Abstract
Circadian rhythms have been demonstrated in organisms across the taxonomic spectrum. In view of their widespread occurrence, the adaptive significance of these rhythms is of interest. We have previously shown that constitutive expression of the CCA1 (CIRCADIAN CLOCK ASSOCIATED 1) gene in Arabidopsis plants (CCA1-ox) results in loss of circadian rhythmicity. Here, we demonstrate that these CCA1-ox plants retain the ability to respond to diurnal changes in light. Thus, transcript levels of several circadian-regulated genes, as well as CCA1 itself and the closely related LHY, oscillate robustly if CCA1-ox plants are grown under diurnal conditions. However, in contrast with wild-type plants in which transcript levels change in anticipation of the dark/light transitions, the CCA1-ox plants have lost the ability to anticipate this daily change in their environment. We have used CCA1-ox lines to examine the effects of loss of circadian regulation on the fitness of an organism. CCA1-ox plants flowered later, especially under long-day conditions, and were less viable under very short-day conditions than their wild-type counterparts. In addition, we demonstrate that two other circadian rhythm mutants, LHY-ox and elf3, have low-viability phenotypes. Our findings demonstrate the adaptive advantage of circadian rhythms in Arabidopsis.
First described in plants more than 270 years ago, circadian rhythms have been found in the vast majority of eukaryotes examined and in many prokaryotes. The basic mechanisms that are responsible for circadian rhythms are being elucidated in several model species. At the core of a circadian system is a molecular oscillator that generates a period of rhythmicity of about 24 h. The oscillator is self-sustaining, but it is influenced by environmental cues such as changes in light and temperature conditions (Dunlap, 1999). The oscillator controls a plethora of biological processes including nitrogen fixation in cyanobacteria (Johnson et al., 1996), scent emission in plants (Kolosova et al., 2001), conidiation in Neurospora crassa. (Pittendrigh et al., 1959), olfactory responses of Drosophila melanogaster (Krishnan et al., 1999), luteinizing hormone levels in birds (Follett et al., 1974), and wheel running activity in hamsters (Mesocricetus auratus; Ralph and Menaker, 1988). The fact that circadian regulation is ubiquitous across the taxonomic spectrum and that its importance was even recognized in the Hebrew Bible (Ancoli-Israel, 2001) suggests that it is important for optimizing an organism's response to its environment and enhancing its fitness.
The circadian clock plays a number of different roles. One is in the regulation of photoperiodism, which is the detection and response to changes in the duration of days and nights that enables organisms to adapt to seasonal changes in their environment (Thomas, 1998). Photoperiodism controls reproduction in many organisms, including flowering time in many plants (Thomas and Vince-Prue, 1997). This photoperiodic response ensures that plants flower during the appropriate season. For example, plants growing in lower latitudes tend to flower in response to short days, thus avoiding the extreme summer heat. In contrast, plants in more temperate climates, which must flower and set seeds before the onset of winter, initiate flowering in response to long days. Arabidopsis is a facultative long-day plant, in which flowering is initiated earlier in response to long days, but Arabidopsis will eventually flower, albeit much later, even under short days (Thomas and Vince-Prue, 1997). An additional advantage of photoperiodic control is that, by ensuring coincident flowering of a population of plants, the chances of outcrossing are increased.
Another function that has been suggested for the clock is to allow organisms to program activities so that they occur at a specific part of the diurnal cycle. This programming may serve to ensure that incompatible reactions, such as nitrogen fixation and photosynthesis in cyanobacteria (Mitsui et al., 1986), are temporally spaced. Circadian clocks may also allow the organism to anticipate night/day changes. For example, some molecular processes important for photosynthesis are initiated before dawn so that by sunrise the plants are ready to take maximum advantage of available light for photosynthesis (Kreps and Kay, 1997). In addition, organisms may produce screening pigments before the sun rises to avoid damage by visible and UV light (Pittendrigh, 1993; Nikaido and Johnson, 2000).
Several approaches have been used to try to determine whether circadian clocks affect the fitness of organisms. One approach has been to examine whether organisms are adversely affected when they are grown in a light/dark cycle with a period that does not match that of their endogenous circadian clock. The growth of some organisms, for example tomato (Lycopersicon esculentum) plants and blowflies (Sarcophaga argyostoma), was found to be extremely sensitive to the period of the light/dark cycle. Both organisms grow poorly in cycles that deviated significantly from their endogenous 24-h rhythms (Highkin and Hanson, 1954; Went, 1960; Saunders, 1972). Furthermore, period length mutants of hamsters (Hurd and Ralph, 1998) and D. melanogaster (Klarsfeld and Rouyer, 1998) that have non-24-h endogenous rhythms show small (less than 20%) reductions in life span when they are kept in 24-h cycles. However, the mechanisms underpinning these deficiencies are unknown. More recently, the effect of light/dark cycle periods on reproductive success in the cyanobacterium Synechococcus elongatus was studied (Ouyang et al., 1998). Pairs of strains of S. elongatus with different period lengths were mixed and grown in culture under light cycles of different periods. It was found that the strain of S. elongatus that had an endogenous period most closely matching that of the light cycle in which they were grown rapidly out-competed the other strain and became the dominant strain in the culture. This study showed convincingly that an organism might benefit from having an endogenous oscillator that matches the environmental cycle, although the physiological mechanisms for this enhanced fitness are unknown. In contrast, similar studies on mixed populations of wild-type and period mutant D. melanogaster showed no selective advantage to the population growing under a light/dark cycle that matched their endogenous oscillator (Klarsfeld and Rouyer, 1998).
