gene cluster of L. rhamnosus and other related species of LAB.
Acknowledgments
We thank the Natural Sciences and Engineering Research Council of Canada (Research Partnerships Program-Research Network on Lactic Acid Bacteria), Agriculture and Agri-Food Canada, Novalait of Quebec, Dairy Farmers of Canada, and Institut Rosell for financial support.
We are also grateful to Daniel Vincent and Pierre Ward for their technical and scientific advice.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourgoin, F. A., A. Pluvinet, B. Gintz, B. Decaris, and G. Guédon. 1999. Are horizontal transfers involved in the evolution of the Streptococcus thermophilus exopolysaccharide synthesis loci? Gene 233:151-161. [DOI] [PubMed] [Google Scholar]
- 4.Cerning, J. 1990. Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol. Rev. 87:113-130. [DOI] [PubMed] [Google Scholar]
- 5.Chabot, S., H.-L. Yu, L. de Léséleuc, D. Cloutier, M.-R. Van Calsteren, D. Roy, M. Lacroix, and D. Oth. 2001. Exopolysaccharides from Lactobacillus rhamnosus RW-9595M stimulate TNF, IL-6 and IL-12 in human and mouse cultured immunocompetent cells, and IFN-γ in mouse splenocytes. Lait 81:683-698. [Google Scholar]
- 6.De Vuyst, L., and B. Degeest. 1999. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol. Rev. 23:153-177. [DOI] [PubMed] [Google Scholar]
- 7.Dupont, I., D. Roy, and G. LaPointe. 2000. Comparison of exopolysaccharide production by strains of Lactobacillus rhamnosus and Lactobacillus paracasei grown in chemically defined medium and milk. J. Ind. Microbiol. Biotechnol. 24:251-255. [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limit on phylogenies: an approach using bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Germond, J. E., G. Lamothe, and F. Stingele. December 1999. Lactic acid bacteria producing exopolysaccharide. Genbank accession numbers AX009415, AX009404, AX009427. International Patent 1999, WO9962. 316-A44.
- 10.Griffin, A. M., V. J. Morris, and M. J. Gasson. 1996. The cpsABCDE genes involved in polysaccharide production in Streptococcus salivarius spp. thermophilus strain NCFB 2393. Gene 183:23-27. [DOI] [PubMed] [Google Scholar]
- 11.Griffin, A. M., V. J. Morris, and M. J. Gasson. 1996. Identification, cloning and sequencing the aceA gene involved in acetan biosynthesis in Acetobacter xylinum. FEMS Microbiol. Lett. 137:115-121. [DOI] [PubMed] [Google Scholar]
- 12.Grimaldi, C., M. Dutertre, and J.-M. Simonet. 2000. Genetic organization of the guaA gene encoding the FGMP synthetase in Lactobacillus rhamnosus. Curr. Microbiol. 40:245-249. [DOI] [PubMed] [Google Scholar]
- 13.Kleerebezem, M., R. van Kranenburg, R. Tuinier, I. C. Boels, P. Zoon, E. Looijesteijn, J. Hugenholtz, and W. M. de Vos. 1999. Exopolysaccharides produced by Lactococcus lactis: from genetic engineering to improved rheological properties? Antonie van Leeuwenhoek 76:357-365. [PubMed] [Google Scholar]
- 14.Knopf, P. M. 2000. Immunomodulation and allergy. Allergy Asthma Proc. 21:215-220. [DOI] [PubMed] [Google Scholar]
- 15.Kolkman, M. A. B., D. A. Morrison, B. A. M. van der Zeijst, and P. J. M. Nuijten. 1996. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J. Bacteriol. 178:3736-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolkman, M. A. B., B. A. M. van der Zeijst, and P. J. M. Nuijten. 1998. Diversity of capsular polysaccharide synthesis gene clusters in Streptococcus pneumoniae. J. Biochem. 123:937-945. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 18.Low, D., J. A. Ahlgren, D. Horne, D. J. McMahon, C. J. Oberg, and J. R. Broadbent. 1998. Role of Streptococcus thermophilus MR-1C capsular exopolysaccharide in cheese moisture retention. Appl. Environ. Microbiol. 64:2147-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morona, J. K., R. Morona, and J. C. Paton. 1999. Comparative genetics of capsular polysaccharide biosynthesis in Streptococcus pneumoniae types belonging to serogroup 19. J. Bacteriol. 181:5355-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morona, J. K., R. Morona, and J. C. Paton. 1999. Analysis of the 5′ portion of the type 19A capsule locus identifies two classes of cpsC, cpsD, and cpsE genes in Streptococcus pneumoniae. J. Bacteriol. 181:3599-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaoka, M., S. Hashimito, T. Watanabe, T. Yokokura, and Y. Mori. 1994. Anti-ulcer effects of lactic acid bacteria and their cell wall polysaccharides. Biol. Pharm. Bull. 17:1012-1017. [DOI] [PubMed] [Google Scholar]
