Abstract
The joining of different genomes in allotetraploids played a major role in plant evolution, but the molecular implications of this event are poorly understood. In synthetic allotetraploids of Arabidopsis and Cardaminopsis arenosa, we previously demonstrated the occurrence of frequent gene silencing. To explore the involvement of epigenetic phenomena, we investigated the occurrence and effects of DNA methylation changes. Changes in DNA methylation patterns were more frequent in synthetic allotetraploids than in the parents. Treatment with 5-aza-2′-deoxycytidine, an inhibitor of DNA methyltransferase, resulted in the development of altered morphologies in the synthetic allotetraploids, but not in the parents. We profiled mRNAs in control and 5-aza-2′-deoxycytidine-treated parents and allotetraploids by amplified fragment length polymorphism-cDNA. We show that DNA demethylation induced and repressed two different transcriptomes. Our results are consistent with the hypothesis that synthetic allotetraploids have compromised mechanisms of epigenetic gene regulation.
Allotetraploids are formed by hybridization between two species and inherit a complete diploid set of chromosomes from each parental species. Although many established wild and cultivated allopolyploids are fertile, well adapted, and genetically stable, allopolyploids of more recent origin commonly display genomic and phenotypic instability (Soltis and Soltis, 1995; Pikaard, 1999; Comai, 2000).
As a consequence of the union of two genomes, abnormal phenotypes have been reported (Comai, 2000; Schranz and Osborn, 2000). The causes of these phenotypes are largely unknown. McClintock (1984) described similar phenomena as “genomic shock,” which she defined as a preprogrammed response to an unusual challenge resulting in extensive restructuring of the genome. This “unusual challenge” may involve epigenetic gene silencing, which results from homologous DNA-DNA or DNA-RNA interactions. The hybridization of redundant and diverged homeologous sets of genes in allopolyploids might trigger widespread gene silencing and changes in chromatin structure and DNA methylation patterns.
Recent molecular data are consistent with the gene silencing hypothesis. Previously, we have reported about 1% changes in gene expression by comparing synthetic allotetraploids derived by hybridizing 4x Arabidopsis and 4x Cardaminopsis arenosa (also known as Arabidopsis arenosa; Comai et al., 2000). These changes can involve both normal genes and genes related to transposons. The corresponding natural allotetraploid, Arabidopsis suecica, was examined by Lee and Chen (2001), who demonstrated similar silencing levels. Furthermore, they found that silencing was related to methylation and could be reversed by treatment with the DNA demethylating agent 5-aza-2′-deoxycytidine (azadC). Instability can also be manifested by genomic rearrangements. Synthetic hybrids of wheat (Triticum aestivum) displayed rapid and widespread loss of DNA sequences and changes in DNA methylation (Ozkan et al., 2001; Shaked et al., 2001). These results corroborate McClintock's hypothesis of genomic shock as a major factor during allotetraploidization. The understanding of this phenomenon, however, is still rudimentary.
Here, we explore the relationship between phenotypic instability and epigenetic determinants. We show that in Arabidopsis, allotetraploidization leads to nonrandom changes in methylation patterns throughout the genome. Further, we demonstrate that demethylating the genome with the methylation inhibitor azadC greatly exaggerates phenotypic abnormalities in the allotetraploids, but not in the parents. Concomitantly, decreased methylation correlates with more gene expression changes in the allotetraploids than in the parents. Taken together, these results suggest that the formation of allotetraploid genomes destabilizes chromatin regulation.