In one instance, mathematical modeling was used to try to demonstrate the significance of circadian rhythms. However, modeling of two circadian-controlled processes, photosynthesis and stomatal conductance, in the wetland perennial plant, Suarurus cernuss, failed to demonstrate significant effects of circadian rhythms on carbon gain or water loss under field conditions (Williams and Gorton, 1998).
An alternative approach for examining the adaptive significance of circadian regulation is to compare the viability of organisms lacking functional clocks with their wild-type counterparts. In two reports, in which ground squirrels (Ammospermophilus leucurus) and chipmunks (Tamias striatus) with ablated circadian systems were studied under field conditions, it was shown that loss of a functional circadian system can reduce viability of an organism by increasing susceptibility to predators (DeCoursey et al., 1997, 2000). However, although clock-less organisms are available from a range of species, there is no published evidence that organisms lacking functional circadian systems are affected in viability under laboratory conditions.
We have a system for examining whether the circadian clock contributes to the fitness of Arabidopsis under laboratory conditions. Two genes, CCA1 and LHY, encoding closely related proteins associated with the central oscillator in Arabidopsis, have been isolated (Wang and Tobin, 1998; Schaffer et al., 1998; Green and Tobin, 1999). Both CCA1 and LHY have been constitutively expressed in transgenic plants (CCA1-ox and LHY-ox) at levels close to their maximum in wild-type plants. Under continuous light (LL) or dark (DD), the CCA1-ox and LHY-ox plants are arrhythmic in all aspects of circadian control that have been examined, including leaf movements, hypocotyl elongation, and gene expression (Schaffer et al., 1998; Wang and Tobin, 1998; E.M. Tobin, Z.-Y. Wang, and A.J. Millar, unpublished data). CCA1 is also involved in a signal transduction pathway from the plant photoreceptor phytochrome (Wang and Tobin, 1998; Green and Tobin, 1999; Martínez-García et al., 2000). Here, we demonstrate that although CCA1-ox plants have lost circadian rhythmicity, they retain the ability to respond to diurnal changes in light. However, they have no ability to anticipate the light/dark transitions. We show that these CCA1-ox and LHY-ox plants that lack functional circadian systems are less fit than their wild-type counterparts under laboratory conditions. Our findings support the hypothesis that the circadian clock contributes to the fitness of Arabidopsis plants.
RESULTS
Clock-Controlled Genes Can Still Be Regulated by Light in CCA1-ox Plants Grown under Diurnal Light/Dark Conditions
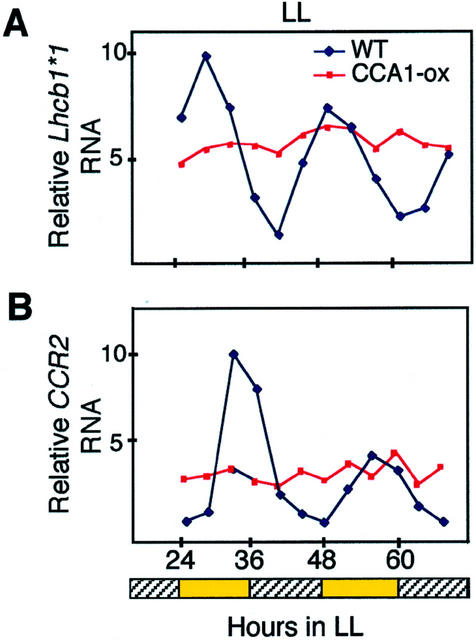
We have previously shown that CCA1 is involved in both light and circadian regulation of gene expression (Wang and Tobin, 1998; Green and Tobin, 1999). In CCA1-ox plants that are grown in LL or DD, the circadian regulation of gene expression is completely disrupted (Wang and Tobin, 1998). This disruption is shown for two genes in Figure 1.
Figure 1.
Circadian oscillations of transcript accumulation from clock-controlled genes in wild-type and CCA1-ox lines. Wild-type and CCA1-ox plants were transferred to constant light (LL) after entrainment in light:dark conditions (12L:12D). The relative RNA levels of Lhcb1*1 (A) and CCR2 (B) are shown relative to the maximum levels of expression. Methylene blue staining was used to check equal loading. Experiments were performed twice with similar results. The hatched bars indicate subjective dark periods in LL.