- 22.Oda, M., S. Nakamura, S. Komatsu, M. Kambe, F. Tsuchiya, K. Komiyama, and I. Umezawa. 1982. Physiological activities of polysaccharide produced by Lactobacillus spp. Jpn. J. Antibiot. 35:2748-2754. [PubMed] [Google Scholar]
- 23.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez, M., and A. Tomasz. 1998. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J. Bacteriol. 180:5273-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 26.Rose, T. M., E. R. Schultz, J. G. Henokoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Sau, S., N. Bhasin, E. R. Wann, J. C. Lee, T. J. Foster, and C. Y. Lee. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143:2395-2405. [DOI] [PubMed] [Google Scholar]
- 29.Saxena, I. M., R. M. Brown, Jr., M. Fevre, R. A. Geremia, and B. Henrissat. 1995. Multidomain architecture of beta-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 177:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stiles, M. E., and W. H. Holzapfel. 1997. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 36:1-29. [DOI] [PubMed] [Google Scholar]
- 31.Stingele, F., J.-R. Neeser, and B. Mollet. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster of Streptococcus thermophilus Sfi6. J. Bacteriol. 178:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stingele, F., J. W. Newell, and J.-R. Neeser. 1999. Unraveling the function of glycosyltransferases in Streptococcus thermophilus Sfi6. J. Bacteriol. 181:6354-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutherland, I. W. 1982. Biosynthesis of microbial exopolysaccharides. Adv. Microbiol. Physiol. 23:79-150. [DOI] [PubMed] [Google Scholar]
- 34.Sutherland, I. W. 1998. Novel and established applications of microbial polysaccharides. Trends Biotechnol. 16:41-46. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. G. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzianabos, A. O. 2000. Polysaccharide immunomodulators as therapeutic agents: structural aspects and biologic function. Clin. Microbiol. Rev. 13:523-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Calsteren, M.-R., C. Pau-Roblot, A. Bégin, and D. Roy. 2002. Structure determination of the exopolysaccharide produced by Lactobacillus rhamnosus strains RW-9595M and R. Biochem. J. 363:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Kranenburg, R., I. I. van Swam, J. D. Marugg, M. Kleerebezem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 39.van Kranenburg, R., H. J. Vos, I. C. Boels, M. Kleerebezem, and W. M. de Vos. 1999. Genetics and engineering of microbial exopolysaccharides for food: approaches for the production of existing and novel polysaccharides. Curr. Opin. Biotechnol. 10:498-504. [DOI] [PubMed] [Google Scholar]
- 40.van Kranenburg, R., H. J. Vos, I. I. van Swam, M. Kleerebezem, and W. M. de Vos. 1999. Functional analysis of glycosyltranferase genes from Lactococcus lactis and other gram-positive cocci: complementation, expression, and diversity. J. Bacteriol. 181:6347-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vincent, D., D. Roy, F. Mondou, and C. Déry. 1998. Characterization of bifidobacteria by random DNA amplification. Int. J. Food Microbiol. 43:185-193. [DOI] [PubMed] [Google Scholar]
- 42.Wang, L., D. Liu, and P. R. Reeves. 1996. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J. Bacteriol. 178:2598-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]