Abstract
SNAREs are important components of the vesicle trafficking machinery in eukaryotic cells. In plants, SNAREs have been found to play a variety of roles in the development and physiology of the whole organism. Here, we describe the identification and characterization of a novel plant-specific SNARE, NPSN11, a member of a closely related small gene family in Arabidopsis. NSPN11 is highly expressed in actively dividing cells. In a subcellular fractionation experiment, NSPN11 cofractionates with the cytokinesis-specific syntaxin, KNOLLE, which is required for the formation of the cell plate. By immunofluorescence microscopy, NSPN11 was localized to the cell plate in dividing cells. Consistent with the localization studies, NSPN11 was found to interact with KNOLLE. Our results suggest that NPSN11 is another component of the membrane trafficking and fusion machinery involved in cell plate formation.
SNAREs are a family of proteins involved in vesicle trafficking that share similar secondary structures. Most SNAREs, with the exception of the SNAP25 family, are type II membrane proteins with a stretch of hydrophobic residues at the C terminus of the protein that form a transmembrane domain. Typical SNAREs contain a cytosolically exposed coiled-coil domain that is important for interacting with other SNAREs and that contributes to the specificity of the SNARE interactions (Sutton et al., 1998). SNAREs can be divided into two groups: v-SNAREs are localized on the transport vesicles membrane, and t-SNAREs are on the target membrane. Another important feature of SNARE proteins is that they have distinct intracellular distributions. Often, the specific membrane fusion event that they play a role in can be inferred by their subcellular location.
During vesicle fusion, the energy required for the consolidation of the two opposing membrane bilayers is thought to be provided by the assembly of a highly stable heterogeneous SNARE complex through interactions of the coiled-coil domain of SNAREs localized on the vesicle and acceptor membranes. Structural analysis has shown that the final SNARE complex is composed of four different helices: one that is contributed from a v-SNARE on the transport vesicle, one from a syntaxin-type SNARE localized on the target membrane, and two additional helices from a SNAP-25 molecule or two additional t-SNAREs on the target membrane (Antonin et al., 2000). Although it is possible to form arbitrary SNARE complexes in vitro, it is believed that the formation and composition of specific SNARE complexes are highly regulated in vivo. Most likely, other secretory compartment-specific factors, such as Rab-GTPases, Sec 1 family proteins, and other “tethering factors” such as Uso1p and p115, are required for the precise docking and fusion of vesicles with their appropriate target compartment (for review, see Pfeffer, 1999). In particular, the Sec1 family of proteins is well conserved between different eukaryotic organisms. Sec1 proteins are peripheral membrane proteins that interact with syntaxin type SNAREs and recruit a special set of tethering factors. They play an important role in regulating the function of the syntaxins and the formation of the SNARE complex (Hanson, 2000). The basic machinery for membrane fusion is universal to all eukaryotic organisms, although each organism uses a distinct set of components depending on their particular requirements. Recent research indicates a role for this machinery in plant physiology and development, for example, in processes like cytokinesis.
Cytokinesis describes a process that partitions the cytoplasm between two daughter nuclei at the end of the cell division. One unique feature of plant cytokinesis is the requirement of cell wall formation at the cell plate. During cytokinesis, many secretory vesicles carrying cell wall material are directed to the division plane by a specialized cytoskeletal apparatus known as the phragmoplast, where they fuse to form the cell plate (Staehelin and Hepler, 1996). Continued growth of the cell plate leads to its eventual fusion with the plasma membrane and formation of a new cross wall between the two daughter cells. The fusion between cell plate-forming secretory vesicles and between the cell plate and the plasma membrane was found to require the cytokinesis-specific syntaxin-type SNARE, KNOLLE (Lukowitz et al., 1996; Lauber et al., 1997). The gene encoding KNOLLE was identified in a forward genetic screen for mutations affecting the body organization of Arabidopsis seedlings (Mayer et al., 1991; Lukowitz et al., 1996). In knolle (kn) mutants, which die as seedlings, cell division in the embryos is retarded, and defects in cell plate membrane consolidation are often observed (Lukowitz et al., 1996). The KNOLLE protein is produced only in dividing cells and is localized on the cell plate, consistent with the phenotype of the mutation (Lauber et al., 1997). Therefore, it is very likely that KNOLLE facilitates cell plate vesicle fusion during plant cytokinesis. SNAP33, a SNAP25-like SNARE that interacts with KNOLLE and that is localized on the plasma membrane and cell plate, was recently also found likely to be involved in cell division. Plants that carry mutations in SNAP33 (snp33) are able to proceed through development to the mature leaf stage with accumulation of necrotic spots and eventually die before flowering (Heese et al., 2001). In necrotic cells of the leaf, incomplete cell walls were observed, consistent with a role in cytokinesis (Heese et al., 2001). In contrast to kn mutants, snp33 mutants mainly show defects later in development (Heese et al., 2001). This is probably because in Arabidopsis, SNAP33 is one member of a three-member SNAP25 gene family and because the other family members may perform partially redundant roles during plant growth and development (Heese et al., 2001). Aside from these SNAREs, KEULE, a Sec1 family member that interacts with KNOLLE (Assaad et al., 2001), is also likely to be involved in the same pathway, because keule (keu) mutants have a similar phenotype to kn, and the mutation is synthetic lethal with both kn and snp33 (Waizenegger et al., 2000; Heese et al., 2001). Similar to that found in secretion of synaptic vesicles in the mammalian brain, the syntaxin KNOLLE and SNAP33 form a three-helix t-SNARE bundle. What remains to be found is a v-SNARE that would contribute the fourth helix necessary for the formation of the SNARE complex that drives cell plate vesicle fusion. The identity of this v-SNARE, however, has yet to be reported.