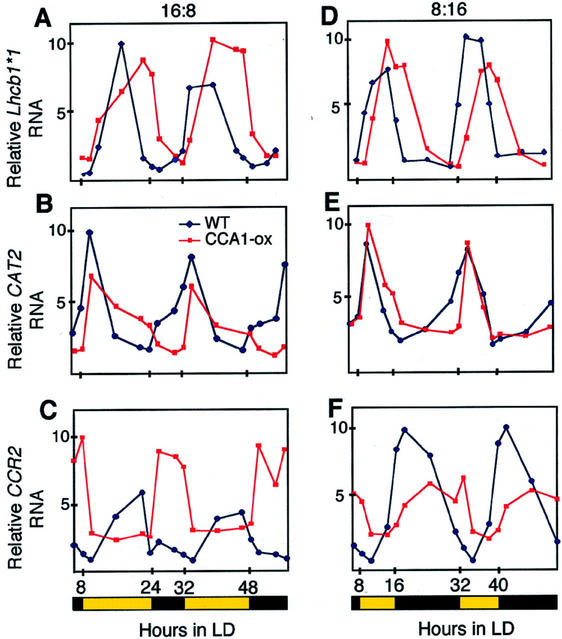
Figure 2 compares the expression of several clock-controlled genes in wild-type and CCA1-ox plants grown under two different photoperiods. Transcript levels of three genes, Lhcb1*1 (also known as CAB2; 24), CCR2 (also known as AtGRP7; Carpenter et al., 1994; Heintzen et al., 1997), and CAT2 (Zhong and McClung, 1996), show robust oscillations in both the wild-type and CCA1-ox plants. However, the CCA1-ox plants have lost their ability to modulate the expression of these genes in anticipation of light/dark changes. Thus, in wild-type plants, the levels of RNA from the two morning-specific genes, CAT2 and Lhcb1*1, start to rise well before the lights come on (Fig. 2, A, B, D, and E). In contrast, in the CCA1-ox plants the transcript levels from these two genes start to increase only after the lights come on. The effect of constitutive CCA1 expression on transcript accumulation from the evening-specific gene, CCR2, is even more pronounced. Figure 2, C and F, show that in the CCA1-ox plants, CCR2 RNA starts to accumulate only after the lights go off and the transcript levels remain high until the lights come on again. As a result, in 16L:8D the oscillations of CCR2 RNA accumulation in the CCA1-ox plants are in the opposite phase from those in wild-type plants (Fig. 2C). Transcript levels of two other genes, GI (Fowler et al., 1999; Park et al., 1999) and CAT3 (Zhong and McClung, 1996), also showed strong oscillations in CCA1-ox plants, but no anticipation of light/dark changes in 16L:8D and 8L:16D (data not shown).
Figure 2.
Diurnal oscillations of transcript accumulation from clock-controlled genes in wild-type and CCA1-ox lines. Plants were grown in photoperiods of 16 h of light and 8 h of dark (16L:8D; A–C) and 8 h of light and 16 h of dark (8L:16D; D–F). The relative RNA levels of Lhcb1*1 (A and C), CAT2 (B and E), and CCR2 (C and F) are shown relative to the maximum levels of expression. Methylene blue staining was used to check equal loading. Experiments were performed twice with similar results. The black and yellow bars beneath the graphs represent dark and light photoperiods.
Transcript Levels of LHY and Endogenous CCA1 Oscillate in CCA1-ox Plants Grown under Light/Dark Conditions
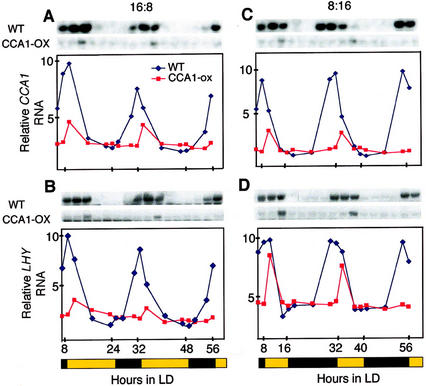
When CCA1-ox or LHY-ox plants are grown in LL, the expression of both LHY and the endogenous CCA1 gene is suppressed, possibly via an autoregulatory negative feedback loop (Schaffer et al., 1998; Wang and Tobin, 1998). To see whether this feedback loop was affected by light/dark changes, we examined the expression of the LHY and the endogenous CCA1 genes in CCA1-ox plants growing in two different photoperiods. Figure 3 shows that under these light/dark conditions, both LHY and endogenous CCA1 genes respond to light in the CCA1-ox plants. Thus, the pathways mediating light regulation of CCA1 and LHY expression appear to be functioning in the CCA1-ox plants. However, in the CCA1-ox plants the transcripts from both genes start to increase only after lights come on, whereas in wild-type plants the increases precede lights on. Taken together, our results demonstrate that the CCA1-ox line of plants, which has lost circadian control of gene expression, retains responsiveness to light.
