Abstract
It has been shown previously that the unfolded N-terminal domain of the prion protein can bind up to six Cu2+ ions in vitro. This domain contains four tandem repeats of the octapeptide sequence PHGGGWGQ, which, alongside the two histidine residues at positions 96 and 111, contribute to its Cu2+ binding properties. At the maximum metal-ion occupancy each Cu2+ is co-ordinated by a single imidazole and deprotonated backbone amide groups. However two recent studies of peptides representing the octapeptide repeat region of the protein have shown, that at low Cu2+ availability, an alternative mode of co-ordination occurs where the metal ion is bound by multiple histidine imidazole groups. Both modes of binding are readily populated at pH 7.4, while mild acidification to pH 5.5 selects in favour of the low occupancy, multiple imidazole binding mode. We have used NMR to resolve how Cu2+ binds to the full-length prion protein under mildly acidic conditions where multiple histidine co-ordination is dominant. We show that at pH 5.5 the protein binds two Cu2+ ions, and that all six histidine residues of the unfolded N-terminal domain and the N-terminal amine act as ligands. These two sites are of sufficient affinity to be maintained in the presence of millimolar concentrations of competing exogenous histidine. A previously unknown interaction between the N-terminal domain and a site on the C-terminal domain becomes apparent when the protein is loaded with Cu2+. Furthermore, the data reveal that sub-stoichiometric quantities of Cu2+ will cause self-association of the prion protein in vitro, suggesting that Cu2+ may play a role in controlling oligomerization in vivo.
Keywords: Copper (II), fluorescence, metal binding, NMR, prion, prion protein (PrP)
Abbreviations: GdnHCl, guanidinium chloride; HSQC, heteronuclear single quantum coherence; p.p.m., parts per million; PrP, prion protein; PrPC, cellular prion protein isoform; PrPSc, pathogenic (scrapie) isoform of prion protein; PrP–Cu2+, complex of the full-length prion protein with one Cu2+ ion; PrP–2Cu2+, complex of the full-length prion protein with two Cu2+ ions
INTRODUCTION
Prion diseases are a group of fatal neurodegenerative disorders that includes Creutzfeldt–Jakob disease in humans, scrapie in sheep and bovine spongiform encephalopathy in cattle. In humans such diseases can arise sporadically, be inherited or occur through infection with prion-contaminated material [1,2]. The ‘protein only’ hypothesis states that the particles that cause infection, termed prions, are composed principally or entirely of a misfolded isoform of a host-encoded protein, termed the PrP (prion protein) [3–5], with prion propagation occurring by conformational conversion of the normal cellular isoform, PrPC (cellular prion protein isoform) to the disease related isoform PrPSc [pathogenic (scrapie) isoform of prion protein]. Mammalian PrPC is a glycolipid anchored cell surface glycoprotein found at high levels in the central nervous system, lymphatic tissue, activated lymphocytes and at neuromuscular junctions [6]. The physiological function of PrP remains unclear; ablation of the Prn-P gene in mice results in viable animals without gross developmental or behavioral abnormalities [7], although more detailed studies revealed some phenotypic effects, including altered synaptic physiology [8]. Biophysical and biochemical studies have demonstrated that PrP specifically binds Cu2+, and this has led to the proposal that PrP is a metalloprotein in vivo [9–21].
PrPC comprises a flexibly disordered N-terminal domain (residues 23–125) [22] and a globular, mainly α-helical, C-terminal domain (residues 126–231) [23]. Residues 60–91 of the protein comprise four tandem repeats of the sequence PHGGGWGQ. It is this region, known as the octapeptide repeats, which was first discovered to bind Cu2+ [10,11], and a considerable body of work has since been devoted to identifying the number of binding sites, their affinities and the nature of co-ordination in both the octapeptide repeats and the full-length prion protein, PrP23–231 [9,12–21]. Until recently, the accepted model of Cu2+ binding to the full-length prion protein, at pH 7.4, was that it binds six Cu2+ ions at separate sites, one in each of the four octapeptide repeats and at two additional sites involving His96 and His111, and each with co-ordination from a single histidine imidazole group and amide nitrogen atoms from nearby glycine residues [19].
Two recent studies have shown that the model described above provides an incomplete picture of Cu2+ binding in the prion protein, by demonstrating that a peptide representing the four octapeptide repeats displays multiple modes of Cu2+ binding at pH 7.4 [24,25]. The peptide can bind a single Cu2+ ion through multiple histidine co-ordination; however, in the presence of more Cu2+, binding of up to four Cu2+ ions occurs, with all of these co-ordinated in identical sites – each involving deprotonated backbone amide nitrogen atoms and a single histidine residue. Perhaps counter-intuitively, these four Cu2+ binding sites replace a single site of higher affinity. Peptides representing the octapeptide repeats cannot, however, be considered a complete model for Cu2+ binding to the prion protein. Cu2+ binding sites outside the octapeptide repeats have also been reported, involving His96 and His111 [12,19,26,27]. It is thus clear that, to fully understand the binding of Cu2+ to the prion protein, binding to the full-length protein needs to be investigated, and in particular the involvement of multiple histidine–Cu2+ co-ordination events. Given the low availability of Cu2+ in the extracellular mileu (examined in more detail in the Discussion), this higher-affinity binding mode is more likely to be substantially populated in vivo.
Although the full-length prion protein is poorly soluble above pH 6 [22], it can, fortunately, be studied at lower pH values where multiple histidine–Cu2+ co-ordination events should be favoured. In aqueous solution, H+ ions compete for the basic co-ordinating groups that form a metal binding site, and thus the pKa of the co-ordinating groups and the pH of the solution are both determinants of the stability of a metal complex. The pKa of the histidine imidazole group is around 6, which is substantially lower than that of a backbone amide nitrogen, and thus co-ordination by multiple histidines will be maintained at lower pH compared with co-ordination modes involving deprotonated backbone amide groups. We find that binding of two Cu2+ ions is maintained in the full-length prion protein at pH 5.5, and we have also investigated the nature of the co-ordination in this complex. Here we report the locations of these binding sites and the identity of the co-ordinating groups. We demonstrate that these binding sites are of sufficient affinity to be maintained in the presence of substantial concentrations of exogenous histidine, which is known to bind Cu2+ in vivo [28]. Finally, we show how Cu2+ binding is altered in a truncated form of PrP, comprising residues 91–231 (PrP91–231), which lacks the octapeptide repeat regions, but which can still transmit infection.
EXPERIMENTAL
Expression, purification and metal stripping of PrP91–231
Human PrP91–231, with a methionine residue at position 129, was expressed and purified as described previously [29,30]. The PrP23–231 and PrP91–231 constructs used here both have the additional three-residue sequence SFR present at the N-terminus, which forms part of the thrombin cleavage site. To ensure the protein was free of metal ions, the protein was repeatedly buffer exchanged with 8 M urea, 20 mM EDTA, 25 mM Tris/acetate, pH 8.0, and then refolded by dilution in 5 mM Mes, pH 5.5, in an Amicon ultrafiltration unit (3 kDa molecular mass cut-off). The protein concentration for refolding was less than 1 mg/ml. All buffers used during and after the metal stripping step were prepared using, wherever possible, Aristar grade reagents. In addition all buffers were made using AnalR grade water and treated with Chelex resin (Bio-Rad Laboratories).