In an attempt to identify and characterize the v-SNAREs that may be involved in vesicle trafficking required for different cellular functions in plants, we examined the many groups of SNAREs found in Arabidopsis (Sanderfoot et al., 2000). A particular family of plant SNAREs have no homolog in the mammalian or yeast genomes and were called the novel plant SNAREs (NPSN; Sanderfoot et al., 2000). Three NPSN gene family members have been identified in Arabidopsis (NPSN11, -12, and -13) and their orthologs are found in other plants. We characterized one of these genes, NPSN11. Using NPSN11-specific antiserum, we showed that NPSN11 is a 36-kD membrane protein that was highly expressed in tissues containing actively dividing cells. NPSN11 was localized primarily on the cell plate in dividing cells. Moreover, NSPN11 was shown by co-immunoprecipitation experiments to interact with KNOLLE. These results suggest that NPSN11 may be involved in cytokinesis. We also report an npsn11 mutant that shows no obvious phenotype, indicating that the other members of the NPSN family may have redundant functions.
RESULTS
NPSN Gene Family of Arabidopsis
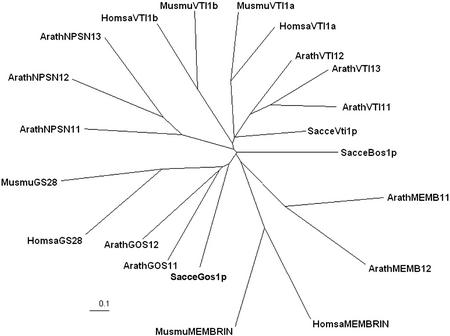
Extensive searches of the Arabidopsis genome identified 55 genes encoding putative SNAREs. One gene family, NPSN, is composed of three genes. The proteins encoded by these genes share approximately 61% to 92% identity among each other and with predicted members from other plants. However, this family has no close relatives in other kingdoms. In phylogenetic analysis, these proteins lie closest to the mammalian VTI1b group of SNAREs, although they share only a low level of homology (Fig. 1). On the other hand, other putative Arabidopsis SNARE families, such as the MEMBRIN- or GOS1-like groups, share branches with their likely orthologs (Fig. 1; for further discussion, see Sanderfoot et al., 2000). For these reasons we felt this group was unique to the plant kingdom and decided to study these SNAREs in more detail.
Figure 1.
NPSN group SNAREs are novel to the plant kingdom. Representative SNARE protein sequences were acquired from GenBank (Arath, Arabidopsis; Sacce, budding yeast; Homsa, human; Musmu, mouse; see “Materials and Methods” for accession nos.) and aligned with the CLUSTALW algorithm. A phylogenetic tree was visualized with TreeView and was prepared for the figure with Adobe Photoshop.
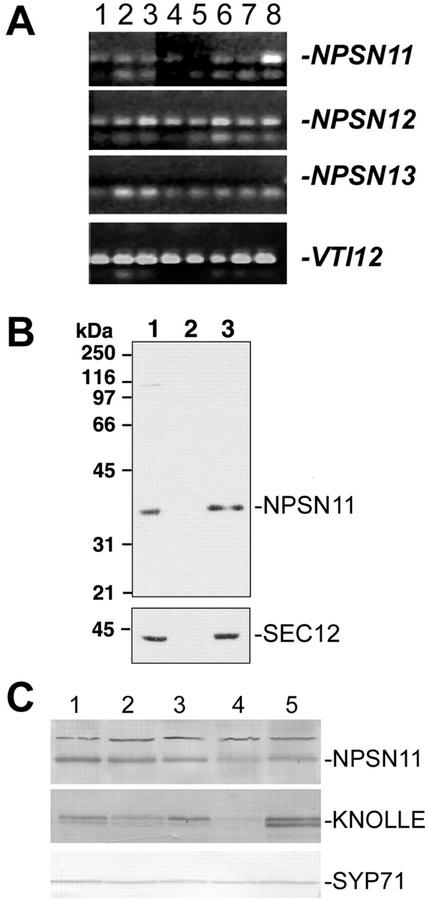
We first determined whether members of this gene family were expressed and, if so, whether there was any tissue-specific distribution of the messages. Using equal amounts of total RNA extracted from various tissues as a template, we performed reverse transcriptase (RT)-PCR analysis with primers specific for each NPSN gene. NPSN11 and NPSN12 cDNAs can be amplified as 800- to 1,000-bp fragments from most tissues, although no expression of NPSN13 was detected (Fig. 2A). Similar amounts of NPSN12 RT-PCR products were found in all these tissues. It is likely that NPSN12 was expressed at nearly equal levels in all tissues that we examined. The highest level of NPSN11 expression was found in the upper most portion of stems (without flowers: Fig. 2A, column 8). In addition, NPSN11 expression was detected in actively dividing tissues including roots, young leaves, flowers, and siliques (Fig. 2A, columns 3 and 6). In contrast, only low levels of NPSN11 were detected in mature rosette leaves (Fig. 2A, column 5). We were unable to detect NPSN13 expression in any of the tissues that we examined, although a full-length cDNA for this gene (Ceres_114054) is present in the Arabidopsis sequence database. One explanation for this difference is that NPSN13 may be expressed at a low level or in a restricted tissue distribution not covered in our analysis.
Figure 2.