Figure 3.
Diurnal oscillations of LHY and CCA1 mRNAs in wild-type and CCA1-ox lines. Plants were grown in 16L:8D and 8L:16D photoperiods. Methylene blue staining was used to check equal loading. Representative northerns are shown. Relative levels of CCA1 and LHY RNA were plotted on the graphs. Experiments were performed twice with similar results. The black and yellow bars beneath the graphs represent dark and light photoperiods.
The Constitutive Expression of CCA1 Affects Photoperiodic Flowering
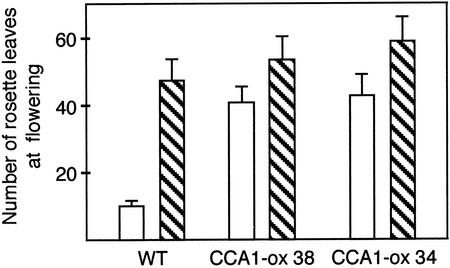
To assess the effect of constitutive expression of CCA1 on sensitivity to photoperiod, we compared the flowering time of wild-type and CCA1-ox plants grown under two different photoperiods, 16L:8D (long days) and 8L:16D (short days). The light intensity was set so that plants received equivalent fluence per 24-h period under each photoperiod. To avoid the influence of different growth rates on the flowering time measurement, we measured the total numbers of rosette leaves of the plants at bolting. Leaf number reflects the developmental stage, and there is a good correlation between leaf number and flowering time in Arabidopsis (Koorneef et al., 1991; Karlsson et al., 1993). Figure 4 compares the flowering time of two CCA1-ox lines, which express similarly high levels of CCA1, with wild-type plants. As expected, the wild-type plants flower much earlier under long days than under short days. By contrast, the CCA1-ox lines show little difference in flowering time between long and short days demonstrating that the CCA1-ox plants are much less sensitive to photoperiod than wild-type plants.
Figure 4.
Flowering times of wild-type and CCA1-ox plants. Seeds of wild-type plants and two CCA1-ox lines, CCA1-ox 34 and CCA1-ox 38, were sown onto soil. Plants were grown in 16L:8D and 8L:16D photoperiods. The numbers of rosette leaves at bolting were counted. The results were plotted on a graph ± sd. White boxes represent 16L:8D and hatched boxes represent 8L:16D.
CCA1-ox Plants Are Less Viable Than Wild-Type Plants in Very Short Days
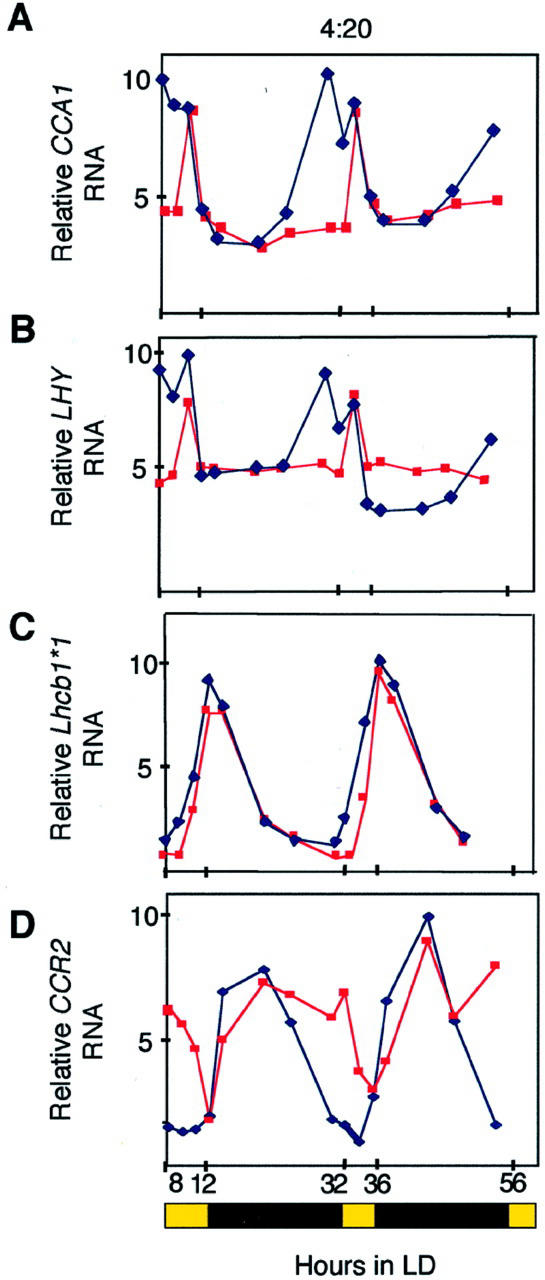
Our observations that the CCA1-ox plants are unable to anticipate diurnal changes led us to ask whether a functional circadian system might confer an advantage to plants under conditions of limited light duration by allowing anticipation of day/night changes. Therefore, we examined the expression of clock-controlled genes under very short-day conditions of 4 h of light:20 h of dark (4L:20D). Figure 5 shows that, consistent with our previous results, transcript accumulation in CCA1-ox plants did not anticipate light/dark transitions. Furthermore, in wild-type plants under 4L:20D, a bimodal peak of RNA accumulation can be seen for both LHY and endogenous CCA1 (Fig. 5, A and B). Such bimodal peaks have been reported for Lhcb1*1 expression in wild type in a range of photoperiods and interpreted to represent expression driven directly by the circadian system and by a transient induction in expression from the acute response to the lights coming on (Millar and Kay, 1996). In the CCA1-ox plants, the part of the bimodal peak that occurs in wild type before the lights come on is missing, suggesting that this is the part of the peak resulting from circadian regulation. The peaks that occur immediately after the lights come on are still present in the CCA1-ox plants, indicating that they are the result of light induction.
