Abstract
NKX2–5 is a homeodomain-containing transcription factor important in cardiac development. Familial mutations in the NKX2–5 gene are associated with cardiac abnormalities, but mutations are rare in sporadic cases. We studied the pathology and molecular genetics of NKX2–5 in diseased heart tissues of 68 patients with complex congenital heart disease (CHD), particularly atrial (ASD), ventricular (VSD), and atrioventricular septal defects (AVSD). We also studied DNA extracted from 16 normal hearts, as well as lymphocytic DNA from 50 healthy volunteers, 7 families, and 4 unrelated individuals with CHD. Direct sequencing revealed 53 NKX2–5 mutations in the diseased heart tissues, including nonsynonymous substitutions in the homeodomain of NKX2–5. We found common mutations among unrelated patients, but certain mutations were specific to VSDs and AVSDs. Many patients had multiple NKX2–5 mutations, up to 14 nonsynonymous mutations per patient in VSDs. Importantly, these nonsynonymous mutations were mainly absent in normal heart tissues of the same CHD patients, thus indicating somatic origin and mosaicism of mutations. Further, observed mutations were completely absent in normal hearts and lymphocytic DNA of healthy individuals. Our findings provide new insights for somatic NKX2–5 mutations to be of importance in congenital heart disease.
Nkx2–5 is a transcription factor which contains a highly conserved 60-amino-acid homeodomain.1 This homeodomain (HD) interacts with DNA through a helix-turn-helix DNA-binding motif of three α helices, with helix 3 providing binding specificity. Nkx2–5 contains additional conserved regions, the TN domain and the NK2-specific domain (NK2-SD). Functions of these two conserved domains are not known, but NK2-SD may serve as a transactivation regulator of Nk-2 family members.2 The human NKX2–5 (CSX1) gene maps to chromosome 5q34 and consists of two exons that encode a 324-amino-acid protein.3,4
Nkx2–5 occupies a crucial position in the hierarchy of cardiac determinants5 and this factor is important to heart development in many organisms including zebra fish, frog, chick, mouse, and human.6 In knockout mice, lack of Nkx2–5 is lethal or results in impaired cardiac development, thus demonstrating an essential role for Nkx2–5 in normal heart morphogenesis and function.7–10 Mutations in the human NKX2–5 gene are associated with cardiac anomalies,11–18 but the molecular mechanisms are still unclear. Familial cases studied have different mutations and no mutation can be associated with a specific clinical phenotype.
The high incidence of severe congenital heart disease (CHD), which is about 3/1000 live births19 necessitates an understanding of the mechanism of disease. We therefore searched for disease-associated NKX2–5 mutations in the heart tissues of patients with cardiac malformations. We identified 53 NKX2–5 mutations in diseased heart tissues and found common mutations in unrelated patients. Certain mutations were specific to ventricular and atrioventricular septal defects. Notably, in matched normal heart tissues of same patients (such as in patients with VSD, tissue samples were taken from unaffected atria), mutations detected in diseased tissues were mainly absent, indicating somatic origin and mosaicism of mutations. Besides germline mutations, our findings are suggestive for NKX2–5 somatic mutations to cause congenital heart disease as well.
Materials and Methods
Materials
Sixty-eight formalin-fixed hearts from unrelated patients with cardiac malformations were obtained from the Institute of Anatomy, University of Leipzig, Germany. The hearts were collected by Dr. F. Spreer from 1954 to 1982 at the Institute of Pathology, University of Leipzig and District Hospital Borna, Germany. We classified them primarily by their septal defects using sequential segmental analysis.20 Most of the patients died in early infancy. As control, we analyzed DNA from n = 16 hearts, eg, 10 formalin-fixed heart tissues collected from 1955 to 1963, in which 7 individuals died at birth or early infancy, and 6 frozen normal heart tissues. We also analyzed blood samples of 4 unrelated individuals and 7 families with CHD (8 family members with cardiac malformations), as well as blood samples of 50 unrelated healthy individuals. Defects in blood samples with CHD included ASD, VSD, hypoplastic left heart syndrome (HLHS), transposition of the great arteries (TGA), subpulmonary stenosis (SPS), and heterotaxy.
DNA Isolation and Amplification of NKX2–5 Fragments from Formalin-Fixed Tissues
Excess formalin in fixed tissues was removed with 1X GTE21 and genomic DNA was isolated with NucleoSpinTissue Kit (Macherey-Nagel, Düren, Germany). The quality and quantity of isolated genomic DNA were checked on 1% agarose gel using known lambda DNA concentration (Amersham, Freiburg, Germany). The primer sequences for the amplification of NKX2–5 fragments are known.12,13 A PCR reaction consisted of 20 to 50 ng of genomic DNA, 12.5 μl of 2X Hot StarTaq Master Mix Kit (Qiagen, Hilden, Germany), 5 μl of 5X Q Solution (Qiagen), 1 μl (10 pmol/μl) of each primer pair, to a volume of 25 μl with distilled water. Except for a 10-minute final extension at 68°C, the PCR conditions were carried out as earlier described.12 PCR reactions were done on Biometra thermocyclers (Biometra, Göffingen, Germany) and the PCR products were analyzed on 1% agarose gels. A negative control was always included in PCR experiments.