Expression, purification and metal stripping of full-length prion protein, PrP23–231
Full-length human prion protein, with a methionine residue at position 129, was expressed with an N-terminal His6-tag containing a thrombin cleavage site, using standard molecular biology protocols. Expression of uniformly 15N and 13C labelled protein and the initial purification by Ni2+-nitrilotriacetate chromatography was carried out using the same procedures described previously for PrP91–231 [29,30]. Oxidation of the disulfide bond was checked by HPLC [30] and was complete after 24 h without the aid of a catalyst. The protein was diluted to less than 1 mg/ml in 6 M GdnHCl (guanidinium chloride) and 50 mM Tris/HCl, pH 8.0, and then refolded by dialysis against 150 mM acetate/Tris and 0.02% NaN3, pH 3.5. In preparation for the thrombin cleavage step, the pH was gradually increased by successive dialysis steps against 10 mM Tris/acetate and 0.02% NaN3 at pH 4.5, 5.5, 6.5 and 7.0. The fusion protein was cleaved by 0.6 units of thrombin (Novagen) per mg of protein with 2.5 mM CaCl2. The reaction mixture was incubated at 37 °C for 24 h. Cleavage was halted by the addition of the Complete™ protease inhibitor cocktail (Roche). For the final purification step a higher pH was required and this had to be carried out under denaturing conditions for the protein to be soluble. Urea was added to the reaction mixture to a final concentration of 8 M and the pH was adjusted to 7.5. The denatured protein solution was then loaded on to an SP-Sepharose™ Fast Flow column (Amersham), pre-equilibrated with equilibration buffer (8 M urea, 150 mM NaCl and 50 mM Tris/HCl, pH 7.5). The column was washed extensively with the equilibration buffer. A salt gradient from 150 to 600 mM NaCl in 8 M urea, 50 mM Tris/HCl, pH 7.5 was run over 8 column volumes and the protein was eluted by approx. 300–400 mM NaCl. To remove any metal ions, the purified protein was repeatedly buffer exchanged against 8 M urea, 25 mM EDTA and 20 mM Tris/acetate, pH 8.0, and then refolded by dilution in 150 mM acetate/Tris, pH 3.5, in an Amicon ultra-filtration unit (3 kDa molecular mass cut-off). The protein concentration for refolding was less than 1 mg/ml. The buffer was exchanged to 20 mM acetate/TRIS pH 4.5 and then finally 10 mM Mes pH 5.5. All buffers used during and after the metal stripping step were prepared using, wherever possible, Aristar grade reagents. In addition all buffers were made using AnalR grade water and treated with Chelex resin (BioRad Laboratories).
NMR sample preparation
For backbone and side-chain assignment experiments, metal-free full-length prion protein in 20 mM Mes, pH 5.5 was concentrated to approx. 1 mM. NaN3 (to a final concentration of 1 mM), EDTA-free Complete™ protease inhibitor (Roche) and 10% 2H2O were added, and the pH readjusted to 5.5 by the addition of Mes. The final Mes concentration was approx. 30 mM. The sample was centrifuged at 16000 g for 5 min to remove any precipitated material. For titrations with Cu2+ the samples were prepared in the same way with the following exceptions, the protein was concentrated to approx. 0.5 mM or 0.1 mM and no NaN3 was added. An absorbance measurement at 280 nm was taken on an aliquot of the sample diluted into 6 M GdnHCl and 50 mM Tris/HCl, pH 8.0. The protein concentration was determined on the basis a calculated molar absorption coefficient [31]. Inductively coupled plasma MS analysis showed that no significant quantities of transition metals were present in the NMR sample (transition metals above 1 μM were: Cd, 5.5 μM; Cu, 4.7 μM; Zn, 3.6 μM; and Fe, 1.8 μM). For titration with Cu2+, a sample of 1.2 mM PrP91–231 was prepared in 5 mM Mes, pH 5.5, with 10% 2H2O. Measurement of the protein concentration was carried out by the method described above. Nitric acid washed NMR tubes were used for metal titration samples.
NMR resonance assignment experiments for the full-length prion protein
See online Supplementary Materials and methods at http://www.BiochemJ.org/bj/399/bj3990435add.htm for details.
Cu2+ titrations monitored by NMR
The PrP91–231 sample was titrated with four aliquots of 4 mM CuSO4, 10 mM glycine and 5 mM Mes, pH 5.5, to give molar fractions of Cu2+ relative to protein of 0.25, 0.50, 0.75 and 1.00. NMR spectra were recorded at 303 K on a Bruker Avance DRX-600 spectrometer. At each point in the titration a 1H15N HSQC (heteronuclear single quantum coherence) and aromatic and aliphatic 1H13C HSQCs were recorded. The full-length prion protein sample was titrated with 13 aliquots of 4 mM CuSO4, 10 mM glycine and 5 mM Mes, pH 5.5, to give molar fractions of Cu2+ relative to protein of 0.05, 0.10, 0.20 and, in steps of 0.20, to 2.20. NMR spectra were recorded at 303 K on a Bruker Avance AV-800 spectrometer. Again at each point in the titration a 1H15N HSQC and aromatic and aliphatic 1H13C HSQCs were recorded.
Determination of dissociation constants
Samples of full-length prion protein (3.5 μM) in 5 mM MES, pH 5.5, were prepared containing either 10 or 100 mM histidine. CuSO4 was added as a glycine complex in 5 mM Mes at pH 5.5, to give the appropriate final Cu2+ concentrations. Fluorescence measurements were made at 30 °C with an excitation wavelength of 291 nm and an emission wavelength of 345 nm, using a Jasco FP-750 spectrofluorimeter. The contribution of collisional quenching effects was evaluated by titrating samples of 20 μM N-acetyltryptophanamide (matching the concentration of tryptophan residues in the prion protein sample) with Cu2+ under the same conditions. The prion protein data was then corrected for the measured collisional quenching effect.
Analysis of the data and least-squares curve-fitting was carried out using the Grafit program (Erithacus Software). To obtain the apparent dissociation constant for the second Cu2+ binding event, the fluorescence data recorded in the presence of 10 mM histidine were fitted to a discontinuous function describing two phases of metal binding (eqns 1 and 2), where F0 is the initial fluorescence and FM is the fluorescence at a given metal concentration, A1 and A2 are the amplitudes of the fluorescence changes associated with each event, M is the total metal concentration, P is the total protein concentration and Kd2 is the apparent dissociation constant of the second binding event. A linear function describes the first event where the apparent Kd value is much less than the concentration of sites (M≤P; eqn 1), whilst the second event, for which the apparent Kd value is similar to the concentration of sites, is described by the quadratically-derived ligand binding equation (M>P; eqn 2).
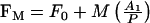 |
(1) |
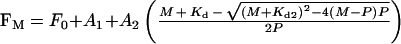 |
(2) |
The data acquired in the presence of 100 mM histidine were fitted to eqn 3 which describes metal binding to a single site, with a dissociation constant of Kd1. The apparent Kd value of the second site, under these conditions, will be too large to make a significant contribution to the observed fluorescence quenching and hence the contribution of this site was ignored in the fitting procedure.
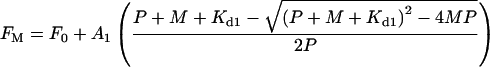 |
(3) |
The true Kd values of first and second Cu2+ binding events at pH 5.5 were calculated using eqn (4), where Kd(app), is the measured apparent dissociation constant, Kd1(His) and Kd2(His) are the stepwise dissociation constants for formation of a Cu(His)2 complex at pH 5.5, (1.2 μM and 160 μM respectively; [30]) and the concentration of histidine is [His].