Figure 5.
Diurnal oscillations of transcript accumulation in wild-type and CCA1-ox lines in very short-day conditions. Plants were grown in 4L:16D photoperiods. Methylene blue staining was used to check equal loading. Relative levels of RNA were plotted on the graphs. Experiments were performed twice with similar results. The black and yellow bars beneath the graphs represent dark and light photoperiods.
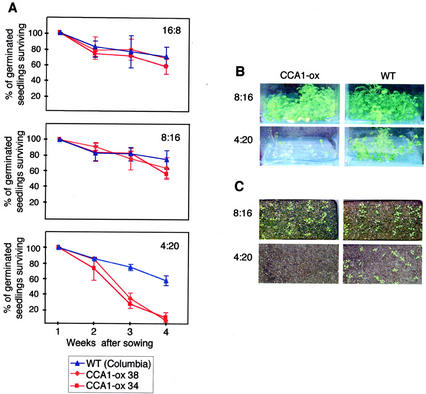
We then compared the viability of wild-type and two lines of CCA1-ox plants grown under three different photoperiods (16L:8D, 8L:16D, and 4L:20D). The light intensities for each treatment were adjusted so that the total fluence received by the plants under each condition was identical. Seeds were sown on soil, and the number of plants that survived after germination was counted each week. Figure 6A shows that under 16L:8D and 8L:16D, there is little difference in the survival of CCA1-ox and wild-type plants. Furthermore, under these conditions, as well as in LL, the CCA1-ox plants grow as vigorously as wild-type plants (Fig. 6, B and C). The survival rate for wild-type plants in 4L:20D is similar to that of all three lines under 16L:8D and 8L:16D, and although the wild-type plants grow more slowly under these very short-day conditions, they appear to be otherwise normal and eventually flowered. In contrast, after 2 weeks in 4L:20D, the seedlings of both CCA1-ox lines began to die off with less than 5% surviving for 4 weeks.
Figure 6.
Viability of wild-type and CCA1-ox plants. A, Approximately 100 seeds of wild-type (Columbia) plants and a CCA1-ox line, CCA1-ox 34, were sown onto soil. Plants were grown in 16L:8D, 8L:16D, and 4L:20D photoperiods. The number of plants surviving was counted each week and expressed as a percentage of the number of seeds that had germinated by 1 week after sowing. The results were plotted on a graph ± sd. Experiments were performed four times with similar results. B, Forty seeds of wild-type plants and a CCA1-ox line, CCA1-ox 34, were sown onto Murashige and Skoog medium in magenta pots and then grown in 8L:16D and 4L:20D photoperiods for 6 weeks. C, Twenty-five seeds of wild-type plants and a CCA1-ox line, CCA1-ox 34, were sown onto soil in a 105- × 155-mm pot and grown 8L:16D (2 weeks) and 4L:20D (4 weeks) photoperiods.
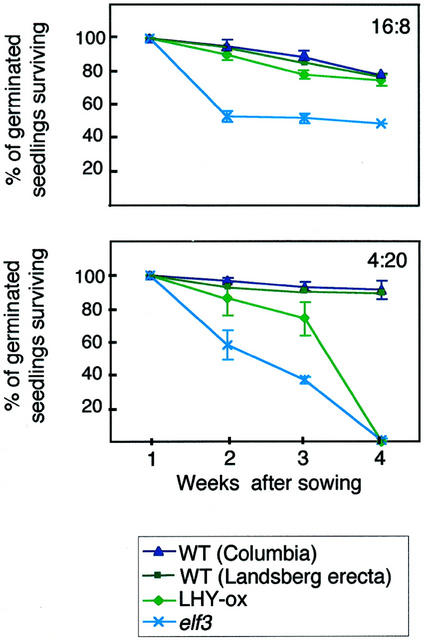
To see whether the lack of fitness we observed in the CCA1-ox plants under very short days could be attributed to pleiotropic effects of constitutively expressing CCA1, we examined the viability of another line that has a defective circadian system. LHY-ox plants (also called Tn120, Schaffer al., 1998) constitutively expresses LHY and are arrhythmic under constant conditions (Schaffer et al., 1998). Figure 6 shows that, under 16L:8D conditions, there is little difference between the survival of LHY-ox plants and the corresponding wild-type control, Arabidopsis ecotype Landsberg erecta. However, in 4L:20D growth conditions, LHY-ox plants fail to survive longer than 4 weeks.