Detection and Confirmation of Mutations
Mutations were analyzed by double-strand direct sequencing using NKX2–5 specific primers. PCR fragments were purified with QIAquick PCR Purification Kit (Qiagen), subjected to cycle sequencing using BigDyeTerminator v3.1 Kit and injected to ABI 3100 Genetic Analyzer (Applied Biosystems, Darmstadt, Germany). Sequences were analyzed using SeqMan (DNASTAR, Madison, WI). For uniformity of numbering of nucleotide changes in both coding and untranslated regions, we used the reference sequence NM004387 throughout the text. Corresponding numbering of NKX2–5 mutations in the coding region starts with A of start codon ATG.
We confirmed mutations by digestion with restriction enzymes (PCR-RFLP). If restriction assay was not possible, we cloned the PCR fragments with heterozygous genotypes, and re-sequenced the clones, allowing the detection of two different alleles. PCR fragments were cloned into TOPO TA Cloning Kit for Sequencing (Invitrogen, Karlsruhe, Germany). Plasmid DNA was isolated using QIAprep Spin Miniprep Kit (Qiagen) and inserts were analyzed by digestion with EcoRI (New England Biolabs, Frankfurt, Germany).
We compared nonsynonymous mutations in normal and diseased heart tissues of the same patient (such as in a patient with VSD, tissue samples were taken from an unaffected atrium). We analyzed 23 mutation loci in both normal and diseased heart tissues in 27 patients with VSDs, 2 mutation loci in 6 ASDs, and 7 mutation loci in 13 AVSDs. For this comparison, we analyzed a total of 92 samples, 146 PCR fragments, and 77,598 nucleotides.
Results
Cardiac Malformations of Patients
The explanted hearts with CHD were classified primarily by their septal defects (Figure 1, A–F). Sequential segment analysis resulted in 29 VSDs, 16 ASDs, and 23 AVSDs. Four patients within VSDs and 14 patients within AVSDs had Down syndrome. We also examined these patients to determine whether cardiac defects associated with Down syndrome would be positive for NKX2–5 mutations.
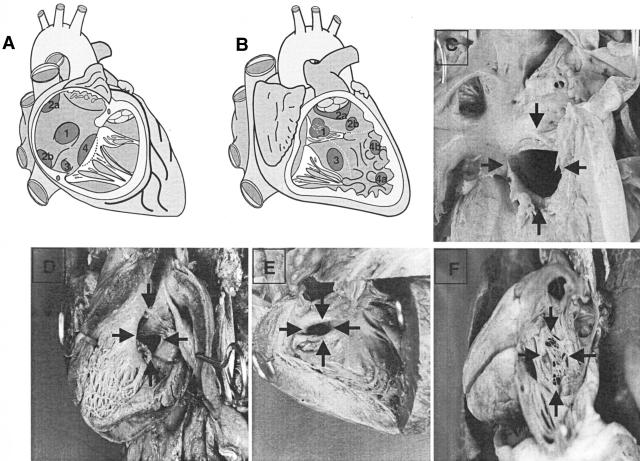
Figure 1.
Classification of cardiac malformations in patients. A: Diagram showing classification of defects with interatrial shunt. 1, ASD at fossa ovalis (ostium secundum type); 2a, ASD at superior vena cava ostium (sinus venosus type); 2b, ASD at inferior vena cava ostium (sinus venosus type); 3, ASD at coronary sinus ostium (sinus coronarius type); 4, atrioventricular septum defect (ostium primum type). B: Diagram showing classification of defects with interventricular shunt. Inlet, trabecular, and infundibular septal components are separated by dotted lines. 1, VSD at membranous septum, subaortic (perimembranous septal defect); 2a, VSD of outlet septum, subarterial, subvalvar; 2b, VSD of outlet septum not related to arterial valves; 3, VSD of inlet septum (central muscular septal defect); 4a, VSD of apical septum, central (centroapical muscular septal defect); 4b, VSD of apical septum, marginal. C, D: Large atrioventricular septal defects (arrows). E: Muscular ventricular septal defect (arrows). F: Arrows, atrial septal defect (ostium secundum type).
Cardiac malformations in VSDs consisted of subaortic (76%) and muscular VSD (28%) (Table 1). Many patients were affected by patent foramen ovale (79%) and patent ductus arteriosus (28%). Several patients had further cardiac defects. For example, patient E39 had muscular and subaortic VSD with overriding aorta and persistent left superior vena cava draining into coronary sinus. The most prevalent septal anomaly in ASDs was septum secundum (88%), followed by septum primum (13%) and septum sinus (13%) (Table 1). Additionally, 12 other cardiac defects were present in ASDs and all patients but one had multiple defects. For example, patient C75 had septum secundum and septum primum defects, patent foramen ovale, patent ductus arteriosus, preductal aortic isthmus stenosis, mitral valve stenosis, hypoplastic left ventricle, and left isomerism. All 23 patients diagnosed as AVSD had subaortic VSD, primum septum defect and a common atrioventricular canal (Table 1). Patients had patent foramen ovale (70%) and patent ductus arteriosus (39%). Except for one, all patients had a minimum of four defects. For example, patient F13 had subaortic VSD, septum primum defect, patent foramen ovale, aortic stenosis, and pulmonary stenosis.
Table 1.