NPSN11 and -12 have different tissue distribution patterns. A, RT-PCR was performed with primers specific for NPSN11, NPSN12, NPSN13, and VTI12, using total RNA prepared from the roots of a 3-week-old liquid-cultured plants (lane 1), the leaves of the same liquid-cultured plants (lane 2), flowers from mature soil-grown plants (lane 3), expanding rosette leaves (lane 4), mature rosette leaves (lane 5), green siliques (lane 6), the lower 3 cm of stem (lane 7), and the top 3 cm of the stem (lane 8). Amplified products were separated on agarose gels and visualized with ethidium bromide. Bands specific to each gene are indicated with a dash. No product could be observed with the NPSN13-specific primers. B, Extracts of Arabidopsis suspension-cultured cells were fractionated by differential centrifugation at 150,000g. Twenty micrograms of the resulting fractions of total protein (lane 1), supernatant (lane 2), and microsomal fractions (lane 3) were separated by SDS-PAGE and immunoblotted with affinity-purified NPSN11 antibodies (see “Materials and Methods”) or with the microsomal marker SEC12 (Bar-Peled and Raikhel, 1997). C, NPSN11 protein distribution. Equal amounts of total protein extracted from stems (lane 1), siliques (lane 2), roots (lane 3), leaves (lane 4), and flowers (lane 5) were separated by SDS-PAGE and blotted with antisera to NPSN11, KNOLLE, or SYP71. Bands specific for each protein are indicated with a dash.
To further aid our studies, we also raised an antiserum to the N-terminal portion of the NPSN11 protein. NSPN11 is predicted to encode a protein of 265 amino acids with estimated molecular mass of 29.7 kD. In protein extracts of wild-type plants separated by SDS-PAGE, these antibodies, however, detected a polypeptide of approximately 36 kD. This is typical of the Arabidopsis SNAREs, which tend to run as larger proteins (Conceição et al., 1997; Bassham et al., 2000; Sanderfoot et al., 2001b). In addition, the NSPN11 antisera also recognized an approximately 41-kD polypeptide by immunoblotting that was, however, not immunoprecipitated by the NPSN11 antibodies (see Fig. 6). The 41-kD polypeptide was also not observed by immunoblotting after the antiserum was affinity purified for immunolocalization studies (Fig. 2B). Further evidence that the 36-kD protein corresponds to NPSN11 is provided in the next section (see Fig. 3B). The NPSN11 protein was found in protein extracts from most tissues, with the lowest levels found in mature leaves, consistent with the mRNA expression results (Fig. 2B). This pattern of protein expression is similar to that of the cytokinesis-specific syntaxin, KNOLLE, which is highly expressed in tissues that contain actively dividing cells (Lukowitz et al., 1996; Lauber et al., 1997). As a control, SYP71, a syntaxin type SNARE previously shown to be expressed in most tissues (Sanderfoot et al., 2001b), was found at approximately equal levels in these extracts.
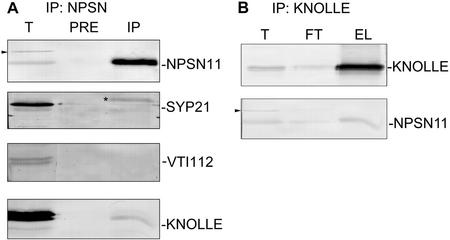
Figure 6.
NPSN11 interacts with the syntaxin KNOLLE. A, NPSN11 protein was immunoprecipitated from total membrane proteins prepared from 5-d-old Arabidopsis suspension-cultured cells. Total protein (1/300; T), protein eluate from preimmune column (1/10; Pre) or from NPSN11-immune column (1/10; IP) was separated by SDS-PAGE. NPSN11, SYP21, VTI12, and KNOLLE were detected by western blots. Note that the 41-kD band observed with the crude NPSN11 antiserum (indicated by an arrowhead) is not immunoprecipitated. The non-specific band recognized by anti-SYP21 is indicated by an asterisk. B, Immunoprecipitation of KNOLLE from 5-d-old Arabidopsis suspension cells using rabbit anti-KNOLLE cross-linked to protein A beads. Total membrane (1/300; T), flow-through (1/300; FT), and eluted protein (1/10; EL) were separated by SDS-PAGE and detected by western blot using antisera against KNOLLE or NPSN11.
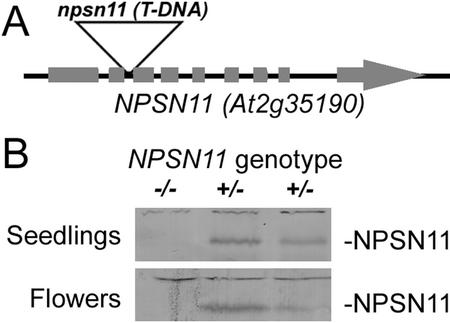
Figure 3.
NPSN11 T-DNA insertion mutant. A, A T-DNA insertion in the NPSN11 (At2g35190) locus was identified from the sequence-tagged database created by the SALK Institute. The insertion was confirmed and characterized as described in “Materials and Methods.” B, Protein extracts from seedlings and flowers of representative siblings of known genotype were separated by SDS-PAGE and blotted with NPSN11 antiserum to show the absence of the NPSN11 protein in the homozygous npsn11 plants.
NPSN11 T-DNA Insertion Mutant
We have identified one npsn11::T-DNA (npsn11-1) insertion line in the SIGnAL (SALK Institute Genomic Analysis Laboratory) database of sequenced T-DNA mutagenized Arabidopsis lines (See Fig. 3A). The T-DNA was inserted in the first intron of NPSN11. Homozygous npsn11-1 plants showed no obvious phenotypes and were completely fertile. RT-PCR analysis confirmed that the expression of NPSN11 was abolished in homozygous plants (data not shown), and western-blot analysis of protein extracts prepared from mutant and wild-type siblings showed that only the 36-kD band recognized by NSPN11 antisera was absent from the homozygous npsn11 lines (Fig. 3B). These results confirmed the specificity of the NPSN11-antibodies for the 36-kD protein and that the antibodies did not cross-react with the other members of the NPSN protein family.