RESULTS
We reported previously that the crossing of tetraploid Arabidopsis with the natural tetraploid C. arenosa produced allotetraploids whose hybrid phenotype was described in detail (Comai et al., 2000; Fig. 1). We established inbreeding allotetraploids lines derived from four F1s, each the product of pollinating a tetraploid of Arabidopsis ecotype Ler with the same C. arenosa individual. To determine whether gross changes in cytosine methylation were associated with allotetraploidization, we compared percent methylation of TaqI sites (TCGA) in diploid Arabidopsis Ler (one individual, 18.3% ± 2.4%), in tetraploid Arabidopsis Ler (two individuals, 20.6% ± 1.5% and 23.8% ± 1.8%, respectively), in C. arenosa (two individuals, 28.8% ± 1.5% and 30.9% ± 1.5%, respectively) and in F3 synthetic allotetraploids (three individuals, 21.4% ± 1.3%, 22.8% ± 1.4%, and 25.9% ± 1.7%, respectively). The results suggest that no major changes in the level of CG methylation occurred upon allotetraploidization. However, the method used could not reveal whether methyl groups were relocated to other sequences within the genome upon allotetraploidization. To test this possibility, we employed methylation-sensitive amplification polymorphism (MSAP) analysis, which assesses cytosine methylation at specific restriction sites throughout the genome.
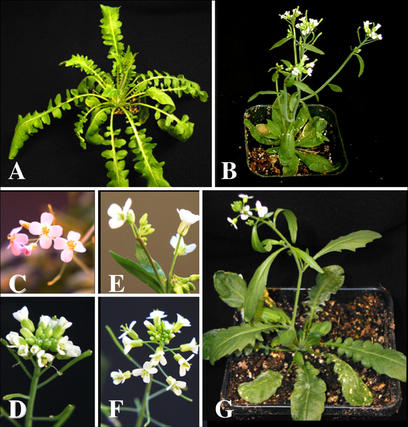
Figure 1.
Phenotype of parents and allotetraploid progeny: tetraploid C. arenosa:whole plant (a) and inflorescence (c); tetraploid Arabidopsis ecotype Landsberg erecta (Ler) LC612:whole plant (b) and inflorescence (d); allotetraploid offspring from Arabidopsis cross with C. arenosa:whole plant (g), allotetraploid inflorescences from different sibling lines resembling more closely either the C. arenosa (e) or the Arabidopsis (f) parent.
Allotetraploidization Leads to Genome-Wide Changes in the Cytosine Methylation State
For MSAP analysis, we used the isoschizomeric methylation-sensitive enzymes HpaII and MspI, which cut at CCGG sites. An example of this analysis is presented in Figure 2. Table I summarizes the different types of cytosine methylation patterns at the CCGG sites found in the parental lines. Interestingly, one-sixth of the methylated sites detected by comparative HpaII/MspI digestion suggested hemimethylation (mCCGG/GGCC; Roberts and Macelis, 2001), whose occurrence has been indicated previously in carrot (Daucus carota; Zhou et al., 1998). We compared 623 MSAP products of four F3 plants with those displayed in the parental lanes (Fig. 2). Differences between F3 plants and parents were frequent. However, only some of these differences were scored as methylation changes as exemplified in Figure 2. Because C. arenosa is an outbreeding species with a relatively high level of polymorphisms, we disregarded changes involving C. arenosa products if they could also be interpreted as polymorphisms that were inherited as heterozygous loci and were segregating in the F3 generation. In addition, we disregarded subtle changes in band intensity (see Fig. 2).
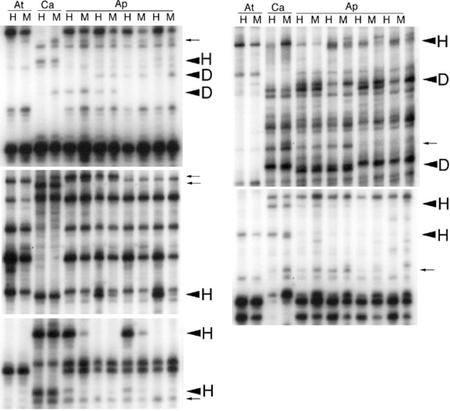
Figure 2.