Cardiac Malformations of Patients Analyzed for NKX2-5 Mutations
| No. | Cardiac malformations | VSD group (29 patients) | ASD group (16 patients) | AVSD group (23 patients) |
|---|---|---|---|---|
| 1 | Subaortic VSD | 22 | 23 | |
| 2 | Subaortic VSD with overriding aorta | 21 | 6 | |
| 3 | Muscular VSD | 8 | ||
| 4 | Septum primum defect | 2 | 23 | |
| 5 | Septum secundum defect | 14 | 1 | |
| 6 | Septum sinus defect | 2 | ||
| 7 | Common atrioventricular canal | 23 | ||
| 8 | Overriding common atrioventricular valve | 8 | ||
| 9 | Overriding common quadricuspid atrioventricular valve | 6 | ||
| 10 | Patent foramen ovale | 23 | 6 | 16 |
| 11 | Patent ductus arteriosus | 8 | 4 | 9 |
| 12 | Aortic stenosis | 1 | 1 | 2 |
| 13 | Preductal aortic isthmus stenosis | 3 | 3 | 3 |
| 14 | Pulmonary stenosis | 3 | 1 | |
| 15 | Mitral valve stenosis | 1 | ||
| 16 | Partial anomalous venous return | 1 | ||
| 17 | Double-outlet left ventricle | 1 | ||
| 18 | Double-outlet right ventricle | 1 | ||
| 19 | Persistent left superior vena cava draining into coronary sinus | 2 | 2 | |
| 20 | Hypoplastic left heart | 1 | ||
| 21 | Hypoplastic left ventricle | 3 | 3 | |
| 22 | Hypoplastic right ventricle | 1 | ||
| 23 | Hypoplastic ascending aorta | 2 | 2 | |
| 24 | Total anomalous pulmonary venous return | 1 | 3 | |
| 25 | Isomerism of right atrial appendages | 1 | ||
| 26 | Bicuspid right atrioventricular valve | 1 | ||
| 27 | Bicuspid aortic valve | 1 |
Amplification of NKX2–5 Fragments from Formalin-Fixed Tissues
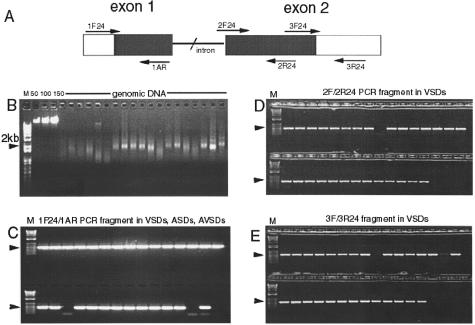
Using known primer sequences, we amplified NKX2–5 fragments from genomic DNA isolated from formalin-fixed heart tissues (Figure 2A). DNA yield from 25 mg of tissue ranged from 0.5 to 1.0 μg, with an average size of about 2 kb (Figure 2B). We were able to amplify all three fragments in the two exons of NKX2–5 except in two patients. We obtained a 489-bp fragment with primers 1F24 and 1AR (Figure 2C), 472-bp fragment with 2F24/2R24 (Figure 2D), and 573-bp fragment with 3F24/3R24 (Figure 2E).
Figure 2.
Amplification of NKX2–5 fragments from formalin-fixed tissues. A: Location of PCR primers on the genomic sequence of NKX2.5. B: Quality and concentration of genomic DNA isolated from formalin-fixed heart tissues in 1% ethidium bromide gel, M (kb plus ladder); 50, 100, 150 ng (lambda DNA standard). C: Amplified 1F24/1AR fragments (489 bp) in VSDs, ASDs, AVSDs (2 failed PCR). D: 2F/2R24 fragments (472 bp) in 29 VSDs (1 failed PCR). E: 3F/3R24 fragments (573 bp) in 29 VSDs (2 failed PCR).
Detection of NKX2–5 Mutations
We identified by direct sequencing 53 mutations in the diseased heart tissues of patients consisting of 35 nonsynonymous, 13 synonymous, and 3′-UTR and 2 intronic (Table 2). Three mutations (Arg25Cys, Thr178Met and Ala219Val) are known. We found 4 patients in VSDs who were positive for both Thr178Met and Ala219Val. We also detected NCBI dbSNPs rs2277923 (A239G, Glu21Glu), and rs703752 (T1212G) in patients (Table 2). Only heterozygous genotypes were obtained. We confirmed mutations by PCR-RFLP assays or by cloning fragments with heterozygous loci, allowing detection of two different alleles (Figure 3, A and B). We obtained in normal lymphocytic DNA, the genotypic frequency 22 AA:17 AG: 6 GG for dbSNP rs2277923 in 45 samples; and for dbSNP rs703752, the frequency 19 GG: 20 GT:11 TT, in 50 samples. Except for these two dbSNPs, which were readily detected in all analyzed material, the 53 nucleotide changes were absent in DNA from six frozen, normal heart tissues and 50 blood samples of healthy individuals. We found 3 nucleotide alterations (G833A, T984A, A1205T) in the 10 formalin-fixed, normal hearts, but these were different from those of patients with CHD. A synonymous change G833A (219 Ala) was found in 5, while T984A (Cys270Ser) and A1205T (3′-UTR) were present in 2 individuals. These 2 individuals, who were compound heterozygous for T984A (Cys270Ser) and A1205T (3′-UTR), died in early infancy. The normal formalin-fixed hearts were basically absent of mutations.
Table 2.