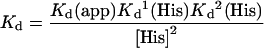 |
(4) |
RESULTS
NMR-detected effects of Cu2+ addition
Observation of NMR signals as a protein is titrated with Cu2+ provides a site-specific probe for the identification of nuclei close to the metal ion in a Cu2+–protein complex. Proximity to the paramagnetic Cu2+ ion causes acceleration of the T2 relaxation of nuclei, and hence broadens NMR signals, leading to a loss of peak height [32]. Nuclei close to the metal binding site can therefore be identified by a reduction in the height of their NMR signals as Cu2+ is titrated into the protein. Backbone and side-chain signals were monitored on addition of Cu2+ bisglycinate to PrP23–231 using 1H15N and 1H13C HSQC NMR spectra. The presence of glycine allows the Cu2+ to be added as a pH buffered solution by preventing the precipitation of Cu2+ as Cu(OH)2, which would otherwise occur at pH 5.5 [33]. The overlap in chemical shift of the signals from the octapeptide repeat region meant that these residues were represented by a single set of resonances, each of which was a combination of four or five signals, one from each repeating unit and from a pseudo-octapeptide repeat sequence at positions 52–59.
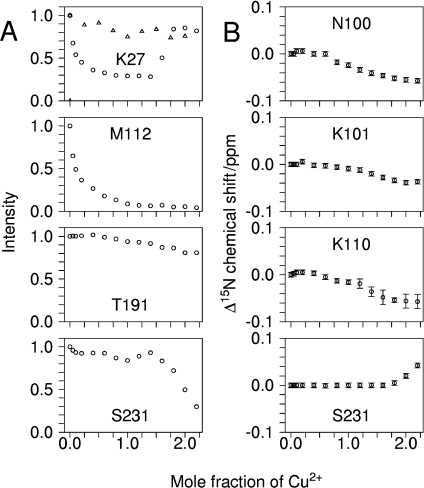
The effects of titration with Cu2+ on the height of the protein's NMR signals fall into four broad categories, of which representative examples are shown in Figure 1(A). Certain signals were almost completely unaffected, while others were initially unaffected but are attenuated after approx. 2 molar equivalents of Cu2+ have been added. A third group show substantial attenuation after addition of sub-stoichiometric quantities of Cu2+ and the final group behave similarly to these but begin to recover after approx. 1.4 molar equivalents of Cu2+ have been added.
Figure 1. Titration of full-length human prion protein with Cu2+ bis-glycinate monitored by 1H15N HSQC.
(A) NMR signal heights for residues Lys27, Met112, Thr191 and Ser231. (B) 15N chemical shift changes for residues Asn100, Lys101, Lys110 and Ser231. All data is from a titration of 440 μM of protein, with the exception of the data shown by triangles in the plot for Lys27, which is from a titration of 110 μM of protein.
The full-length prion protein binds two Cu2+ ions at pH 5.5
Numerous backbone [N,HN] (Throughout 2D 1H15N HSQC and 1H13C HSQC crosspeaks will be referred to by giving the two coupled nuclei that produce the crosspeaks in brackets, for example [Cβ, Hβ].) signals, for example those of Thr191 (Figure 1A), show very limited reduction in height throughout the titration, indicating that these residues are remote from any metal binding site. In contrast, there are a groups of signals from residues such as Ser231 (Figure 1A) for which, initially, the backbone [N,HN] signal peak heights are largely unaffected by Cu2+ addition but which rapidly lose height after approx. 2 molar equivalents of Cu2+ have been added. This behaviour occurs at multiple sites in the vicinity of residues that have a functional group with a propensity for metal binding (e.g. His140, Asp144, Asp178, Glu207 and Ser231). Similarly, in terms of chemical shift, some [N,HN] signals exhibit changes only after approx. 2 molar equivalents of Cu2+ have been added, including those of Ser143, Asp144, Tyr145 and Ser231 (Figure 1B). The affected residues are positioned at several distinct sites on the surface of the protein, so the effects cannot be due to a single specific Cu2+ binding site. Instead they are the result of several different transient interactions with Cu2+ ions, at separate locations on the protein's surface. The most noteworthy feature of these observations is that the NMR signals from residues involved in these Cu2+ binding sites are completely unaffected by the first two mole equivalents of Cu2+ added to the sample. The only explanation for this is that the protein completely sequesters two mole equivalents of Cu2+ in binding sites of substantially higher affinity. This shows that higher affinity Cu2+ binding in the full-length prion protein saturates at an apparent Cu/PrP stoichiometry of 1.8±0.2, which is compatible with a stoichiometry of 2:1.
Identification of the Cu2+ co-ordinating groups in the PrP–Cu2+ (complex of the full-length prion protein with one Cu2+ ion) and PrP–2Cu2+ (complex of the full-length prion protein with two Cu2+ ions) complexes
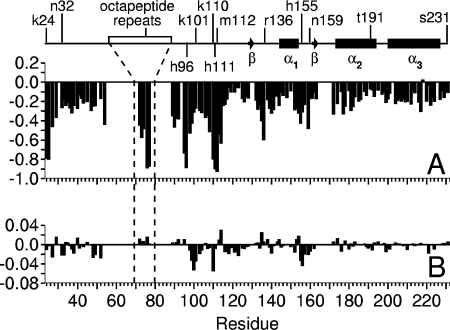
The behaviour of the third category of NMR signals, of which Met112 is typical (Figure 1A), reveals the identity of the co-ordinating groups that form the two higher affinity Cu2+ binding sites. This group of [N,HN] signals show a substantial loss of peak height on addition of the first aliquots of Cu2+ to the sample, with complete loss of the signal occurring prior to the addition of two molar equivalents of Cu2+. The largest reductions in peak heights are in signals from the six histidine residues of the N-terminal domain and from residues nearby in the sequence. Figure 2(A) shows the reductions in peak heights at a Cu/PrP stoichiometry of 1.8:1. At this stoichiometry, any reductions in peak heights due to the the weaker binding sites, described above, can be eliminated. The 1H15N HSQC data thus point to all six histidine residues of the N-terminal domain being involved in the formation of the higher affinity complexes. Importantly, no large changes in behaviour are observed for any of the NMR signals on passing a Cu/PrP stoichiometry of 1:1, which suggests that the first and second binding sites are not distinct from each other – a point that is discussed in more detail below. The behaviour of signals such as Ser231 [N,HN] (Figure 1A), which are largely protected from attenuation by Cu2+ binding up to a Cu/PrP stoichiometry approaching 2:1, allows us to rule out the possibility that the effects we attribute to formation of a Cu2+–protein complex are instead due to non-specific interactions with free Cu2+ ions.
Figure 2. Changes in the backbone [N,HN] NMR signals induced by Cu2+ binding to full-length human prion protein plotted against residue number.
The degenerate octapeptide repeat signals are plotted in the order (GG)WGQ(P)HG and no data is presented for the bracketed glycine residues due to signal overlap. Signal height changes are shown as a fraction of the starting value, for the addition of 1.8 mole equivalents of Cu2+ bisglycinate (A). The large reductions at residues 24–25, 95–96, 110–112 and the octapeptide repeat His-Gly sequence arise from paramagnetic quenching due to the proximity of these residues to Cu2+ in the PrP(Cu2+)2 complex. (B) The 15N chemical shift changes after addition of 1.8 mole equivalents of Cu2+ bisglycinate in p.p.m.