Examples of MSAP analysis. Selected portions of autoradiographs from MSAP polyacrylamide gels. At, Arabidopsis; Ca, C. arenosa; Ap, allopolyploids; H, EcoRI-HpaII products; M, EcoRI-MspI products. Large arrows indicate differential MSAP products due to hypermethylation (H) or demethylation (D). Small arrows indicate products that, although differential, were not scored because they appeared weak or could be explained as C. arenosa polymorphisms. Bands appearing in the allopolyploids that are absent in both parents result from a loss of methylation at the restriction enzyme cut site (D). In cases where bands are present in the parents but absent in the allopolyploids, methylation occurred at the cut site, preventing the display (H).
Table I.
Methylation patterns at CCGG sites for Arabidopsis, C. arenosa, and A. suecica
The boxes represent the double-stranded, four-base HpaII-MspI recognition site (CCGG). The black boxes represent methylated cytosine.
Schematic representation of the MSAP gel band pattern diagnostic of the associated CCGG methylation pattern.
The last two patterns are resistant to both enzymes and are not distinguishable (N.D.) by the MSAP technique.
Of the 623 products scored, 52 (8.3%) showed polymorphic methylation patterns between the parents and the F3 progeny. Of these, 40 were invariable across the four F3 individuals analyzed, whereas 12 varied in pattern between the allotetraploids. Depending on their status in the progeny, the first 40 were classified into either a demethylation or hypermethylation group (Table II). Twenty-five of 40 (62.5%) that underwent demethylation were subgrouped in nine different classes, D1 through D9 (Table II). Fifteen of 40 (37.5%) displayed an increase in methylation levels and were subgrouped into six classes, H1 through H6 (Table II). The analysis indicates that the methylation pattern of the allotetraploid genome is subject to considerable changes and that many of these changes are not random because the same changes occurred in multiple allotetraploid individuals grown under different conditions (see “Materials and Methods”).
Table II.
Methylation changes in the F3 allopolypoids
The boxes represent the four bases (CCGG) of the recognition site of isoschizomer enzymes HpaII and MspI. Black boxes indicate methylated cytosines, white boxes indicate unmethylated cytosines, and gray boxes represent cytosines whose methylation state varies from sequence copy to sequence copy (partial methylation). Slashed boxes represent cytosines whose state cannot be determined using this method.
Schematic representation of the MSAP gel band pattern diagnostic of the associated CCGG methylation pattern. The thinner bands represent lower concentration products. See Figure 2 for examples.
Fragment not present in one of the parents.
Fragments that differed in methylation status between parents and F3s were eluted from the gel, re-amplified, and sequenced. Some fragments yielded poor sequence quality presumably due to the co-amplification of background fragments. In 23 cases, satisfactory sequence was obtained and compared with the GenBank database using the BLASTN and BLASTX programs (Altschul et al., 1990). Of the 23 tested, nine had no similarity to GenBank entries, whereas 14 had high similarity to characterized regions of the Arabidopsis genome (Table III). Eight of these were within genes, whereas the rest were in intergenic regions. One of them displayed structural similarity to miniature inverted-repeat transposable elements.
Table III.