NKX2-5 Mutation Summary
| Nucleotide change (NKX2-5 NM004387) | Nucleotide reference (coding region) | Amino acid change | Location | No. of positive patients (VSD) | No. of positive patients (ASD) | No. of positive patients (AVSD) | PCR-RFLP assay |
|---|---|---|---|---|---|---|---|
| Nonsynonymous | |||||||
| 196 T→C | 20 | 7 Leu→Pro | Exon 1 | 1 | |||
| 232 A→G | 56 | 19 Asn→Ser | Exon 1, TN domain | 18 | |||
| 249 C→T | 73 | 25 Arg→Cys | Exon 1 | 1 | HhaI, create site | ||
| 309 T→C | 133 | 45 Ser→Pro | Exon 1 | 1 | |||
| 327 T→C | 151 | 51 Phe→Leu | Exon 1 | 1 | |||
| 382 T→C | 206 | 69 Leu→Pro | Exon 1 | 1 | |||
| 406 C→T | 230 | 77 Pro→Leu | Exon 1 | 1 | |||
| 516 T→A | 340 | 114 Cys→Ser | Exon 2 | 3 | 2 | ||
| 516 T→C | 340 | 114 Cys→Arg | Exon 2 | 8 | 6 | 8 | |
| 529 A→G | 353 | 118 Lys→Arg | Exon 2 | 4 | 1 | ||
| 547 A→G | 371 | 124 Lys→Arg | Exon 2 | 6 | |||
| 553 A→T | 377 | 126 Glu→Val | Exon 2 | 4 | 2 | 5 | |
| 573 C→T | 397 | 133 Pro→Ser | Exon 2 | 6 | |||
| 579 G→A | 403 | 135 Ala→Thr | Exon 2 | 1 | 8 | AvaI, abolish site | |
| 607 T→C | 431 | 144 Leu→Pro | Exon 2 | 3 | 8 | ||
| 709 C→T | 533 | 178 Thr→Met | Exon 2, HD | 5 | NlaIII, create site | ||
| 723 A→G | 547 | 183 Lys→Glu | Exon 2, HD, 3rd helix | 7 | 22 | TaqI, create site | |
| 735 C→T | 559 | 187 Gln→Ter | Exon 2, HD, 3rd helix | 6 | XbaI, create site | ||
| 751 A→C | 575 | 192 Lys→Thr | Exon 2, HD, 3rd helix | 6 | TaiI, create site | ||
| 751 A→G | 575 | 192 Lys→Arg | Exon 2, HD, 3rd helix | 2 | BfmI, create site | ||
| 757 A→G | 581 | 194 Lys→Arg | Exon 2, HD, 3rd helix | 2 | BsgI, create site | ||
| 790 T→A | 614 | 205 Val→Glu | Exon 2 | 6 | |||
| 832 C→T | 656 | 219 Ala→Val | Exon 2, NK2-SD | 10 | |||
| 852 G→A | 676 | 226 Asp→Asn | Exon 2 | 2 | |||
| 918 T→C | 742 | 248 Tyr→His | Exon 2 | 5 | |||
| 1011 T→C | 835 | 279 Ser→Pro | Exon 2 | 3 | |||
| 1012 C→T | 836 | 279 Ser→Phe | Exon 2 | 11 | 13 | ||
| 1018 C→T | 842 | 281 Ala→Val | Exon 2 | 24 | 5 | 5 | BmrI, create site |
| 1033 C→T | 857 | 286 Ala→Val | Exon 2 | 3 | 11 | 8 | |
| 1056 A→C | 880 | 294 Asn→His | Exon 2 | 14 | |||
| 1072 A→G | 896 | 299 Asp→Gly | Exon 2 | 9 | 6 | 21 | |
| 1089 A→G | 913 | 305 Ser→Gly | Exon 2 | 1 | |||
| 1134 G→A | 958 | 320 Gly→Ser | Exon 2 | 8 | 4 | 5 | |
| 1141 G→A | 965 | 322 Arg→Gln | Exon 2 | 2 | |||
| 1149 T→C | 973 | Stop→Gln | Exon 2 | 3 | 2 | 7 | |
| Synonymous | |||||||
| 239 A→G | 63 | 21 Glu | Exon 1, dbSNP | 2AA: 26 AG | 12 AG | 6AA: 17 AG | BpmI, create site |
| 560 C→T | 384 | 128 Asp | Exon 2 | 6 | 19 | ||
| 614 G→T | 438 | 146 Ser | Exon 2 | 3 | 10 | ||
| 629 T→C | 453 | 151 Tyr | Exon 2, HD | 22 | 5 | 1 | AccI, create site |
| 677 A→G | 501 | 167 Glu | Exon 2, HD | 2 | |||
| 680 C→T | 504 | 168 Arg | Exon 2, HD | 5 | |||
| 704 A→G | 528 | 176 Lys | Exon 2, HD | 6 | 18 | ||
| 779 T→C | 603 | 201 Thr | Exon 2 | 2 | |||
| 902 C→G | 726 | 242 Gly | Exon 2 | 10 | |||
| 995 T→C | 819 | 273 Ala | Exon 2 | 28 | 4 | 6 | |
| 1034 C→T | 858 | 286 Ala | Exon 2 | 2 | 17 | ||
| 1079 T→C | 903 | 301 Asn | Exon 2 | 1 | |||
| 1118 A→G | 942 | 314 Gly | Exon 2 | 4 | 6 | 8 | |
| 1142 A→G | 966 | 322 Arg | Exon 2 | 28 | 1 | 1 | MspI, create site |
| Untranslated region | |||||||
| 113 C→T | Exon 1, 5′ UTR | 1 | |||||
| 141 C→T | Exon 1, 5′ UTR | 1 | |||||
| 1156 G→A | Exon 2, 3′ UTR | 13 | 13 | 23 | |||
| 1212 T→G | Exon 2, 3′ UTR, dbSNP | 28 GT | 1GG: 14 GT | 12 GG: 11 GT | |||
| Intronic region | |||||||
| −24 IVS C→A | Intron | 28 | 4 | 1 | |||
| −11 IVS T→C | Intron | 1 |
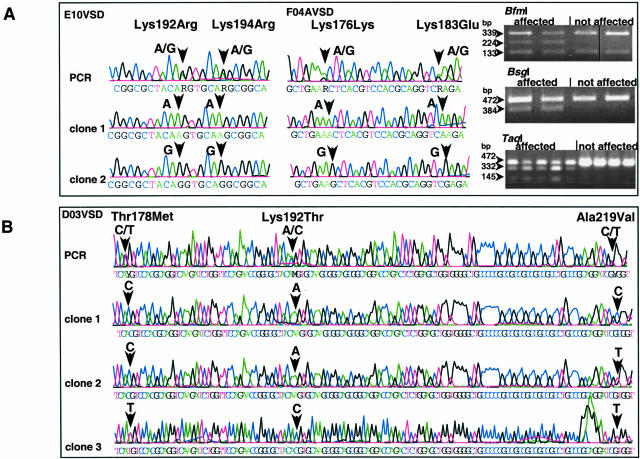
Figure 3.