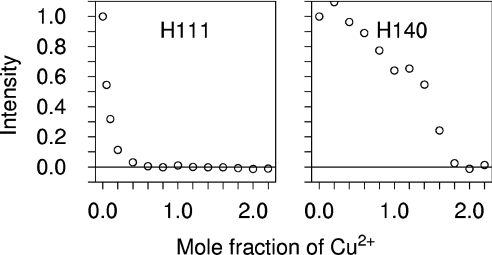
To confirm that all six histidine residues of the N-terminal domain are the co-ordinating residues, and to determine if any additional co-ordinating groups are involved, the behavior of [C,H] NMR signals on titration with Cu2+ was also measured. Signals completely attenuated by the addition of sub-stoichiometric amounts of Cu2+ included the [Cβ,Hβ], [Cδ2,Hδ2] and [Cϵ1,Hϵ1] signals of His96, His111 and all of the octapeptide repeat histidine residues, but not those of other histidine residues such as His140 (Figure 3). Attenuation of signals in this way is expected for nuclei that are close to Cu2+ in a protein complex, where the exchange rate of Cu2+ between protein molecules is fast on an NMR timescale (i.e. where the dissociation rate exceeds the chemical-shift change induced by binding in Hz). Thus all six histidines (i.e. four from the octapeptide repeats plus His96 and His111) must co-ordinate Cu2+ in the PrP(Cu2+)1 complex, and exchange on a 10 ms or faster timescale. The only other signals reduced in the same fashion were the [Cα,Hα] and [Cβ,Hβ] signals from the N-terminal Ser20, which is the first of the three residues added to the N-terminus of the full-length prion protein sequence to form a thrombin cleavage site. The N-terminal amine is thus also a co-ordinating group. The hydroxy group of Ser20 may be an additional ligand, though the affinity of this group for Cu2+ is predicted to be orders of magnitude weaker than that of the amine [34,35].
Figure 3. Reductions in the heights of histidine side-chain [Cδ,Hδ] NMR signals induced by proximity to Cu2+ in PrP23–231.
Plots show the signal height as a fraction of its starting value during titration with Cu2+ for His111 (A), part of the higher affinity binding site, and His140 (B), which only binds Cu2+ after higher affinity binding is saturated.
In summary, the data indicate that at least seven functional groups (the six histidine residues of the N-terminal domain and the N-terminal amine) contribute to co-ordinating a single Cu2+ ion in the PrP(Cu2+)1 complex. Cu2+ generally co-ordinates with three, four or five groups in proteins (http://metallo.scripps.edu/PROMISE/CUMAIN.html), and hence these seven co-ordinating groups cannot all be simultaneously involved in a single Cu2+ complex. The higher-affinity Cu2+ binding in the full-length prion protein must instead be derived from a rapidly exchanging ensemble of different co-ordination geometries, each of which has comparable stability, involving different combinations of the seven co-ordinating groups.
On increasing the Cu/PrP stoichiometry beyond 1:1, no new protein signals become substantially broadened until the second binding site is saturated. Therefore, the second binding site must be formed from a subset of the co-ordinating groups that take part in the ensemble of binding modes, which collectively form the highest-affinity site.
Self-association of the PrP–Cu2+ complex
In the fourth group of NMR signals another distinct behaviour is observed, peak height is initially reduced by the addition of Cu2+ but the signals recover towards their initial height as a Cu/PrP stoichiometry of 2:1 is approached (e.g. those of residue Lys27 in Figure 1A). This group of [N,HN] signals includes those from residues in several stretches of the N-terminal domain, namely: 24–48, 100–108 and 125–126. When the experiment was repeated with the protein concentration reduced from 440 μM to 110 μM (Lys27; Figure 1A), signals in this group were not dramatically affected by the addition of Cu2+, indicating that their behaviour at the higher protein concentration must be caused by an intermolecular process. Thus at sub-saturating levels of Cu2+ and 440 μM protein, a self-associated form of the prion protein must be at least transiently present. Concentration dependent effects are confined to the disordered N-terminal domain and thus show that it is this section of the protein that self-associates. The most probable cause of this self-association is that metal ions are readily co-ordinated by histidine side-chains from more than one protein molecule, since the concentration of the protein and the effective concentration of histidine residues in the same chain are likely to be similar. This self-associated protein species causes an acceleration of the relaxation of the nuclei in residues such as Lys27, either by placing them in closer proximity to the Cu2+ ion or through localized increases in the correlation time. At a Cu/PrP stoichiometry of 1.8:1, where these signals have fully recovered, we can infer that no significant amount of the self-associated species remains. At this point in the titration the effects on signals that arise solely from proximity to Cu2+ in the monomer can therefore be clearly distinguished and are illustrated in Figure 2(A).
Chemical-shift changes in the PrP–Cu2+ and PrP–2Cu2+ complexes
Large chemical-shift changes were not observed on formation of either PrP–Cu2+ or PrP–2Cu2+ complexes, indicating that the electronic environment for the paramagnetic ion must be close to isotropic in both cases [37]. However, smaller chemical-shift changes of less than 0.1 p.p.m. (parts per million) in the 15N dimension and less than 0.02 p.p.m. in the 1H dimension are present, confirming the rapid exchange of Cu2+ between prion protein molecules. The distribution of 15N chemical-shift changes for the PrP–2Cu2+ complex is plotted in Figure 2(B). Certain signals change in chemical shift up to a Cu/PrP stoichiometry of approx. 2:1 and then stop. This saturable behaviour shows that these chemical-shift changes must be associated with the formation of the PrP–Cu2+ and PrP–2Cu2+ complexes. The largest shift changes in this category are seen in signals from residues in the region between His96 and His111 (for example Asn100, Lys101 and Lys110 in Figure 1B), plus some from residues in the first helix (namely His155 and Arg156) and at the N-terminus (namely Lys24, Arg25 and Lys27). Similar but less substantial changes are seen in signals from other residues close to the N-terminus and a few residues near His155 and Arg156 in the C-terminal domain. The locations of these chemical-shift changes do not correlate with the changes in peak height, for example the height of signals like Lys101 and Asn100 are not substantially affected by Cu2+ binding, so these changes are unlikely to be a direct result of proximity to Cu2+. The shifts observed in the region between His96 and His111 (Figure 2B) are most likely reporting a conformational redistribution associated with the formation of a partially ordered loop between these residues. However the lack of substantial 1H chemical-shift-dispersion in the PrP–Cu2+ and the PrP–2Cu2+ complexes indicates Cu2+ binding does not bring about a highly ordered, side-chain-immobilized structure in the N-terminal domain of the full-length prion protein. Instead, Cu2+ binding at this second site uses the excess metal co-ordinating residues present in the ensemble of PrP–Cu2+ complexes to produce a poorly ordered structure.
Interaction of the Cu2+ binding site with the C-terminal domain
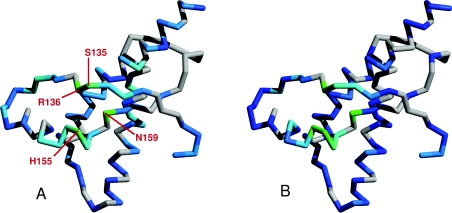
Although the C-terminal domain does not contribute directly to Cu2+ co-ordination at the higher-affinity site, the heights of certain [N,HN] NMR signals from the C-terminal domain are substantially reduced in the PrP–2Cu2+ complex. The largest of these reductions map to a region of the protein surface formed by residues close to the C-terminal end of helix 1 (His155 and Asn159) and the nearby loop between the first β-strand and helix 1 (Ser135 and Arg136; Figure 4A). The reductions can be explained if this area of the C-terminal domain surface comes into close proximity with the section of the N-terminal domain that co-ordinates Cu2+. An alternative explanation, where these reductions in peak height were due to direct weak Cu2+ binding at a site on the C-terminal domain can be eliminated. In this scenario the reductions in signal height would be expected to become more dramatic after the higher-affinity Cu2+ binding was saturated, in the same way as is seen at the Ser231 [N,HN] signal (Figure 1A). This does not happen and instead the changes stop at the 2:1 point of the titration showing that they must be related to the saturable higher-affinity binding.
Figure 4. The Cu2+-bound N-terminal domain contacts the C-terminal domain.