NPSN11 Cofractionates with KNOLLE
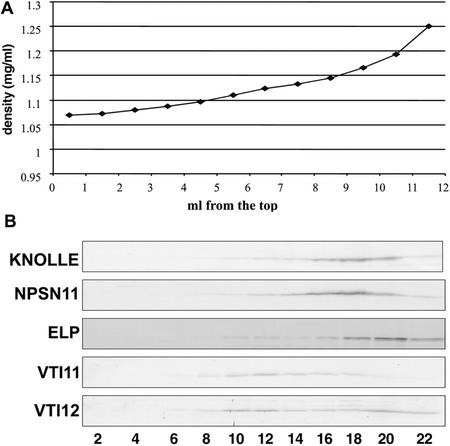
To further characterize NPSN11, we analyzed the intracellular distribution of the protein by subcellular fractionation. A post-nuclear supernatant was prepared from 21-d-old Arabidopsis roots and fractionated on a discontinuous Accudenz density gradient (see “Materials and Methods”). The gradient was equilibrated by ultracentrifugation at 100,000g for 16 h at 4°C, and 22 fractions were collected from the top to the bottom. These fractions were separated by SDS-PAGE and analyzed by immunoblotting with antibodies specific to NPSN11 and various endomembrane markers. The density gradient fractionation conditions were chosen to differentiate between the trans-Golgi network (TGN) and the prevacuolar compartment (PVC), using marker syntaxins for each compartment (SYP21 and SYP41, respectively; Bassham et al., 2000). For example, the SNARE VTI11, previously shown to cofractionate with PVC markers (Zheng et al., 1999), has a single peak at a density of 1.125 mg mL−1. On the other hand, a related SNARE, VTI12 was detected in two peaks; one corresponding to the PVC and another that equilibrated at a density of approximately 1.175 mg mL−1 (Fig. 4). This second peak cofractionated with ELP, a TGN marker as shown by previous subcellular fractionation experiments (Sanderfoot et al., 1998). As shown in Figure 4, membranes containing NPSN11 did not cofractionate with VTI11, VTI12, or ELP. NPSN11 also did not cofractionate with the vacuolar marker aleurain (data not shown). The highest level of NSPN11 was found in fractions with an approximate density of 1.15 mg mL−1 (Fig. 4). These results indicate that NPSN11 is not associated with the TGN, the PVC, or the vacuole. The fractionation pattern of NPSN11 closely resembles that of KNOLLE, suggesting that NSPN11 and KNOLLE may reside on the same membrane compartment.
Figure 4.
NPSN11 and KNOLLE cofractionate on an Accudenz gradient. Total membranes prepared from 21-d-old liquid-cultured Arabidopsis roots were separated by a discontinuous Accudenz gradient. Fractions of 0.5 mL were collected from top to the bottom. The density of each fraction was plotted against the fraction number (A). Fractions were collected, and equal amounts were separated by SDS-PAGE and then blotted with the indicated antisera (B). NPSN11 cofractionated with KNOLLE but not with AtELP, the TGN marker; VTI11, which labels the PVC; or VTI12, which labels both the PVC and the TGN.
NPSN11 Is Localized at the Division Plane during Cytokinesis
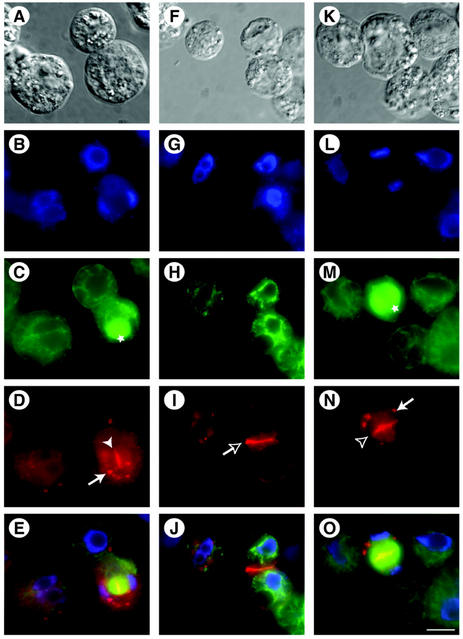
Using immunofluorescence microscopy, we further examined the localization of NSPN11 in dividing and non-dividing Arabidopsis cells. Protoplasts from actively dividing Arabidopsis suspension-cultured cells were fixed and processed for indirect immunofluorescence microscopy using affinity-purified NSPN11 and KNOLLE antibodies. Western-blot analysis of total Arabidopsis protein extracts confirmed that affinity-purified NSPN11 and KNOLLE antibodies are highly specific and cross-react only with their appropriate 36- and 34-kD polypeptides, respectively (data not shown). During cytokinesis, cell plate vesicles are targeted to and fuse within the equatorial plane of the phragmoplast. Previous localization studies have demonstrated that KNOLLE, which is required for cell plate membrane fusion, is localized within the phragmoplast mid-plane in dividing Arabidopsis cells (Lauber et al., 1997; see also Fig. 5N). Similarly, we have observed that NSPN11 is targeted to cell plate during cytokinesis (Fig. 5D). NSPN11 was also found to be associated with newly completed cross wall plasma membranes that separate two recently divided cells (Fig. 5I). However, only low levels of NSPN11 were detected in the mature plasma membrane. In addition to their association with the cell plate, we have also observed significant anti-NSPN11 and anti-KNOLLE immunolabeling of punctate subcellular organelles. These structures do not correspond to Golgi stacks as shown by immunolabeling with affinity-purified antibodies to the Arabidopsis Golgi-resident marker protein α-mannosidase (data not shown). The identity of these NSPN11 and KNOLLE positive subcellular structures remains to be defined.
Figure 5.