Detection and confirmation of NKX2–5 mutations. Heterozygous loci as detected by direct sequencing (electropherograms) are confirmed by PCR-RFLP assay or cloning and re-sequencing of clones allowing detection of two alleles. A: Analysis of two patients (E10VSD, F04AVSD) who are compound heterozygous for mutations in the third helix of the homeodomain. BfmI and BsgI are PCR-RFLP assays for Lys192Arg and Lys194Arg, respectively; while TaqI is for Lys183Glu. B: Analysis of a patient (D03VSD) showing multiple known mutations and more than two haplotypes. Clone 1: C/A/C (all reference alleles); clone 2: C/A/T (recombinant type); clone 3: T/C/T (all mutant alleles).
Furthermore, we did not detect any NKX2–5 mutations in blood samples of patients with CHD except for a synonymous nucleotide change (G719A, 181 Gln) in an unaffected family member. This substitution was confirmed by BspMI restriction assay.
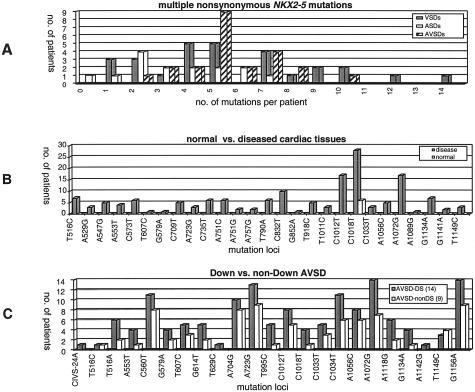
Together with the three known mutations, we obtained 35 nonsynonymous mutations; distributed as 29 in VSDs, 12 in ASDs, and 12 in AVSDs. Certain mutations were specific for ventricular and atrioventricular septal defects. Most patients had multiple nonsynonymous mutations, with VSDs carrying up to 14 mutations per patient (Figure 4A). We identified 6 nonsynonymous mutations in the homeodomain, and 5 would affect the third helix of the homeodomain. Prevalent nonsynonymous mutations in VSDs were A232G (Asn19Ser) and C1018T (Ala281Val) with frequencies 64% and 86%, respectively. Prevalent nonsynonymous mutations in AVSDs were A723G (Lys183Glu) and A1072G (Asp299Gly) with frequencies 96% and 91%, respectively. The nonsynonymous NKX2–5 mutations were identified in diseased heart tissues of patients, but not in matched normal tissues of the same patients’ hearts (eg, patients with VSD, tissue samples were taken from unaffected atria). Except for one (C1018T, Ala281Val), none of the mutations was detected in normal heart tissue after comparing normal versus disease cardiac tissues of 27 distinct mutation loci (Figure 4B). Furthermore, mutation spectrum did not differ basically in AVSD patients with and without Down syndrome (Figure 4C).
Figure 4.
Evidence of somatic nature of identified NKX2–5 mutations in diseased heart tissues. A: Multiple nonsynonymous mutations in VSDs, ASDs, and AVSDs. B: Mutations are mainly absent in matched normal and diseased cardiac tissues of the same patient. C: No difference in NKX2–5 mutation spectrum in Down and non-Down syndrome AVSD.
Cluster Analysis of Mutations
We used a mathematical algorithm to allow clustering of mutations by response.22 Clustering by response tests all possible combinations among all patients and mutations and reveals the best constellation ranked by statistical significance. We asked two questions: 1) which among the prevalent mutations would be highly associated with Down syndrome; 2) which prevalent mutations would be of significance in predicting septal defects, independently of Down syndrome. For the first question, the test found Lys183Glu highly associated with Down syndrome. For the second question, we excluded the18 patients with Down syndrome and used prevalent nonsynonymous mutations as predictors. These predictors included Asn19Ser, Lys183Glu, Ser279Phe, Ala281Val and Asp299Gly. Permutation tests gave a P value of < 0.001, which shows that these five mutations are predictive of septal defects. Two mutations eg, Asn19Ser and Lys183Glu were selected as highly predictive (P value 6.07 × 10−9) This constellation predicts 100% VSD if Asn19Ser is present, but Lys183Glu is absent (Table 3).
Table 3.