Sequence of methylated fragments and database search
| MSAP Fragmenta | Len. Rdb | Primersc Eco/M-H | Patternd | GenBank Accession No. | Chromosome (Bacteria Artificial Chromosome Clone)e | Homology Product, Gene | Nucl. id.f (%) | BLAST E Score | TE Linkg |
|---|---|---|---|---|---|---|---|---|---|
| bp | |||||||||
| RM10-5-24 | 322 | TCAA/ACC | D1 | AL138638.2 | 3 (F23N14) | Nodulin-like protein, F23N14.40 | 88 | 4e-47 | Y |
| RM10-6-31 | 265 | CAA/AAA | H3 | AC015450 | 1 (F14G6) | The end of unknown protein, F14G6.10 | 91 | 9e-52 | N |
| RM10-5-42 | 205 | CAA/ACC | D1 | AB009048.1 | 5 (K15E6) | FrnE protein-like, K15E6.12 | 91 | 2e-25 | Y |
| RM10-6-II | 80 | CAA/AAA | H5 | AB025635.1 | 5 (MUF8) | Related to disease resistance protein, MUF8.2 and MUF8.3 | 96 | 5e-04 | Y |
| RM10-4-A | 214 | TCAA/CAG | H1 | AC007764.2 | 1 (F22C12) | Similar to wheat serpin protein, F22C12.21 | 89 | 4e-38 | N |
| RM10-4-B | 199 | TCAA/CAG | D1 | AC009398.6 and AC007354.2 | 1 (F20B24 and T6B5) | Non-coding region, between F20B24.12 and F20B24.13 | 93 | 6e-49 | N |
| RM10-6-34 | 282 | TCAA/CAA | D2 | AC022520 and AC019018.7 | 1 (F14G24) | Putative GTP-binding protein, F14G24.25 | 100 | 1e-26 | Y |
| RM11-3-23 | 293 | TCCA/ACA | D4 | AB005244.2 | 5 (MR011) | Amino acid transporter, MR011.15 | 97 | 4e-140 | N |
| RM10-7-I | 109 | TCAA/AAC | H2 | AC007357.2 | 1 (F3F19) | Non-coding region, between F3F19.15 and F3F19.16 | 94 | 1e-05 | N |
| RM11-6-34 | 160 | – | D2 | AB006697.1 and Z12120.1 | 5 (MAH20) | Ser/Thr-protein kinase, MAH20.15 ATPROKIN | 89 | 9e-26; 1e-24 | N |
| RM11-6-27 | 141 | – | D8 | AB026657.1 | 3 (MX021) | Repeated element, perhaps a MITE | 87 | 3e-22 | Y? |
| RM11-6-44 | 160 | – | H1, H3 | AB023029.1 | 5 (K24C1) | S-adenosyl-Met:2-demethylmenaquinone methyltransferase like, K24Cl.2 | 81 | 7e-05 | N |
| RM11-7-50 | 160 | – | H2 | AF175991.1 and AB010695 | 5 (MDK4) | Myb-related transcription factors-like protein, MYB49 | 94 | 8e-48 | N |
| Rm11-7-31 | 160 | – | Controlh | AL161591.2 and AL035605.1 | 4 (F19F18 and F6G17) | Non-coding region, between AT4g37560 and AT4g37570 | 95 | 2e-70 | N |
Working designation.
Length of sequence read.
Nucleotides' extensions of primers used in amplification listed with the EcoRI primers first and HpaII or MspI second.
Refer to Table 2; chromosome 1 through 5, bacteria artificial chromosome clone no. pinpoints physical map location.
GenBank BAC no.
Nucleotide identity to BLASTN hit; because the MSAP products were identified from unidirectional sequencing of gel-eluted and amplified DNA, the percent identity reflected also the sequencing quality.
Linkage (less than 6 kb) to annotated transposable element; Y, yes; N, no; Y?, the putative transposable element was not annotated as such in GenBank.
Positive control, methylated non-differential product from 612.
Demethylation Increases Phenotypic Instability
The joint occurrence of phenotypic abnormalities, gene silencing (Comai et al., 2000) and cytosine methylation of previously un-methylated sites (Fig. 2; Tables I–III), suggested that demethylation might restore the allotetraploids to a stable phenotype similar to that of A. suecica, if hypermethylation was the main reason for the observed abnormalities.