NPSN11 is localized at the cell plate during cytokinesis. Arabidopsis suspension-cultured cell protoplasts (A, F, and K) were double immunolabeled with antibodies directed against α-tubulin (α-tub) to visualize phragmoplast (marked by stars), cortical microtubules (C, H, and M), and either affinity-purified NPSN11 (D and I) or KNOLLE antisera (N). Nuclei in dividing and non-dividing cells were revealed by staining with 4′,6′-diamidino-2-phenylindole (DAPI; B, G, and L). Electronically merged images of B through D, G through I, and L through N are shown in E, J, and O, respectively. NSPN11 positive cell plate (solid arrowhead) and new plasma membrane (empty arrow) are visible in D and I, respectively. N shows the localization of KNOLLE at the cell plate (empty arrowhead). Arrows in D and N indicate NSPN11- and KNOLLE-containing intracellular organelles. Scale bar in A through O = 10 μm.
NPSN11 Forms a SNARE Complex with KNOLLE
The above observations suggest that both KNOLLE and NSPN11 are targeted to the division plane during cytokinesis. A critical question remaining to be addressed was whether KNOLLE and NPSN11 physically interact. First, we immunoprecipitated NPSN11 from Triton X-100 solubilized membranes prepared in the presence of EDTA from 5-d-old Arabidopsis suspension-cultured cells. As shown in Figure 6A, the 36-kD NPSN11 polypeptide was detected by immunoblotting in the eluate from the anti-NSPN11 column but not from the control preimmune column. The 41-kD polypeptide that cross-reacted with anti-NSPN11 in the total protein extract did not immunoprecipitate with the 36-kD NPSN 11 polypeptide, again indicating that it has a distinct origin than the NPSN11 protein. Interestingly, KNOLLE was detected by immunoblotting in the NSPN11 antibody column eluate (Fig. 6A). The amount of KNOLLE immunoprecipitated in these experiments is small, although this is typical of SNARE-SNARE interactions as reported by many researchers in many eukaryotes (Antonin et al., 2000; Bassham et al., 2000; Gurunathan et al., 2000; Sanderfoot et al., 2001b). In contrast, SYP21, a PVC syntaxin, was not precipitated by either the preimmune or anti-NPSN11 column. The polypeptide observed in the anti-NSPN11 eluate that was recognized by anti-SYP21 is likely to be a non-specific band because this polypeptide is not the correct size for SYP21 and was not reproducibly observed in other experiments (Fig. 6A). Furthermore, immunoprecipitation experiments using SYP21 antiserum confirmed that SYP21 did not interact with either NPSN11 or KNOLLE (Sanderfoot et al., 2001b; A.A. Sanderfoot, unpublished data). Two other proteins, VTI12 (Fig. 6A) and VPS45 (data not shown), were also not found to be immunoprecipitated with NPSN11. To confirm that the physical interaction between NPSN11 and KNOLLE is genuine, we performed the reverse experiment: KNOLLE was first immunoprecipitated from a detergent-solubilized Arabidopsis membrane extract, and the eluate was analyzed by immunoblotting. As shown in Figure 6B, NPSN11 co-immunoprecipitated with KNOLLE, whereas another SNARE, VTI12 (data not shown), was not detected in the anti-KNOLLE column eluate. These complementary experiments suggest that NPSN11 specifically interacts with KNOLLE, the cytokinesis-specific syntaxin in vivo.
DISCUSSION
Extensive research over the past several years has shown that SNAREs play essential roles in vesicle trafficking and membrane fusion throughout the secretory pathway. Likewise, SNAREs have been shown to be required for the de novo assembly of the cell plate during plant cytokinesis, a highly critical process involving extensive vesicle trafficking and fusion. Specifically, the cytokinesis-specific syntaxin KNOLLE has been shown to be required for cell plate membrane fusion and the completion of the plant cell division process (Lukowitz et al., 1996; Lauber et al., 1997). A second SNARE, the SNAP25-like protein SNAP33, has recently also been shown to be involved in plant cytokinesis (Heese et al., 2001). The commonality between the canonical neuronal exocytic t-SNARE complex, which is composed of syntaxin and SNAP25, and the cell plate t-SNARE complex of KNOLLE and SNAP33 suggests that a v-SNARE similar to the exocytic synaptobrevin will be involved in cell plate membrane fusion. Surprisingly, however, synaptobrevin homologs have not been identified in the Arabidopsis (or any other plant) genome (Sanderfoot et al., 2000). It is, therefore, likely that other plant-specific SNAREs function in place of the exocytic synaptobrevins in membrane fusion during plant cytokinesis.
In this work, we have characterized NPSN11, a SNARE protein that is a part of a three-member family of plant-specific SNAREs. Based on its interaction with KNOLLE and its cell plate localization, we speculate that NPSN11 is another component of the cell plate membrane fusion machinery. NPSN11 may be the above-mentioned v-SNARE for a KNOLLE/SNAP33 t-SNARE complex, and we are currently investigating this possibility using a combination of biochemical and genetic approaches. As an alternative, the NPSN11/KNOLLE complex may function in cell plate and other vesicle trafficking steps that do not involve SNAP33. We have recently found that both NPSN11 and KNOLLE are capable of interacting with other SNAREs that are not associated with the cell plate (H. Zheng, A.A. Sanderfoot, and N.V. Raikhel, unpublished data) and, thus, may indicate multiple roles for these proteins in vesicle trafficking. In support of this idea, SNAP33 has been found to interact not only with KNOLLE but also with SYR1 (SYP121), possibly as part of the general exocytic pathway in plant cells (Kargul et al., 2001). In addition to SNAP33, the Arabidopsis genome encodes two other SNAP25 homologs (Sanderfoot et al., 2000) that may function in place of SNAP33 with the NPSN11/KNOLLE cell plate SNARE complex. Cell plate membrane fusion is highly dynamic and involves not only transport vesicle fusion, but also homotypic consolidation of the initial tubular vesicular membrane network and fusion of the mature cell plate with the parental plasma membrane (Staehelin and Hepler, 1996). One or more of these cell plate membrane fusion events may require distinct cell plate SNARE complexes. Future work will be required to identify all of the different SNAREs and SNARE complexes that are likely to be involved in cell plate formation.