The reductions in the heights of backbone [N,HN] signals in the C-terminal domain of PrP, induced by proximity to the Cu2+ bound to the N-terminal domain, are illustrated on the backbone structure by a colour scale, ranging from dark blue to cyan (0–30% peak height reduction) and cyan to green (30–60% reduction). Data is shown for the full-length prion protein loaded with two Cu2+ ions (A) and the truncated PrP91–231 construct, loaded with one Cu2+ ion (B). Residues for which there are no data are coloured grey. Signals from the area of the C-terminal domain that contacts the unfolded N-terminal domain see the largest reductions (coloured green) because they are closer to the Cu2+ ion bound to the unfolded domain. The unfolded domain of the protein and the Cu2+ ion bound to it are omitted for clarity.
Chemical-shift changes are also observed for the same section of helix 1 (Figure 2B), arising either from the interaction between domains forming in a Cu2+ dependent fashion, or from Cu2+ binding to a region of the N-terminus, which was already interacting with the C-terminal domain. Strikingly, the changes observed for full-length prion protein, PrP23–231, bound to two Cu2+ ions are very similar to the changes we observe for the truncated construct PrP91–231 bound to a single Cu2+ in the same conditions (Figure 4B; experiments on this construct are discussed in more detail below). Since signals for residues 115–134 are not greatly affected by Cu2+ binding, the sequence responsible for the interaction with the C-terminal domain must lie between the residues at positions 91 and 114.
Measurement of Cu2+ binding affinity
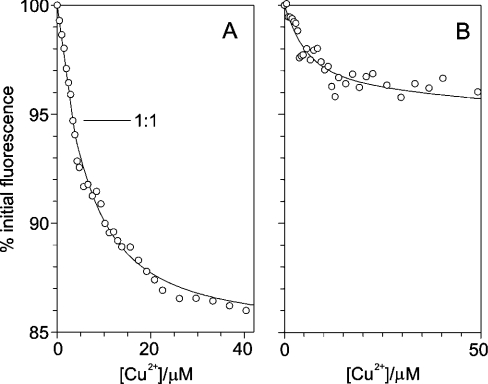
There are two highly specific Cu2+ binding events in the full-length prion protein that are apparent from the NMR observations. These events can be monitored at much lower protein concentration by measuring the quenching of the intrinsic tryptophan fluorescence, revealing that the two sites have significantly different affinities. In these experiments, the apparent affinities of the two metal binding events are reduced to measurable values by the inclusion of a competitive chelator that competes for free metal ions. With knowledge of the affinity of the competition reagent for the metal, a true dissociation constant for the protein can be calculated from the measured apparent dissociation constant. The affinities of both sites were measured using competition by histidine, which forms a Cu2+–bishistidinate complex. The affinity of the second of the two Cu2+ binding sites was measured in the presence of 10 mM histidine. Under these conditions, fluorescence quenching was linear to a stoichiometry of 1:1, indicating that the first binding event was at the strong binding limit. Throughout, we use ‘strong binding’ to describe a situation where the dissociation constant is much less than the concentration of binding sites and ‘weak binding’ for situations where it is similar to or greater than the concentration of sites. Weak binding is a condition necessary for measuring a dissociation constant in a titration experiment. In contrast, the second binding event had a measurable apparent dissociation constant of 6 μM (Figure 5A), showing that the affinities of the two binding events are not equal, as might be predicted from the similarity of the co-ordinating groups in each. To reduce the apparent affinity of the first site to a measurable value it was necessary to use competition with 100 mM histidine. Under these conditions an apparent dissociation constant of 4 μM was measured, with no initial strong binding phase (Figure 5B). In combination, these fluorescence data also show that the first and second binding events are not co-operative, since the data shown in Figures 1(A) and 1(B) are not compatible with a single dissociation constant process.
Figure 5. Cu2+ binding to full-length human prion protein monitored by tryptophan fluorescence quenching.
The graphs show the tryptophan fluorescence, as a percentage of the starting fluorescence, as Cu2+ bisglycinate is titrated into samples of 3.5 μM prion protein at pH 5.5 in the presence of two different competition systems, (A) 10 mM histidine and (B) 100 mM histidine. The point at which one mole equivalent of Cu2+ has been added to the protein, and the highest affinity site is therefore saturated, is labelled 1:1 in (A). The lines of best fit to the binding equations (described in the Experimental section) are shown.
The absolute affinity of histidine for Cu2+ can be used to calculate the affinity at pH 5.5 [30] and using this information we can make estimates of the true dissociation constants for the two binding events in the full-length prion protein at this pH. The interpretation of these competition experiments is, however, ambiguous, because it is not known if the protein is interacting with a Cu2+ ion or a Cu2+–monohistidinate complex. If the second binding event in the protein involves a Cu2+–mono-histidinate complex, log10 (Kd/M) for the Cu2+–histidine–protein complex would be −7.0±0.4, however if the protein is binding a free Cu2+ ion then the log10 (Kd/M) has a value of −10.8±0.4 (the Kd values calculated here are Kd values at pH 5.5. To calculate intrinsic Kd values it would be necessary to take into account the protonation state of the metal binding site of the protein.). For the first binding event, the apparent dissociation constant translates to a true Kd value of log10 (Kd/M) of −8.2±0.5 for binding of a Cu2+–mono-histidinate complex, or to log10(Kd/M) of −13.1±0.5 for binding of a free Cu2+ ion. Despite the ambiguities, it can be concluded that the affinities of the two Cu2+ binding events at pH 5.5 are both in the submicromolar range and are at least an order of magnitude apart.
Identification of Cu2+ co-ordinating groups in PrP91–231
The integrated nature with which all of the histidine side-chains of the N-terminal domain are involved in Cu2+ co-ordination in the full-length prion protein is somewhat surprising, given that a truncated form of the protein without the octapeptide repeat regions (PrP91–231) is also capable of submicromolar affinity Cu2+ binding [12]. In PrP91–231, the Cu2+ binding site was proposed to involve His96 and His111 on the basis of broadening of NMR signals from backbone NH moieties and X-ray absorption data [12,38]. To investigate the relationship between the two Cu2+ binding modes, we have carried out further characterization of the PrP91–231–Cu2+ complex under equivalent conditions to our investigation of the full-length protein.
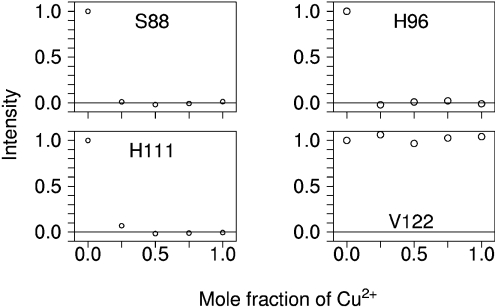
The identity of the ligands was determined by following the titration of Cu2+ bisglycinate into a sample of 15N 13C labelled PrP91–231 using heteronuclear NMR methods, as described above. After addition of 0.25 molar equivalents of Cu2+ to PrP91–231, a specific group of [C,H] signals were completely attenuated, whilst the majority of signals, such as Val122, were unaffected by Cu2+ addition (Figure 6). The attenuated signals included the [Cβ,Hβ], [Cδ2,Hδ2] and [Cϵ1,Hϵ1] resonances of His96 and His111. In addition, the [Cα,Hα] and [Cβ,Hβ] signals of the N-terminal Ser88 also became unobservable. As in the case of our PrP23–231 construct, Ser88 was one of three residues inserted on the N-terminal side of the PrP91–231 sequence to form a thrombin cleavage site. The total quenching of these signals at substoichiometric Cu2+ concentrations again shows that exchange of Cu2+ between PrP91–231 molecules occurs rapidly within the timescale of the NMR experiment. None of the side-chain signals for other potential ligands (namely Gln91, Thr95, Ser97, Gln98, Asn100, Ser103, Thr107, Asn108, Met109 and Met112) or signals for backbone amides exhibited this complete quenching at the first addition of Cu2+, thus demonstrating that at pH 5.5 PrP91–231 co-ordinates Cu2+ through the two histidine imidazoles of the N-terminal region and the N-terminal amine.