Cluster Analysis of NKX2-5 Nonsynonymous Mutations in Complex CHD without Down Syndrome
| Cluster | Predictors
|
ASD | VSD | AVSD | Total | ||
|---|---|---|---|---|---|---|---|
| Asn19Ser | Lys183Glu | ||||||
| 1 | 1 0 | + | − | 0 (0.0%) | 17 (100.0%) | 0 (0.0%) | 17 (34.0%) |
| 2 | 1 1 | + | + | 7 (43.8%) | 0 (0.0%) | 9 (56.4%) | 16 (32.0%) |
| 0 1 | + | − | |||||
| 3 | 0 0 | − | − | 9 (52.9%) | 8 (47.2%) | 0 (0.0%) | 17 (34.0%) |
| Total | 16 (32.0%) | 25 (50.0%) | 9 (18.0%) | 50 | |||
Selected predictors: Asn19Ser, Lys183Glu. Predictors used: Asn19Ser, Lys183Glu, Ser279Phe, Ala281Val, Asp299Gly.
Cluster 1, VSD: to obtain 100% VSD, Asn19Ser present, but Lys183Glu absent.
χ2: 4.411443e+001.
P value: 6.074074e-009.
Discussion
Genetic analysis of NKX2–5 in diseased heart tissues of 68 unrelated patients with primarily septal defects, yielded 53 mutations and two known single nucleotide polymorphisms (NCBI dbSNPs). Among these were 35 single nucleotide changes that would result in amino acid change (nonsynonymous mutations). We identified three known mutations, namely Arg25Cys, Thr178Met, and Ala219Val in VSDs. Mutations Arg25Cys and Ala219Val were detected previously in patients with tetralogy of Fallot (TOF).12,13,18 Detection of Arg25Cys and Ala219Val in VSDs is not surprising, as ventricular septal defect is one of the four complications of TOF. Among familial NKX2–5 mutations, Thr178Met was detected in unrelated families with ASD.11,17 All other known mutations are uncommon and specific to single families. We show here that Thr178Met affects ventricular septal defects as well and found four patients in VSDs who were compound heterozygous for Thr178Met and Ala219Val.
Six of the new nonsynonymous mutations are located in highly conserved regions. Mutation Asn19Ser is in the TN domain at the NH2 terminus. The function of this domain remains unclear, but the change from asparagine to serine results in change of amino acid charge and may affect binding properties of NKX2–5. Mutations Lys183Glu, Glu187Ter, Lys192Arg, Lys192Thr, and Lys194Arg are located in the third helix of HD, which is important for DNA binding specificity. These mutations may cause cardiac defects through reduced DNA affinity or affect transactivation activities.23,24 Further studies will be undertaken to clarify the exact molecular mechanisms by which these mutations lead to septal defects.
Independent of their location, clustering by response analysis of NKX2–5 mutations found Asn19Ser and Lys183Glu as highly predictive mutations. Cluster analysis also found that Ser279Pro, Ala281Val, and Asp299Gly predictive of septal defects. How these three mutations will effect cardiac malformations needs further study, because these are located in the COOH terminus of NKX2–5. Whether they cause cardiac malformations through gain-of-function activities or through other mechanisms is unclear. Different results were obtained in carriers of COOH terminus mutations.25,26 The applied genetic algorithm in cluster analysis assumes correlation between primary mutations and disease phenotype. It will, therefore, be interesting to explore further the role of primary mutations in predisposing mutant cardiac tissue to secondary somatic mutations.
While all previous reports on NKX2–5 mutations were based on lymphocytic DNA and mutations are of germline in nature,11–18 our genetic analysis was based on DNA from diseased cardiac tissues of unrelated patients. These earlier studies found familial and private mutations in patients, and mutations were rare in sporadic cases. We obtained a higher rate of mutations among unrelated patients, which may be explained by complex cardiac malformations in patients and somatic nature of mutations. Several lines of evidence suggest that the NKX2–5 mutations may be of somatic nature, originating most likely from cell division errors during early embryogenesis. For instance, we observed multiple nonsynonymous mutations in the same patient, which is an unlikely event in inherited mutations. Multiple mutations are well known in cancer,27 but the occurrence of more than two inherited gene mutations in a patient is rare.28 We found that mutations in diseased heart tissues were mainly absent in matched normal heart tissue (ie, patients with VSD, tissue samples were taken from unaffected atria), indicating somatic nature and mosaicism of mutations.
We observed multiple haplotypes in a patient. Cloning of NKX2–5 fragments containing several heterozygous mutations yielded more than the two expected haplotypes in a patient, including clones with all of the mutant alleles. We obtained three haplotypes in each two patients who were compound heterozygous for known mutations Thr178Met and Ala219Val. Similar results were found in a patient (D03VSD) who had three known mutations (Figure 3B). The presence of multiple haplotypes suggests mixed population of cells or de novo chromosomal rearrangements or duplications affecting NKX2–5 in heart tissues of patients. Congenital heart defects (VSD, TOF) have been observed in trisomy 5 mosaicism29 or tandem duplication mosaicism on the long arm (5q13–33) of chromosome 5.30
We did not find any NKX2–5 mutations after screening of lymphocytic DNA of families and unrelated patients with CHD except dbSNPs and a synonymous substitution in an unaffected family member. Analysis of lymphocytic DNA alone does not reveal chromosomal aberrations occurring in cardiac tissues, as has been found previously.31,32 It is of considerable importance that we did not observe any difference in NKX2–5 mutation spectrum in AVSD Down and non-Down syndrome. This result was unexpected, since atrioventricular septal defect is the most common feature in Down syndrome (trisomy 21).33 The existence of both NKX2–5 mutations (chromosome 5) and Down syndrome in a patient indicates further genomic chaos in the diseased heart tissues of patients with CHD.