Because of the interaction with KNOLLE, we think that it is very likely that NPSN11 is involved in cell plate formation. However, it is important to note that NPSN11 and KNOLLE have some distinctions in their expression and localization. First, we have found that NPSN11 expression, like KEULE (Assaad et al., 2001), is not solely limited to dividing cells, as has been found with KNOLLE (Lauber et al., 1997). Second, NPSN11 is present in the new cross walls for a short time after cytokinesis, whereas KNOLLE appears to rapidly disappear after cytokinesis is finished. As can be seen in Figure 5, some small bright foci of NPSN11-and KNOLLE-labeling are found in the cytoplasm of dividing and non-dividing cells. This labeling is similar to that observed for KNOLLE and other plasma membrane localized proteins (i.e. PIN1) in root cells treated with brefeldin A, which it is believed to cause these proteins to accumulate in an early endosomal compartment (Geldner et al., 2001). As an alternative, the NSPN11- and KNOLLE-positive compartments may represent Golgi stacks, as has been suggested for the punctate distribution of KNOLLE in early mitotic cells before phragmoplast formation in anaphase (Lauber et al., 1997). However, our subcellular fractionation (Fig. 4) and immunofluorescence microscopy localization studies with a Golgi-resident marker protein (data not shown) are not consistent with this idea. Our initial attempts to localize NPSN11 using immunoelectron microscopy have not been successful, perhaps because of problems associated with the processing of samples for electron microscopy. Future experiments using different antibodies and/or epitope-tagged NSPN11 fusion protein should help to precisely identify this non-cell plate organelle.
Finally, our results concerning the gene disruption of NPSN11 are of interest. Plants homozygous for the npsn11 disruption have no observable phenotype. This result is perhaps attributable to redundancy with the other members of the NPSN gene family. RT-PCR analysis has shown that another member of the NSPN gene family, NSPN12, is highly expressed throughout Arabidopsis plants. Thus far, several gene disruptions of single members of Arabidopsis SNARE gene families have been reported. To date, mutations that disrupt the function of single genes encoding syntaxin-type SNAREs have been either seedling or gametophytic lethal. For example, the kn mutation is seedling lethal (Lukowitz et al., 1996). Similarly, mutations in any of the SYP21, SYP22, SYP41, and SYP42 genes are lethal at the male gametophyte stage (Sanderfoot et al., 2001a). On the other hand, mutation of the SNAP33 gene, snp33, disrupts plant growth only very late in development, possibly indicating a partial functional redundancy with the other two members of the SNAP25-like family in Arabidopsis (Heese et al., 2001). Recent results suggest that mutations in the VTI1-family of genes may also show some level of redundancy (Kato et al., 2002; H. Zheng and N.V. Raikhel, unpublished data). Future studies will identify disruptions in the other members of the NPSN gene family, and, thus, define the phenotype of the loss of NPSN gene function.
In conclusion, we have begun to characterize a novel family of plant-specific SNAREs from Arabidopsis, the NPSN group. We have shown that one member of this family, NPSN11, is localized at the cell plate during plant cell division. Furthermore, NPSN11 interacts directly with the cytokinesis-specific syntaxin KNOLLE. These results suggest that this group of plant-specific SNAREs are involved in membrane trafficking and fusion during plant cytokinesis, a process that is highly critical for normal plant growth and development.
MATERIALS AND METHODS
Sequence Analysis
The Arabidopsis Genome Initiative and associated cDNA sequencing projects have produced accessions supported by full-length cDNAs for NPSN11 and NPSN13 that are available in the database (AF439822 and NM_112623, respectively). Because our RT-PCR products matched these sequences, we did not resubmit our sequences. The splicing of the NPSN12 cDNA was incorrectly predicted by the Genome Initiative, thus, we submitted our cDNA as AF487545. The protein sequences of representative SNAREs from several eukaryotes were acquired from GenBank as follows: Arabidopsis (Arath) GOS11, AF357528; GOS12, AF357529; MEMB11, AAD31575; MEMB12, BAB09463; VTI11, AAF24061; VTI12, AAF24062; VTI13, BAB01986; NPSN11, AAL27494; NPSN12, AF487545; and NPSN13, NP_566578. Yeast (Sacce) Gos1p, NP_011832; Bos1p, NP_013179; and Vti1p, NP_013924. Human (Homsa) GS28, NP_004862; Membrin, NP_004278; VTI1a, AAH17052; and VTI1b, NP_006361. Mouse (Musmu) GS28, NP_058090; Membrin, NP_062624; VTI1a, AAC23482; and VTI1b, AAC23483. The protein sequences were aligned by the CLUSTALW algorithm, and a phylogenetic tree was visualized using TreeView (Rod Page, http://taxonomy. zoology.gla.ac.uk/rod/rod.html) and prepared as a figure using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Plasmids, Transgenic Plant, and Arabidopsis Mutant
NPSN11 and NPSN12 cDNA clones were generated by RT-PCR from total RNA extracted from seedlings. The primers used for this purpose were: For NPSN11: forward primer, 5′-CTG GGA ATC TGT GTA AAG ATG-3′; reverse primer, 5′-ATG ACT AAG GG AGG ATC AAG-3′. For NPSP12: forward primer, 5′-GAG CCT GAA ATA ATC CGG CAG AT-3′; reverse primer, 5′-AGT GTA ATA TGC ACC AAA CC-3′. For NPSN13: forward primer, 5′-CTG AAT TGT CTC CGG CGA CAT-3′; reverse primer, 5′-CAT CCA TGA AAT GGA TTG TT-3′. The PCR fragment was blunted by Klenow fragment of DNA polymerase (Invitrogen, Carlsbad, CA) and cloned into EcoRV site of pBluescript (Stratagene, La Jolla, CA). For Escherichia coli overexpression of 6×-His-NPSN11, the BamHI and SacI (blunt) fragments of NPSN11 cDNA were subcloned into pET14b (BamHI and BLP1 [blunt], Novagen, Madison, WI) and transformed into E. coli BL21(DE3) cells for overexpression. For generating a glutathione-S-transferase fusion of NPSN11, the EcoRI-XhoI fragment from pBluescript-NPSN11 was inserted into EcoRI-XhoI sites of pGEX5x-1 (Amersham-Pharmacia Biotech, Uppsala) to generate in frame fusions.