Figure 6. Effects of titration with Cu2+ bisglycinate on the heights of side-chain [Cβ,Hβ] NMR signals in PrP91–231.
Reductions induced by proximity to Cu2+ show the co-ordinating groups for the Cu2+ binding site in this truncated construct. The plots show the signal height as a fraction of its starting value during titration with Cu2+.
In previous work, the affinity of Cu2+ binding in the PrP91–231 construct was over-estimated. The affinities reported for the interaction of the truncated prion protein constructs PrP91–231 (and PrP52–98) with Cu2+ [12] were calculated from apparent dissociation constants measured in the presence of glycine at pH 8, but this did not take into account the extent of protonation of the glycine amino group at this pH, which reduces the stability of the glycinate complex and weakens the competition for Cu2+. The stepwise dissociation constants for the Cu2+ bisglycinate complex at pH 8 are 0.3 and 3 μM, 40-fold greater than the absolute dissociation constants for the formation of this complex from fully deprotonated glycine, which were used in the original calculations. When this is considered, the dissociation constant for free Cu2+ binding to PrP91–231 is in the picomolar range, with a log (Kd/M) of −10.1 and the equivalent value for PrP52–98 is −10.5.
DISCUSSION
A model of Cu2+ binding to the full-length prion protein
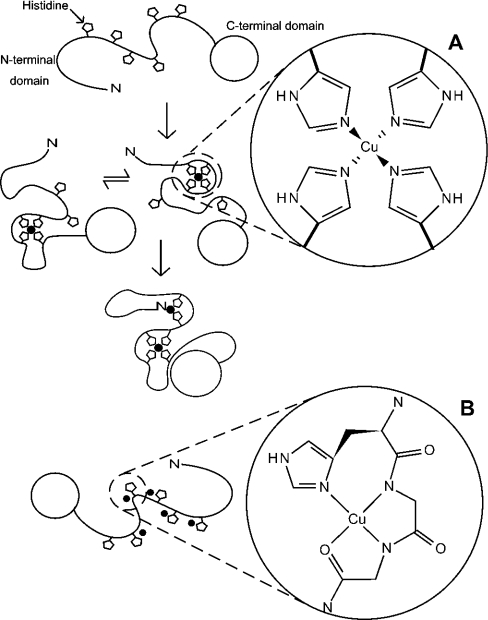
A model of Cu2+ binding to the full-length prion protein is illustrated in Figure 7(A). The PrP–Cu2+ complex is an ensemble of species in which Cu2+ co-ordination is shared between the six histidine residues of the N-terminal domain and the N-terminal amine, with rapid exchange of ligands at the metal centre. Some fraction of the PrP–Cu2+ species exists as an oligomer, which is likely to simply comprise Cu2+ ions co-ordinated by histidine residues from more than one protein molecule. The PrP–2Cu2+ complex co-ordinates Cu2+ through total occupancy of the ligands used in the PrP–Cu2+ ensemble of complexes. With the present approach it cannot be determined which ligands co-ordinate each Cu2+ ion, and it is possible that PrP–2Cu2+ is also a rapidly exchanging ensemble of species with different co-ordination modes. Although the formation of two distinct sites would be a simpler model, this cannot account for the broadening of NMR signals from all seven co-ordinating groups by the first addition of Cu2+ and the observation that no further signals are broadened after the 1:1 point of the titration. In the wild-type protein, lacking the three residue addition in the construct used here, co-ordination by the Ser20 amine would almost certainly be replaced by the Lys23 backbone amine, since the basicity of the backbone amines and the effective molarity of the two amino acids are virtually the same. The backbone amine group in lysine has a pKa of 9.18 and the dissociation constant for formation of a 1:1 Cu2+ complex is 10−7.6 M, whilst for serine these values are 9.60 and 10−7.9 M respectively [30]. The pKa of the sidechain amine in lysine is 10.72, indicating it is far less likely to be involved in metal binding.
Figure 7. An illustration of the two alternative modes of Cu2+ co-ordination in the full-length prion protein with details showing the co-ordination environment of the Cu2+ ion in each case.
Binding of Cu2+ by the full-length prion protein at pH 5.5 involves co-ordination by an exchanging combination of histidine imidazoles (A). The Figure shows apo-PrP (top), the PrP–Cu2+ ensemble (middle) and PrP–2Cu2+ (bottom), which may also be an ensemble of different co-ordinations involving the same ligands. Note that the assignment of particular combinations of ligands to any one complex or metal centre is arbitrary. The histidine Nϵ co-ordination shown is based on an IR study of multiple histidine co-ordination in the octapeptide repeats [13]. At pH 7.4, with saturating concentrations of copper, the full-length prion protein can bind at least five Cu2+ ions, each co-ordinated by a single imidazole and nearby backbone amides (B).
Here we have shown that at pH 5.5 the full-length prion protein is capable of binding two Cu2+ ions with co-ordination from multiple histidine imidazole groups (Figure 7A). At first, this may appear to contradict some other studies that have reported that the protein can bind five or six Cu2+ ions at pH 7.4 with a co-ordination involving deprotonated backbone amides [19] (Figure 7B), but the crucial difference between the experiments is the pH. The mode of Cu2+ co-ordination that occurs at neutral pH, with maximum Cu2+ occupancy, has been extensively studied. The octapeptide repeats can bind four Cu2+ ions, with one ion in each repeat unit, and with co-ordination of each ion by a histidine imidazole Nδ1, two deprotonated backbone amides from adjacent glycine residues and a carbonyl oxygen [16]. Cu2+ ions can also bind at two additional and independent sites, involving the residues His96 and His111, again with co-ordination from the histidine imidazole and nearby backbone amides [19,26]. This brings the total number of sites to six.
However recent studies of peptides, which comprise the four repeats of the octapeptide sequence, have shown that if the peptide binds only a single Cu2+ ion, then co-ordination involves multiple histidine imidazole groups [24,25]. When Cu2+ binding in the peptide is saturated, this form of binding is replaced by the four identical sites involving backbone amides. The latter binding sites are very sensitive to reduction in pH because each involves two deprotonated backbone amide groups and the pKa of this group is well above 7.4. In contrast, multiple histidine co-ordination would be expected to be maintained at lower pH values because the pKa of the histidine group is around 6, and this is indeed what we observe in the full-length protein at pH 5.5.
The peptide studies of Valensin et al. [25] and Chattopadhyay et al. [24] and our own recently published work [38a], suggest that the multiple histidine Cu2+ co-ordination mode we observe in the full-length protein at pH 5.5 will be similar to the binding mode that occurs at neutral pH at low Cu2+ occupancy, even though at maximum Cu2+ occupancy co-ordination will be very different (Figure 7B). Consistent with this proposal EPR investigations also report that, in the presence of 1 or 2 molar equivalents of Cu2+ at pH 7.4, Cu2+ co-ordination is different to that which occurs in the Cu2+ saturated species [19]. It is further supported by an X-ray absorption study, which found evidence that single and multiple imidazole Cu2+co-ordination might both be accessible in the full-length protein at neutral pH [39].