Although there are reservations in the use of formalin-fixed tissues,34,35 our data show that DNA sequence is preserved after more than 40 years of formalin storage. Indeed we demonstrate reliability in applying a well-developed assay for isolating DNA from long-term formalin-fixed tissue. Our study benefits from well-characterized hearts with distinct and definitively determined malformations. We, therefore, overcome the limitation of using surrogate tissue DNA (such as lymphocytic DNA) to correlate phenotype to genotype. Archival material is therefore valuable and should be considered for genetic studies.
The original aim of this study initiated 40 years ago was to improve an understanding of the anatomy of complex malformed hearts. We, therefore, have limited information on the pedigrees of our cohorts. In the case of AVSDs, patient F08 had a prematurely aborted sibling and a further sibling who died 3 months after birth; patient F15 had a prematurely aborted sibling and, patient F17 had a twin sister who died shortly after birth. In addition, patient E27 in VSDs had a prematurely aborted sister. The scarce information on family history is supportive of possible chromosomal abnormalities in patients.
We found three nucleotide changes in formalin-fixed normal hearts, including a nonsynonymous mutation (Cys270Ser) in two patients who died in early infancy. Nonsynonymous NKX2–5 mutations have been observed in normal population (NCBI dbSNPs rs3729754, rs3729938). Cys270Ser has not been found in diseased heart tissues of patients with CHD and thus, this alteration may simply reflect genetic polymorphism. We also detected the germline mutation Arg25Cys, which was reported previously in patients with CHD,12,13,18 but was later detected in control population13 or in unaffected family members.17 It is, therefore, not impossible to have nonsynonymous NKX2–5 mutations in disease-free individuals. Further, comparison of mutations in normal versus diseased heart tissues shows that mutations may differ in different regions of the same heart. After analysis of 27 distinct nonsynonymous mutation loci in normal and diseased heart tissues, only one (C1018T) was detected in both tissues of some patients. Such mutation may simply have occurred during early embryogenesis.
The diseased heart tissues may, in turn, contain a mixture of cells carrying different mutations. We cannot discount the possibility of not detecting all types of NKX2–5 mutations present within the diseased heart tissues. Also, we cannot determine with certainty disease-causing mutations from disease-associated mutations, although specific patterns of mutation loci and frequency of occurrence are linked to patient phenotype (see clustering by response analysis in Table 3, which reports a statistically significant relationship between various constellations of mutations and disease phenotype).
The process of cardiac development is complex, and NKX2–5 participates in protein-protein interaction with other transcription factors. New studies are underway to obtain in-depth information on the role of mutations of other transcription factors in the network of NKX2–5 protein-protein interactions. Perhaps failure of cardiac transcription factor networks resulting from genetic instability in cardiac cells is a plausible pathway to congenital heart disease. We show for the first time that malformed hearts are affected by multiple somatic NKX2–5 mutations. It will be of interest to investigate further whether a single transcription factor mutation leads to genomic instability or whether several transcription factors are affected simultaneously to give rise to genomic chaos. Lastly, a prospective analysis of CHD patients is needed to compare cardiac and non-cardiac tissues to confirm our findings.
Acknowledgments
The financial support of the Lower Saxony Ministry of Science and Culture to J.B. is greatly appreciated. We thank the Department of Cardiac Surgery and Pediatric Cardiology, University of Mainz, for the blood samples from patients with CHD. The technical support of T. Wilmes, Y. Becker, and A. Hiemisch is highly appreciated.
Footnotes
Address reprint requests to Juergen Borlak, Drug Research and Medical Biotechnology, Fraunhofer Institute of Toxicology and Experimental Medicine, Nikolai-Fuchs-Strasse 1, D-30625 Hannover, Germany. E-mail: borlak@item.fraunhofer.de.
Supported by grant 25A.5–76251–99-3/00 (to J.B.) from the Lower Saxony Ministry of Science and Culture, Germany.
S.M.R-B. and J.B. contributed equally to this work.