We also examined the viability of elf3-1 plants (Hicks et al., 1996; McWatters et al., 2000), which are arrhythmic in constant light, but not in constant dark. elf3-1 plants show markedly poorer survival than wild type (Columbia) under both 16L:8D and 4L:20D growth conditions (Fig. 7). Together, our results with the three mutant lines demonstrate that plants lacking functional circadian systems are less fit that their wild-type counterparts. However, the difference between the elf3-1 and the LHY-ox and CCA1-ox lines suggests that elf3-1 might be sensitive to additional stress conditions.
Figure 7.
Viability of wild-type, LHY-ox, and ELF3 plants. Approximately 100 seeds of wild-type (Columbia and Landsberg erecta) plants, LHY-ox and ELF3 lines, were sown onto soil. Plants were grown in 16L:8D and 4L:20D photoperiods. The number of plants surviving was counted each week and expressed as a percentage of the number of seeds that had germinated by 1 week after sowing. The results were plotted on a graph ± sd. Experiments were performed three times with similar results.
DISCUSSION
The growth, development, and metabolic processes of plants need to be well coordinated with environmental day/night cycles. This coordination is regulated both by light and by the circadian system. In this study, we have demonstrated that, although CCA1-ox plants no longer show circadian control of gene expression (Wang and Tobin, 1998), they retain light responsiveness. CCA1 has been shown to be involved in both light and circadian pathways (Wang and Tobin, 1998; Green and Tobin, 1999), and our present results demonstrate that the effect of constitutively expressing CCA1 differs for each pathway. It is possible that CCA1 is regulated at different levels in the light and circadian pathways. We have shown previously that CCA1 can be phosphorylated by the protein kinase CK2 (Sugano et al., 1998), and it is possible that the distinct phosphorylation states of CCA1 may also affect its regulatory role in light and circadian control of gene expression.
Because circadian regulation is disrupted in the CCA1-ox lines, they are a useful system with which to examine the adaptive importance of a functional circadian system for plants. One aspect of plant growth and development under circadian control is the photoperiodic regulation of flowering. Consistent with this function, all the circadian mutants that have been identified thus far in Arabidopsis show an alteration in flowering time (Mizoguchi and Coupland, 2000). For example, plants constitutively expressing LHY are day length insensitive with respect to flowering time (Schaffer et al., 1998). We show here that flowering time in CCA1-ox plants is much less sensitive to photoperiod than in wild-type Arabidopsis. Photoperiodic control of flowering probably optimizes pollination and seed set (Samach and Coupland, 2000). However, it is difficult to prove, under laboratory conditions, that loss of photoperiodic control of flowering affects the reproductive success of Arabidopsis.
In another approach to demonstrate the importance of a functional circadian system for plants, we grew the plants in different day lengths and showed that the CCA1-ox plants are less viable than wild-type Arabidopsis under conditions of limited light duration. This low-viability phenotype is only observed in CCA1-ox plants grown in very short days. It is possible that the CCA1-ox plants are less viable than wild type for reasons, such as pleiotropic effects of expressing the CCA1 gene from a constitutive promoter, that are not related to the plant's circadian defects. However, we have shown previously that in the CCA1-ox plants, CCA1 protein is constitutively present at levels that are no higher than the peak levels seen in wild-type plants (Wang et al., 1998). In addition, all the other phenotypes that we have observed in the CCA1-ox lines, for example long hypocotyls, late flowering time, and loss of circadian regulation of leaf movement (E.M. Tobin, Z.-Y. Wang and A.J. Millar, unpublished data) and gene expression (Wang and Tobin, 1998), can be attributed to the disruption of circadian regulation. Furthermore, in longer day lengths (16L:8D, 8L:16D, and LL), CCA1-ox plants grow robustly, are fully fertile, and produce more seeds than do wild-type plants (data not shown). We have also demonstrated that LHY-ox plants show a lack of viability under very short-day conditions. Therefore, a loss of fitness is the simplest explanation for the phenotypes seen in very short days in plants that have defects in their circadian systems.