The affinity of the prion protein for Cu2+ and the availability of Cu2+ are the key determinants of whether the prion protein binds Cu2+ in vivo. The high capacity, backbone amide co-ordinated form of Cu2+ binding, which is accessible at pH 7.4, produces a substantial CD signal, and this has been used to determine that the affinity of these sites is in the low micromolar range [14]. In vivo, virtually all extracellular Cu2+ is sequestered in complexes with affinities significantly higher than micromolar, for example with amino acids such as histidine or with proteins such as serum albumin, which have affinities in the low nanomolar and low picomolar range respectively [28,40]. A micromolar affinity therefore suggests that the cellular prion protein does not bind Cu2+ in vivo. However, the preference of the prion protein for multiple histidine Cu2+ co-ordination at low Cu2+ concentrations and at lower pH values shows that this is likely to be the highest- affinity form of binding in the protein. The affinity measurements carried out here indicate that this form of co-ordination has an affinity at least in the nanomolar range and potentially higher. While not conclusively showing that the prion protein is an authentic cuproprotein, it is clear that the multiple histidine mode of Cu2+ binding is the most likely to be of biological relevance. In addition, the capability for Cu2+ binding at pH 5.5 indicates that Cu2+ binding could be maintained during the cycling of the prion protein through acidic endosomal compartments [41], which have been postulated to be the site of PrPSc formation [42].
It should be emphasized that the Cu2+ dependent oligomerization we observe is a distinct process from the oligomerization involved in formation of either PrPSc or PrP fibrils. We observe association of the disordered N-terminal domain with no evidence for any structural changes in the globular C-terminal domain. In contrast, formation of PrPSc or PrP fibrils from monomeric PrPC involves a substantial conformational change within residues 90–231, giving an increase in β-sheet type structure [43,44] and does not require residues 23–89 [45], which contain the octapeptide repeats. Nevertheless, our work suggests that metal binding may play a role in controlling self-association of PrP in vivo, which could influence the rate of formation of other PrP oligomers such as PrPSc. Although the Cu2+-dependent self-association is relatively weak at pH 5.5, requiring protein concentrations in the order of 0.5 mM, at pH 7.4 self-association is likely to be stronger. At this pH we have found that Cu2+ binding to a peptide comprising the four tandem octapeptide repeats causes self-association with peptide concentrations in the sub-micromolar range (M. A. Wells and G. S. Jackson, unpublished work). Also, in vivo, effective concentrations of the prion protein may be vastly elevated above the true concentration because the protein is anchored to the cell membrane and is further localized in cholesterol-rich lipid rafts [46], which may be sufficient to allow Cu2+ binding to trigger self-association.
The involvement of His96 and His111 in Cu2+ binding in the full-length protein shows that metal ion occupancy must play a role in determining the conformation of this section of the protein in the PrPC isoform, as has also been shown for PrPSc [47]. Given the known importance of this region of the protein for prion propagation, the role of metal ions in the conversion process should be considered. The formation of a single Cu2+ binding site by histidine residues from the unstructured N-terminal domain also has possible implications for understanding the mechanism by which a unique group of pathogenic mutations, namely the octapeptide repeat insertion mutations, cause disease. Most of the pathogenic mutations seen in inherited prion disease occur within residues 89–231 [1], which form the relatively protease resistant core of PrPSc and are sufficient to support prion replication and the development of pathology [45,48]. Insertional mutations comprising addition of integral numbers of the octapeptide repeat sequence to the four repeats of the wild-type protein are the exception [1]. The mechanism by which the octapeptide repeat region influences the formation of PrPSc is unclear, especially since this section of the protein is unfolded and highly mobile in PrPC, and is rapidly digested by proteinase K in PrPSc [49]. However the Cu2+ mediated interaction demonstrated here between the octapeptide repeats and His96 and His111, illustrates that the insertions would cause alterations in metal co-ordination, which have the potential to modulate PrPSc formation by causing conformational changes within residues 91–231.
Online data
Acknowledgments
This work was funded by the U.K. Medical Research Council and the U.K. Biotechnology and Biological Sciences Research Council. We thank Clare Trevitt for assistance with protein expression and purification, Matt Cliff and Lilia Milanesi for critical reading of the manuscript, Cameron McLeod and Neil Bramall for trace metal analysis of NMR samples, and Accelrys for the provision of Felix.
References
- 1.Collinge J. Prion diseases of humans and animals: Their causes and molecular basis. Annu. Rev. Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S. B. Prions. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith J. S. Self-replication and scrapie. Nature (London) 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner S. B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 5.Oesch B., Westaway D., Walchli M., McKinley M. P., Kent S. B. H., Aebersold R., Barry R. A., Tempst P., Teplow D. B., Hood L. E., et al. A cellular gene encodes scrapie Prp 27-30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 6.Stahl N., Borchelt D. R., Hsiao K., Prusiner S. B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 7.Bueler H., Fischer M., Lang Y., Bluethmann H., Lipp H. P., Dearmond S. J., Prusiner S. B., Aguet M., Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface Prp protein. Nature (London) 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 8.Collinge J., Whittington M. A., Sidle K. C. L., Smith C. J., Palmer M. S., Clarke A. R., Jefferys J. G. R. Prion protein is necessary for normal synaptic function. Nature (London) 1994;370:295–297. doi: 10.1038/370295a0. [DOI] [PubMed] [Google Scholar]
- 9.Brown D. R., Qin K. F., Herms J. W., Madlung A., Manson J., Strome R., Fraser P. E., Kruck T., vonBohlen A., SchulzSchaeffer W., et al. The cellular prion protein binds copper in vivo. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 10.Hornshaw M. P., McDermott J. R., Candy J. M. Copper-binding to the N-terminal tandem repeat regions of mammalian and avian prion protein. Biochem. Biophys. Res. Commun. 1995;207:621–629. doi: 10.1006/bbrc.1995.1233. [DOI] [PubMed] [Google Scholar]
- 11.Hornshaw M. P., McDermott J. R., Candy J. M., Lakey J. H. Copper-binding to the N-terminal tandem repeat region of mammalian and avian prion protein – structural studies using synthetic peptides. Biochem. Bioph. Res. Co. 1995;214:993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 12.Jackson G. S., Murray I., Hosszu L. L. P., Gibbs N., Waltho J. P., Clarke A. R., Collinge J. Location and properties of metal-binding sites on the human prion protein. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8531–8535. doi: 10.1073/pnas.151038498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura T., Hori-i A., Mototani H., Takeuchi H. Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–11569. doi: 10.1021/bi9909389. [DOI] [PubMed] [Google Scholar]
- 14.Viles J. H., Cohen F. E., Prusiner S. B., Goodin D. B., Wright P. E., Dyson H. J. Copper binding to the prion protein: structural implications of four identical cooperative binding sites. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2042–2047. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittal R. M., Ball H. L., Cohen F. E., Burlingame A. L., Prusiner S. B., Baldwin M. A. Copper binding to octarepeat peptides of the prion protein monitored by mass spectrometry. Protein Sci. 2000;9:332–343. doi: 10.1110/ps.9.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns C. S., Aronoff-Spencer E., Dunham C. M., Lario P., Avdievich N. I., Antholine W. E., Olmstead M. M., Vrielink A., Gerfen G. J., Peisach J., et al. Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry. 2002;41:3991–4001. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garnett A. P., Viles J. H. Copper binding to the octarepeats of the prion protein. Affinity, specificity, folding, and cooperativity: insights from circular dichroism. J. Biol. Chem. 2003;278:6795–6802. doi: 10.1074/jbc.M209280200. [DOI] [PubMed] [Google Scholar]