We treated F2 and F3 plants from each of the four original allotetraploid lines as well as their parents, diploid Arabidopsis, and the natural allotetraploid A. suecica with the demethylating agent azadC. Parents and untreated, as well as azadC-treated plants, were grown to maturity and compared phenotypically. Diploid and tetraploid Arabidopsis and A. suecica treated with azadC grew normally (data not shown) and C. arenosa displayed relatively less frequent and less severe abnormalities, whereas the allotetraploids consistently produced abnormal phenotypes (Table IV). Some of the displayed abnormalities were also seen in untreated allotetraploids but at much reduced severity. The experiment was repeated five times with similar results. The most common abnormality was a semidwarf phenotype that displayed many secondary inflorescences with shorter and zigzag internodes. Other frequent abnormalities included fasciation of the shoots and homeotic transformations of the flowers (Fig. 3), including dipetalous and tetrapetalous phylloid flowers, apetala-like flowers, open carpels, or cauliflower-like inflorescences. Abnormal and normal body sectors sometimes appeared on the same plants (Fig. 3). Other examples of phenotypic abnormalities included dwarfism, aberrant branching patterns, and tumor formation (Fig. 3; data not shown). Often, these phenotypes affected only lateral inflorescences and showed occasional reversion during branching or further apical growth. In certain cases, abnormalities changed in intensity along the axis of growth. In contrast, untreated synthetic allotetraploids displayed the usual variability but failed to show most of the aberrant phenotypes displayed by the azadC-treated cohort. Although most of the azadC-treated synthetic allotetraploids were sterile, a few produced seed upon selfing. Plants originating from these seeds generally differed from the parent, displaying either the normal hybrid phenotype or an altered phenotype unrelated to the parents (data not shown).
Table IV.
Induction of abnormal phenotypes by azadC treatment
| Line | Species or F1 Ancestora | X (ploidy) | Affected | n | Pb |
|---|---|---|---|---|---|
| % | |||||
| Maternal (Ler) | Arabidopsis 612 | 4 | 0 | 30 | 9.1E-03 |
| Ler | Arabidopsis | 2 | 0 | 29 | 8.1E-03 |
| Paternal | C. arenosa | 4 | 22c | 27 | – |
| Synth. Allopolyploid α1 | 49-2A | 4 | 73 | 25 | 2.6E-05 |
| Synth. Allopolyploid α2 | 49-2B | 4 | 90 | 33 | 3.2E-04 |
| Synth. Allopolyploid α3 | 605A | 4 | 70 | 10 | 2.4E-03 |
| Synth. Allopolyploid α4 | 605B | 4 | 58 | 20 | 1.1E-02 |
| Natural Allopolyploid | A. suecica | 4 | 0 | 24 | 2.3E-02 |
The synthetic allopolyploids are described in (Comai et al. 2000).
Two-tailed probability of the null hypothesis (no difference) from Fisher exact test of independence in which all lines' responses are compared with that of the paternal C. arenosa.
These plants were considerably less affected than the synthetic allopolyploids (see “Materials and Methods”).
Figure 3.
Induction of abnormal phenotypes by azadC treatment of F2 allotetraploids. Vastly differing degrees of fasciation of the inflorescence of untreated (a) and azadC-treated (b, inflorescence; p, inflorescence stem) plants. Abnormal branching patterns in azadC-treated plants (c and g). Branching was either altered in the entire plant (g) or only in one or more branches (b, k, and m). Flowers displayed homeotic abnormalities (d–f and m–o). The phylloid petal phenotype of the plant shown (m, top view; n, side view; o), accompanied by fasciation (p), affected only two branches, whereas the rest of this plant displayed a normal morphology (l). See Figure 4C for analysis of centromeric repeat methylation in the affected versus unaffected tissue.