The very short-day (4L:20D) conditions that we are used are an extreme of the natural day length for Arabidopsis growing in its native habitats. However, Arabidopsis is a mainly winter annual (i.e. it germinates in the fall, overwinters, and then flowers and produces seed in the spring) that occurs naturally in a range of locations including many high-latitude areas. In these high-latitude areas, the amount of solar radiation that the plans receive may be severely limited (Li et al., 1998). In addition, growth conditions in the laboratory environment are optimized to ensure minimum stress for the plants, and the effect of short day length that we observed under these conditions might be seen in slightly longer day lengths when plants grow under natural conditions.
It is unclear what mechanisms are involved in the reduction of viability that we see in the circadian mutants under very short-day conditions. Interestingly, survival rates of LHY-ox, CCA1-ox, and wild-type seedlings are similar for the first 2 weeks of growth under 4L:20D, suggesting that as long as the seed reserves are able to support growth of the seedling, the LHY-ox and CCA1-ox plants are not less viable than wild type. However, after 2 weeks, the LHY-ox and CCA1-ox plants are significantly less fit than wild type in very short days. This might be caused by reduced photosynthetic capacity in the mutant plants. Microarray analysis of gene expression in Arabidopsis has revealed that a large cluster of genes encoding proteins implicated in photosynthesis showed robust oscillations with mRNA levels starting to rise well before dawn and peaking at midday (Harmer et al., 2000). This has been proposed to ensure that the plant can coordinately assemble its photosynthetic machinery and is ready to take maximum advantage of all the available sunlight (Pittendrigh, 1993; Harmer et al., 2000). Thus, the plants that are unable to anticipate dawn because they lack a functioning circadian regulatory system would be predicted to be at an adaptive disadvantage, especially if the duration of available light for photosynthesis is limited.
However, of the four genes we examined in wild-type plants under 4L:20D, the expression pattern of the photosynthetic gene, Lhcb1*1, shows least anticipation of dawn. In addition, elf3-1 plants that are able to anticipate dawn (Hicks et al., 1996) grow more poorly than wild type under both long- and short-day conditions, suggesting that they are sensitive to stresses other than light duration. Thus, we certainly cannot rule out the possibility that the LHY-ox and CCA1-ox plants are also reacting to other stresses when they are grown under 4L:20D. Consistent with this possibility, the clock has been shown to regulate several genes involved in stress responses in Arabidopsis (Harmer et al., 2000). It would be of interest to investigate whether CCA1-ox and LHY-ox plants are compromised in their ability to grow under other stress conditions.
In summary, we have shown that although plants that constitutively express the CCA1 gene show no circadian rhythmicity, they are able to respond to diurnal changes in light conditions. Furthermore, we have demonstrated that a functioning circadian system is important for plant viability.
MATERIALS AND METHODS
Plant Materials and Growth
The Arabidopsis plants used were: CCA1-ox (Wang and Tobin, 1998) and elf3-1 (Hicks et al., 1996), both in the Columbia ecotype, and LHY-ox (Tn 120, Schaffer et al., 1998) in the Landsberg erecta ecotype. All seeds were imbibed and cold treated at 4°C for 4 d before germination and growth. Plants were grown either in petri dishes on Murashige and Skoog medium from Sigma (St. Louis) supplemented with 2% (w/v) Suc or on soil at a density of 25 seedlings per 105- × 155-mm pot. Plants for LL experiments were grown under 12:12, light (125 μE m−2 s−1):dark cycles at 23°C for 17 d before being transferred to constant light (100 μE m−2 s−1) at 23°C. Plants for the experiments in light:dark photoperiods were grown under 16 h of light (30 μE m−2 s−1) and 8 h of dark (16L:8D), 8 h of light (60 μE m−2 s−1) and 16 h of dark (8L:16D), and 4 h of light (120 μE m−2 s−1) and 20 h of dark (4L:20D). Lighting was provided by a combination of fluorescent and incandescent light bulbs.
Flowering Time Measurements
Arabidopsis plants were grown on soil in 105- × 155-mm pots. The time of bolting was determined as the day when the plant had a bolt of 10 mm, and the number of rosette leaves was counted.
RNA Analysis
RNA extractions and analyses were carried out as previously described (Green and Tobin, 1999).
ACKNOWLEDGMENTS
We thank Drs. May Ong, Simon Barak, David Greenberg, Moshe Kiflawi, Nile Kurashige, Shoji Sugano, and Carl Johnson for their critical reading of this manuscript; Chan Sing Sun and Ana Lozano for their excellent technical assistance; and George Coupland for providing the LHY-ox (Tn120) seeds.
Footnotes
This work was supported by the National Institutes of Health (grant no. GM23167 to E.M.T.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004374.