- 18.Aronoff-Spencer E., Burns C. S., Avdievich N. I., Gerfen G. J., Peisach J., Antholine W. E., Ball H. L., Cohen F. E., Prusiner S. B., Millhauser G. L. Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry. 2000;39:13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burns C. S., Aronoff-Spencer E., Legname G., Prusiner S. B., Antholine W. E., Gerfen G. J., Peisach J., Millhauser G. L. Copper co-ordination in the full-length, recombinant prion protein. Biochemistry. 2003;42:6794–6803. doi: 10.1021/bi027138+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stockel J., Safar J., Wallace A. C., Cohen F. E., Prusiner S. B. Prion protein selectively binds copper(II) ions. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 21.Kramer M. L., Kratzin H. D., Schmidt B., Romer A., Windl O., Liemann S., Hornemann S., Kretzschmar H. Prion protein binds copper within the physiological concentration range. J. Biol. Chem. 2001;276:16711–16719. doi: 10.1074/jbc.M006554200. [DOI] [PubMed] [Google Scholar]
- 22.Zahn R. The octapeptide repeats in mammalian prion protein constitute a pH-dependent folding and aggregation site. J. Mol. Biol. 2003;334:477–488. doi: 10.1016/j.jmb.2003.09.048. [DOI] [PubMed] [Google Scholar]
- 23.Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., Wuthrich K. NMR structure of the mouse prion protein domain PrP(121–321) Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 24.Chattopadhyay M., Walter E. D., Newell D. J., Jackson P. J., Aronoff-Spencer E., Peisach J., Gerfen G. J., Bennett B., Antholine W. E., Millhauser G. L. The octarepeat domain of the prion protein binds Cu(II) with three distinct co-ordination modes at pH 7.4. J. Am. Chem. Soc. 2005;127:12647–12656. doi: 10.1021/ja053254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valensin D., Luczkowski M., Mancini F. M., Legowska A., Gaggelli E., Valensin G., Rolka K., Kozlowski H. The dimeric and tetrameric octarepeat fragments of prion protein behave differently to its monomeric unit. Dalton Trans. 2004:1284–1293. doi: 10.1039/b402090a. [DOI] [PubMed] [Google Scholar]
- 26.Jones C. E., Klewpatinond M., Abdelraheim S. R., Brown D. R., Viles J. H. Probing copper2+ binding to the prion protein using diamagnetic nickel2+ and 1H NMR: the unstructured N terminus facilitates the co-ordination of six copper2+ ions at physiological concentrations. J. Mol. Biol. 2005;346:1393–1407. doi: 10.1016/j.jmb.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 27.Jones C. E., Abdelraheim S. R., Brown D. R., Viles J. H. Preferential Cu2+ co-ordination by His96 and His111 induces β-sheet formation in the unstructured amyloidogenic region of the prion protein. J. Biol. Chem. 2004;279:32018–32027. doi: 10.1074/jbc.M403467200. [DOI] [PubMed] [Google Scholar]
- 28.Linder M. C. Extracellular copper substituents and mammalian copper transport. In: Linder M. C., editor. Biochemistry of Copper. New York: Plenum Press; 1991. pp. 73–134. [Google Scholar]
- 29.Hosszu L. L. P., Jackson G. S., Trevitt C. R., Jones S., Batchelor M., Bhelt D., Prodromidou K., Clarke A. R., Waltho J. P., Collinge J. The residue 129 polymorphism in human prion protein does not confer susceptibility to Creutzfeldt–Jakob disease by altering the structure or global stability of PrPC. J. Biol. Chem. 2004;279:28515–28521. doi: 10.1074/jbc.M313762200. [DOI] [PubMed] [Google Scholar]
- 30.Jackson G. S., Hill S. F., Joseph C., Hosszu L., Power A., Waltho J. P., Clarke A. R., Collinge J. Multiple folding pathways for heterologously expressed human prion protein. BBA-Protein Struct. M. 1999;1431:1–13. doi: 10.1016/s0167-4838(99)00038-2. [DOI] [PubMed] [Google Scholar]
- 31.Gill S. C., Vonhippel P. H. Calculation of protein extinction coefficients from amino-acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 32.Bertini I., Luchinat C., Parigi G. Solution NMR of Paramagnetic Molecules. Amsterdam: Elsevier; 2001. Nuclear relaxation due to dipolar coupling with unpaired electrons; pp. 89–96. [Google Scholar]
- 33.Dawson R. M. C., Elliott D. C., Elliott W. H., Jones K. M. Stability constants for metal complexes. In: Dawson R. M. C., editor. Data for Biochemical Research. Oxford: Oxford University Press; 1986. pp. 399–416. [Google Scholar]
- 34.Shriver D. F., Atkins P. W., Langford C. H. Oxford: Oxford University Press; 1994. Inorganic Chemistry. [Google Scholar]
- 35.Wilkinson G., Gillard R. D., McCleverty J. Stability of complexes. In: Wilkinson G., Gillard R. D., McCleverty J., editors. Comprehensive Co-ordination Chemistry. Oxford: Pergamon; 1987. p. 526. [Google Scholar]
- 36. Reference deleted.
- 37.Bertini I., Luchinat C., Parigi G. Solution NMR of Paramagnetic Molecules. Amsterdam: Elsevier; 2001. Metal centred point-dipole approximation; pp. 37–42. [Google Scholar]
- 38.Hasnain S. S., Murphy L. M., Strange R. W., Grossmann J. G., Clarke A. R., Jackson G. S., Collinge J. XAFS study of the high-affinity copper-binding site of human PrP91–231 and its low-resolution structure in solution. J. Mol. Biol. 2001;311:467–473. doi: 10.1006/jmbi.2001.4795. [DOI] [PubMed] [Google Scholar]
- 38a.Wells M. A., Jelinska C., Hosszu L. L. P., Craven C. J., Clarke A. R., Collinge J., Waltho J. P., Jackson G. S. Multiple forms of copper (II) co-ordination occur throughout the N-terminal region of the prion protein at pH 7.4. Biochem. J. 2006 doi: 10.1042/BJ20060721. Immediate publication doi 10.1042/BJ20060721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morante S., Gonzalez-Iglesias R., Potrich C., Meneghini C., Meyer-Klaucke W., Menestrina G., Gasset M. Inter- and intra-octarepeat Cu(II) site geometries in the prion protein – implications in Cu(II) binding cooperativity and Cu(II)-mediated assemblies. J. Biol. Chem. 2004;279:11753–11759. doi: 10.1074/jbc.M312860200. [DOI] [PubMed] [Google Scholar]
- 40.Masuoka J., Hegenauer J., Vandyke B. R., Saltman P. Intrinsic stoichiometric equilibrium – constants for the binding of zinc(II) and copper(II) to the high-affinity site of serum-albumin. J. Biol. Chem. 1993;268:21533–21537. [PubMed] [Google Scholar]
- 41.Shyng S. L., Huber M. T., Harris D. A. A Prion Protein Cycles between the Cell-Surface and an Endocytic Compartment in Cultured Neuroblastoma-Cells. J. Biol. Chem. 1993;268:15922–15928. [PubMed] [Google Scholar]
- 42.Mayer R. J., Landon M., Laszlo L., Lennox G., Lowe J. Protein processing in lysosomes: the new therapeutic target in neurodegenerative disease. Lancet. 1992;340:156–159. doi: 10.1016/0140-6736(92)93224-b. [DOI] [PubMed] [Google Scholar]
- 43.Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z. W., Fletterick R. J., Cohen F. E., Prusiner S. B. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baskakov I. V., Legname G., Gryczynski Z., Prusiner S. B. The peculiar nature of unfolding of the human prion protein. Protein Sci. 2004;13:586–595. doi: 10.1110/ps.03457204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers M., Yehiely F., Scott M., Prusiner S. B. Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3182–3186. doi: 10.1073/pnas.90.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vey M., Pilkuhn S., Wille H., Nixon R., DeArmond S. J., Smart E. J., Anderson R. G., Taraboulos A., Prusiner S. B. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wadsworth J. D. F., Hill A. F., Joiner S., Jackson G. S., Clarke A. R., Collinge J. Strain-specific prion-protein conformation determined by metal ions. Nat. Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 48.Fischer M., Rulicke T., Raeber A., Sailer A., Moser M., Oesch B., Brandner S., Aguzzi A., Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 49.Prusiner S. B., Groth D. F., Bolton D. C., Kent S. B., Hood L. E. Purification and structural studies of a major scrapie prion protein. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.