References
- Banerjee-Basu S, Baxevanis AD. Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 2001;29:3258–3269. doi: 10.1093/nar/29.15.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watada H, Mirmira RG, Kalamaras J, German MS. Intramolecular control of transcriptional activity by the NK2-specific domain in NK-2 homeodomain proteins. Proc Natl Acad Sci USA. 2000;97:9443–9448. doi: 10.1073/pnas.97.17.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I, Komuro I, Mizuno T, Aikawa R, Akazawa H, Oka T, Yamazaki T, Yazaki Y. Molecular cloning and characterization of human cardiac homeobox gene CSX1. Circ Res. 1996;79:920–929. doi: 10.1161/01.res.79.5.920. [DOI] [PubMed] [Google Scholar]
- Turbay D, Wechsler SB, Blanchard KM, Izumo S. Molecular cloning, chromosomal mapping, and characterization of the human cardiac-specific homeobox gene hCsx. Mol Med. 1996;2:86–96. [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, Harvey RP. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2–5. Circ Res. 2000;87:888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, Seidman CE, Plowden J, Kugler JD. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmuntz E, Geiger E, Benson DW. NKX2.5 mutations in patients with tetralogy of Fallot. Circulation. 2001;104:2565–2568. doi: 10.1161/hc4601.098427. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Hiroi Y, Hosoda T, Utsunomiya T, Matsuo S, Ito T, Inoue J, Sumiyoshi T, Takano H, Nagai R, Komuro I. Novel point mutation in the cardiac transcription factor CSX/NKX2.5 associated with congenital heart disease. Circ J. 2002;66:561–563. doi: 10.1253/circj.66.561. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Benson DW, Yano S, Akagi T, Yoshino M, Murray JC. Two novel frameshift mutations in NKX2.5 result in novel features including visceral inversus and sinus venosus type ASD. J Med Genet. 2002;39:807–811. doi: 10.1136/jmg.39.11.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Roelens I, Sluysmans T, Gewillig M, Devriendt K, Vikkula M. Progressive AV-block and anomalous venous return among cardiac anomalies associated with two novel missense mutations in the CSX/NKX2–5 Gene. Hum Mutat. 2002;20:75–76. doi: 10.1002/humu.9041. [DOI] [PubMed] [Google Scholar]
- Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, Feneley M, Harvey RP. Cardiac homeobox gene NKX2–5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol. 2003;41:2072–2076. doi: 10.1016/s0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- McElhinney DB, Geiger E, Blinder J, Woodrow BD, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003;42:1650–1655. doi: 10.1016/j.jacc.2003.05.004. [DOI] [PubMed] [Google Scholar]
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Devine WA, Webber SA, Anderson RH. Congenitally malformed hearts from a population of children undergoing cardiac transplantation: comments on sequential segmental analysis and dissection. Pediatr Dev Pathol. 2000;3:140–154. doi: 10.1007/s100240050018. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Haygood MG, Pietsch TW, Bentzen P. Enhanced DNA extraction and PCR amplification of mitochondrial genes from formalin-fixed museum specimens. Biotechniques. 1997;22:394–6, 398–400. doi: 10.2144/97223bm03. [DOI] [PubMed] [Google Scholar]
- Hecker H, Wübbelt P. Clustering by response. Computat Stat Data Anal. 1997;24:193–215. [Google Scholar]
- Zhu W, Shiojima I, Hiroi Y, Zou Y, Akazawa H, Mizukami M, Toko H, Yazaki Y, Nagai R, Komuro I. Functional analyses of three Csx/Nkx-2.5 mutations that cause human congenital heart disease. J Biol Chem. 2000;275:35291–35296. doi: 10.1074/jbc.M000525200. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Lee B, Schott JJ, Benson DW, Seidman JG, Seidman CE, Izumo S. Loss of function and inhibitory effects of human CSX/NKX2.5 homeoprotein mutations associated with congenital heart disease. J Clin Invest. 2000;106:299–308. doi: 10.1172/JCI9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Izumo S. Identification of the in vivo casein kinase II phosphorylation site within the homeodomain of the cardiac tissue-specifying homeobox gene product Csx/Nkx2.5. Mol Cell Biol. 1999;19:526–536. doi: 10.1128/mcb.19.1.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Loeb LA. On the origin of multiple mutations in human cancers. Semin Cancer Biol. 1998;8:421–429. doi: 10.1006/scbi.1998.0113. [DOI] [PubMed] [Google Scholar]
- Westphal V, Schottstadt C, Marquardt T, Freeze HH. Analysis of multiple mutations in the hALG6 gene in a patient with congenital disorder of glycosylation Ic. Mol Genet Metab. 2000;70:219–223. doi: 10.1006/mgme.2000.3017. [DOI] [PubMed] [Google Scholar]
- Sciorra LJ, Hux C, Day-Salvadore D, Lee ML, Mandelbaum DE, Brady-Yasbin S, Frybury J, Mahoney MJ, Dimaio MS. Trisomy 5 mosaicism detected prenatally with an affected liveborn. Prenat Diagn. 1992;12:477–482. doi: 10.1002/pd.1970120602. [DOI] [PubMed] [Google Scholar]
- Rauen KA, Bitts SM, Li L, Golabi M, Cotter PD. Tandem duplication mosaicism: characterization of a mosaic dup(5q) and review. Clin Genet. 2001;60:366–370. doi: 10.1034/j.1399-0004.2001.600508.x. [DOI] [PubMed] [Google Scholar]
- Yokoyama Y, Narahara K, Kamada M, Tsuji K, Seino Y. Tissue-specific mosaicism for trisomy 21 and congenital heart disease. J Pediatr. 1992;121:80–82. doi: 10.1016/s0022-3476(05)82547-8. [DOI] [PubMed] [Google Scholar]
- Consevage MW, Seip JR, Belchis DA, Davis AT, Baylen BG, Rogan PK. Association of a mosaic chromosomal 22q11 deletion with hypoplastic left heart syndrome. Am J Cardiol. 1996;77:1023–1025. doi: 10.1016/s0002-9149(97)89165-5. [DOI] [PubMed] [Google Scholar]
- Freeman SB, Taft LF, Dooley KJ, Allran K, Sherman SL, Hassold TJ, Khoury MJ, Saker DM. Population-based study of congenital heart defects in Down syndrome. Am J Med Genet. 1998;80:213–217. [PubMed] [Google Scholar]
- Wong C, DiCioccio RA, Allen HJ, Werness BA, Piver MS. Mutations in BRCA1 from fixed, paraffin-embedded tissue can be artifacts of preservation. Cancer Genet Cytogenet. 1998;107:21–27. doi: 10.1016/s0165-4608(98)00079-x. [DOI] [PubMed] [Google Scholar]
- Williams C, Ponten F, Moberg C, Soderkvist P, Uhlen M, Ponten J, Sitbon G, Lundeberg J. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]