LITERATURE CITED
- Ancoli-Israel S. “Sleep is not tangible” or what the Hebrew tradition has to say about sleep. Psychosom Med. 2001;63:778–787. doi: 10.1097/00006842-200109000-00011. [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey PJ, Krulas JR, Mele G, Holley DC. Circadian performance of suprachiasmatic nuclei (SCN)-lesioned antelope ground squirrels in a desert enclosure. Physiol Behav. 1997;62:1099–1108. doi: 10.1016/s0031-9384(97)00263-1. [DOI] [PubMed] [Google Scholar]
- DeCoursey PJ, Walker JK, Smith SA. A circadian pacemaker in free-living chipmunks: essential for survival? J Comp Physiol. 2000;186:169–180. doi: 10.1007/s003590050017. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Follett BK, Mattocks PW, Farner DS. Circadian function in the photoperiodic induction of gonadotrophin secretion in the white-crowned sparrow. Proc Natl Acad Sci USA. 1974;71:1666–1669. doi: 10.1073/pnas.71.5.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, Kay S. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- Highkin HR, Hanson JB. Possible interaction between light-dark cycles and endogenous daily rhythms on the growth of tomato plants. Plant Physiol. 1954;29:301–302. doi: 10.1104/pp.29.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd MW, Ralph MR. The significance of circadian organization for longevity in the golden hamster. J Biol Rhyth. 1998;13:430–436. doi: 10.1177/074873098129000255. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996;21:5–11. doi: 10.1046/j.1365-2958.1996.00613.x. [DOI] [PubMed] [Google Scholar]
- Karlsson BH, Sills GR, Nienhuis J. Effects of photoperiod and verbalization on the number of leaves at flowering in 32 Arabidopsis thaliana (Brassicaceae) ecotypes. Am J Bot. 1993;80:646–648. [Google Scholar]
- Klarsfeld A, Rouyer F. Effects of circadian mutations and LD periodicity on the life span of Drosophila melanogaster. J Biol Rhyth. 1998;13:471–478. doi: 10.1177/074873098129000309. [DOI] [PubMed] [Google Scholar]
- Kolosova N, Gorenstein N, Kish CM, Dudareva N. Regulation circadian methyl benzoate emission in diurnally and nocturnally emitting plants. Plant Cell. 2001;13:2333–2347. doi: 10.1105/tpc.010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koorneef M, Hanhart CJ, Van Der Veen JH. A genetic and physiological analysis of later flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Kay SA. Coordination of plant metabolism and development by the circadian clock. Plant Cell. 1997;9:1235–1244. doi: 10.1105/tpc.9.7.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- Li B, Suzuki J-I, Hara T. Latitudinal variation in plant size and relative growth rate in Arabidopsis thaliana. Oecologia. 1998;115:293–301. doi: 10.1007/s004420050519. [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail P. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar A. The ELF3 zeitneimer regulates light signaling to the circadian clock. Nature. 2000;408:716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui A, Kumazawa S, Takahashi A, Ikemoto H, Arai T. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature. 1986;323:720–722. [Google Scholar]
- Mizoguchi T, Coupland G. ZEITLUPE and FKF1: novel connections between flowering time and circadian clock control. Trends Plant Sci. 2000;5:409–411. doi: 10.1016/s1360-1385(00)01747-7. [DOI] [PubMed] [Google Scholar]
- Nikaido SS, Johnson CH. Daily and circadian variation in survival from ultraviolet radiation in Chlamydomonas reinhardtii. Photochem Photobiol. 2000;71:758–765. doi: 10.1562/0031-8655(2000)071<0758:dacvis>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS. Temporal Organization-Reflections of a Darwinian Clock-Watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Bruce VG, Rosenzweig NS, Rubin ML. Growth patterns in Neurospora: a biological clock in Neurospora. Nature. 1959;184:169–170. [Google Scholar]
- Ralph MR, Menaker M. A mutation of the circadian system in golden hamsters. Science. 1988;241:1225–1227. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- Samach A, Coupland G. Time measurement and the control of flowering in plants. Bioessays. 2000;22:38–47. doi: 10.1002/(SICI)1521-1878(200001)22:1<38::AID-BIES8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Saunders DS. Circadian control of larval growth rate in Sarcophaga argyrostoma. Proc Natl Acad Sci USA. 1972;69:2738–2740. doi: 10.1073/pnas.69.9.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. Photoperiodism: an overview. In: Lumsden PJ, Millar AJ, editors. Biological Rhythms and Photoperiodism in Plants. Oxford: Bios Scientific Publishers; 1998. pp. 151–165. [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in Plants. Ed 2. San Diego: Academic Press, Inc.; 1997. [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Went FW. Photo- and thermoperiodic effects in plant growth. Cold Spring Harbor Symp Quant Biol. 1960;25:221–230. doi: 10.1101/sqb.1960.025.01.022. [DOI] [PubMed] [Google Scholar]
- Williams WE, Gorton HL. Circadian rhythms have insignificant effects on plant gas exchange under field conditions. Physiol Plant. 1998;103:247–256. [Google Scholar]
- Zhong HH, McClung CR. The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol Gen Genet. 1996;251:196–203. doi: 10.1007/BF02172918. [DOI] [PubMed] [Google Scholar]