An Arabidopsis NPSN11 T-DNA insertion line (JP64_8E02) was identified among a collection of sequence-index T-DNA insertion lines. The mutant was generated in the Columbia-0 background using the binary vector pROK2 (Baulcombe et al., 1986). To identify homozygous plants, the presence of the wild-type and mutant gene was analyzed in the progeny of the individual T1 plants known to contain the insertion. Genomic DNA was prepared from individual T2 plants using the cetyl-trimethyl-ammonium bromide procedure (Sanderfoot et al., 2001a). Two gene-specific primers (5′-end gene-specific primer, 5′-TGA TTT CCC TAT CGA AAT CTT-3′; 3′-end gene-specific primer, 5′-GCT TGA TCT GTG TCT TCC ATC A-3′) flanking the insertion site and the T-DNA specific primer (left border, 5′-GCG TGG ACC GCT TGC TCG AAC T-3′) were used to identify homozygous mutant plants. PCR amplification of genomic DNA was performed using tag polymerase (Invitrogen) following the standard condition recommended by the manufacturer. Homozygous seeds derived from this line have been deposited into the Arabidopsis Biological Resource Center (Ohio State University, Columbus) collection.
Antibody Production, Purification, and Antibody Columns
6×-His-tagged NPSN11 was overexpressed by isopropylthio-β-galactoside induction. The His-tagged protein was purified by nickel-nitrilotriacetic acid agarose (Novagen, Madison, WI) column under standard conditions and injected into a rabbit for antibody production. SYP21 rabbit antiserum was described by Conceição et al. (1997). AtELP rabbit antiserum and preimmune serum were described by Ahmed et al. (1997). AtVTI11 antiserum was described by Zheng et al. (1999). AtVTI12 antiserum was described by Bassham et al. (2000). Preparation of affinity-purified KNOLLE antiserum is described by D.R. Rancour, C. Dickey, S. Park, and S.Y. Bednarek (unpublished data). For immunofluorescence microscopy, NPSN11 antiserum was affinity purified against E. coli-expressed glutathione-S-transferase fusion of NPSN11 as described previously (Bassham et al., 2000).
To make antibody columns for immunoprecipitation, IgG from NPSN11 preimmune serum, anti-NPSN serum, or anti-KNOLLE serum was purified by protein A affinity columns. The protein concentration of purified antibodies was determined by Bradford dye binding procedure (Bradford, 1976). Two milligrams of purified antibodies were incubated with 1.0 mL of immobilized Protein A-6MB (Amersham-Pharmacia Biotech) for 1 h at room temperature. The resin was washed with 10 mL of coupling buffer (200 mm sodium borate, pH 9.0), dimethyl pimelimidate was added (5 mg mL−1 final concentration), and the suspension was incubated at 20°C for 30 min. The coupling reaction was terminated with 0.2 m ethanolamine (pH 8.0), and the beads were stored in phosphate-buffered saline with 0.02% (w/v) sodium azide at 4°C.
Immunoprecipitation
Twenty grams of 5-d-old suspension-cultured Arabidopsis cells was homogenized on ice with 5 mL of extraction buffer (50 mm HEPES-KOH, pH 6.5, 10 mm potassium acetate, 100 mm sodium chloride, 5 mm EDTA, and 0.4 m Suc) with Complete protease inhibitor tablet (Roche, Indianapolis). To prepare total membranes, the homogenate was centrifuged at 1,000g for 15 min, and the supernatant was subjected to ultracentrifugation at 100,000g for 3 h. The total membrane pellet was homogenized in 3 mL of Tris-buffered saline (0.14 m NaCl, 2.7 mm KCl, and 25 mm Tris, pH 8.0) with miniComplete tablet (Roche), solubilized by the addition of Triton X-100 (1% [v/v] final concentration), and cleared of insoluble material by ultracentrifugation at 100,000g for 30 min. The clarified supernatant was incubated with the immobilized antibodies for 2 h at 4°C. The antibody columns were washed five times with 5 mL of Tris-buffered saline plus Tween 20 (TBS + 1% [v/v] Triton X-100), and specifically bound proteins were eluted with 4 mL of 0.1 m Gly (pH 2.5). The eluted proteins were concentrated by precipitation with trichloroacetic acid (10% [w/v] final concentration), and the precipitates were sedimented at 10,000g for 30 min at 4°C. After two acetone washes, the protein pellet was solubilized in 50 μL of 2× Laemmli buffer and analyzed by SDS-PAGE and immunoblotting as described by Bassham et al. (2000).
Subcellular Fractionation
Ten grams of 21-d-old Arabidopsis-cultured roots was used to prepare total membrane using the same procedure for membrane preparation from suspension cells as described above. The total membrane was then layered on top of a discontinuous Accudenz (Accurate Chemicals and Scientific Corp., New York) gradient (1.5 mL of each: 2%, 5%, 9%, 12%, 15%, 20%, and 30% [w/w] from the top to the bottom). The gradient was then equilibrated by ultracentrifugation at 100,000g for 16 h. Fractions of 0.5 mL were collected from the top to the bottom. The densities of different fractions were measured by refractometry and proteins were analyzed by SDS-PAGE and immunoblotting to visualize different subcellular markers.
Epifluorescence Microscopy
Preparation and immunostaining of Arabidopsis suspension-cultured cell protoplasts was performed essentially as described by Kang et al. (2001) with the exception that the fixed protoplasts were permeabilized in microtubule stabilizing buffer (MTSB; Goodbody and Lloyd, 1994; 50 mm PIPES-KOH, pH 6.9, 5 mm MgSO4, and 5 mm EGTA) containing 0.5% (v/v) NP-40 and 10% (v/v) dimethyl sulfoxide in suspension for 5 min at room temperature. The permeabilized cells were collected by centrifugation at 200g for 2 min at room temperature, washed three times with 1.0 mL of MTSB, plated on ProbeOn Plus slides (Fisher Scientific, Pittsburgh), and allowed to air dry. The cells were rehydrated and blocked with MTSB containing 3% (w/v) bovine serum albumin. All subsequent immunolabeling steps, epifluorescence microscopy, and image processing were performed as described (Kang et al., 2001).
Footnotes
This research was supported by the National Science Foundation grant no. MCB–0296080 (to N.V.R.) and by the U.S. Department of Energy, Division of Energy Biosciences (project no. DE–FG02–99ER203 32 to S.Y.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003970.
LITERATURE CITED
- Ahmed SU, Bar-Peled M, Raikhel NV. Cloning and subcellular location of an Arabidopsis receptor-like protein that shares common features with protein-sorting receptors of eukaryotic cells. Plant Physiol. 1997;114:325–336. doi: 10.1104/pp.114.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Holroyd C, Fasshauer D, Pabst S, Fisher von Mollard G, Jahn R. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Huet Y, Mayer U, Jürgens G. The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J Cell Biol. 2001;152:531–543. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV. Characterization of AtSEC12 and AtSAR1: proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol. 1997;114:315–324. doi: 10.1104/pp.114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV. AtVPS45 complex formation at the trans-Golgi network. Mol Biol Cell. 2000;11:2251–2265. doi: 10.1091/mbc.11.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D, Saunders G, Bevan M, Mayo M, Harrison B. Expression of biologically active viral satellite RNA from nuclear genome of transformed plants. Nature. 1986;321:446–449. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Conceição AS, Marty-Mazars D, Bassham DC, Sanderfoot AA, Marty F, Raikhel NV. The syntaxin homologue AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell. 1997;9:571–582. [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Goodbody KC, Lloyd CW. Immunofluorescence techniques for analysis of the cytoskeleton. In: Harris N, Oparka KJ, editors. Plant Cell Biology: A Practical Approach. Oxford: IRL Press; 1994. pp. 221–243. [Google Scholar]
- Gurunathan S, Chapman-Shimshoni D, Trajkovic S, Gerst JE. Yeast exocytic v-SNAREs confer endocytosis. Mol Biol Cell. 2000;11:3629–3643. doi: 10.1091/mbc.11.10.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI. Sec1 gets a grip on syntaxin. Nat Struct Biol. 2000;7:347–349. doi: 10.1038/75103. [DOI] [PubMed] [Google Scholar]
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jürgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol. 2001;155:239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY. The Arabidopsiscell plate-associated dynamin-like protein, Adl1ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001;126:47–68. doi: 10.1104/pp.126.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargul J, Gansel X, Tyrrell M, Sticher L, Blatt MR. Protein-binding partners of the tobacco syntaxin NtSyr1. FEBS Lett. 2001;508:253–258. doi: 10.1016/s0014-5793(01)03089-7. [DOI] [PubMed] [Google Scholar]
- Kato T, Morita MT, Fukaki H, Yamauchi Y, Uehara M, Niihama M, Tasaka M. SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell. 2002;14:33–46. doi: 10.1105/tpc.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. The ArabidopsisKNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsisembryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Mayer U, Torres Ruiz RA, Erleth T, Miséra S, Jürgens G. Mutations affecting body organization in the Arabidopsisembryo. Nature. 1991;353:402–407. [Google Scholar]
- Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. Nat Cell Biol. 1999;1:E17–E22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Ahmed SU, Marty-Mazars D, Rapoport I, Kirchhausen T, Marty F, Raikhel NV. A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsisroots. Proc Natl Acad Sci USA. 1998;95:9920–9925. doi: 10.1073/pnas.95.17.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Assaad FF, Raikhel NV. The Arabidopsis genome: an abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 2000;124:1558–1569. doi: 10.1104/pp.124.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Pilgrim M, Adam L, Raikhel NV. Disruption of individual members of Arabidopsissyntaxin gene families indicates each has essential functions. Plant Cell. 2001a;13:659–666. doi: 10.1105/tpc.13.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell. 2001b;12:3733–3743. doi: 10.1091/mbc.12.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA, Hepler PK. Cytokinesis in higher plants. Cell. 1996;84:821–824. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Waizenegger I, Lukowitz W, Assaad F, Schwarz H, Jurgens G, Mayer U. The Arabidopsis KNOLLE and KEULEgenes interact to promote vesicle fusion during cytokinesis. Curr Biol. 2000;10:1371–1374. doi: 10.1016/s0960-9822(00)00775-2. [DOI] [PubMed] [Google Scholar]
- Zheng H, von Mollard GF, Kovaleva V, Stevens TH, Raikhel NV. The plant vesicle associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol Biol Cell. 1999;10:2251–2264. doi: 10.1091/mbc.10.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]