Treatment with azadC Causes Centromeric Demethylation
To verify that the azadC affected genome methylation equally in all genotypes, we examined cytosine methylation at the CCGG sites in the centromeric repeat region of 180-bp units in parents and allotetraploids in the presence or absence of azadC. If the CCGG sites are hypomethylated, they become susceptible to HpaII digestion and the repeats produce a ladder of variable multimers differing by 180 bp. Figure 4A shows that treatment with azadC decreased methylation in all genotypes, although somewhat less markedly in C. arenosa. The use of species-specific probes on replicate gel blots confirmed that the degree of methylation of the centromeric repeats depended on the genome of origin. When we tested the methylation state of three individual allotetraploids treated with azadC, we noticed that one of the three individuals displayed differential methylation patterns on its two genomes (Fig. 4B). This individual showed normal demethylation in response to azadC on its C. arenosa genome, but the methyl-sensitive enzymes failed to digest the Arabidopsis-specific 180-bp repeat, suggesting heavier methylation in this region of the Arabidopsis-derived genome. We compared the repeat methylation status in a dimorphic plant (Fig. 4C). A branch displaying severely altered flowers and fasciation (Fig. 3, M–P) had strongly hypomethylated DNA (Fig. 4C, “Alt” lane), whereas a morphologically normal branch of the same plant (Fig. 3L) had a distinct pattern of higher methylation (Fig. 4C, “Nor” lane).
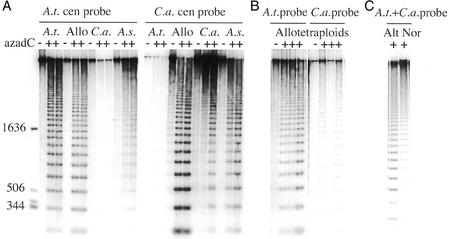
Figure 4.
Effect of azadC and differential methylation of Arabidopsis and C. arenosa centromeric repeats in the hybrid genomes. Shown are replicate gels of HpaII-digested DNA, probed with either the Arabidopsis or the C. arenosa 180-bp centromeric repeat (A). Gel lanes loaded with DNA from control and from azadC-treated plants are marked − and +, respectively. The size in bp of the markers (first lane) is indicated on the left. B, Analysis of replicate blots of four allotetraploid plants either probed with the Arabidopsis or the C. arenosa 180-bp centromeric repeat (one control and three azadC-treated). C, Analysis of altered (Alt, see Fig. 3, M–P) and normal (Nor, see Fig. 3L) branches from the same azadC-treated plant. A.t., Tetraploid Arabidopsis 612; Allo, synthetic allotetraploids; C.a., C. arenosa; A.s., A. suecica.
Genome Demethylation Causes Increased Rates of Transcriptional Changes in Allotetraploids
Demethylation of allotetraploids with azadC did not normalize the phenotype but instead increased the frequency of phenotypic abnormalities when compared with untreated plants (Fig. 3). Therefore, we asked if demethylation caused by azadC would lead to an increased incidence of transcriptional changes in allotetraploids when compared with their parents. To investigate this possibility, we employed cDNA-AFLP display (Fig. 5; Bachem et al., 1996). Differences in the cDNA-AFLP patterns between the genotypes were frequent. We categorized these differences into distinct groups shown in Table V. We observed more transcriptional variation between the untreated and azadC-treated allotetraploids as compared with transcriptional variation between untreated or azadC-treated Arabidopsis parents, suggesting a higher rate of gene activation in azadC-treated allotetraploids than in azadC-treated Arabidopsis. To verify that the increased rate of transcriptional changes was due to the treatment with azadC and not simply a result of the allotetraploidization process, we compared the AFLP patterns of untreated Arabidopsis with those of untreated allotetraploids. Here, the variation was significantly less pronounced than in the azadC-treated allotetraploids (Table V). Changes in the AFLP patterns often differed among the four tested allotetraploid individuals, either azadC treated or untreated. Changes in the AFLP pattern that could be explained by differentially inherited polymorphic markers from C. arenosa were not scored; therefore, a comparison between C. arenosa and the allotetraploids was not performed. Similarly, we did not include the natural allopolyploid A. suecica in the cDNA-AFLP analysis because the maternal ecotype is not available for comparison. We did include in our changes, however, AFLP products that appeared in the allotetraploids but not in either parent. The comparison did not use the C. arenosa individual that was the ancestor of the allotetraploids. Thus, it is possible that the allotetraploid-specific bands may have resulted from polymorphisms that were not present in the reference C. arenosa but were inherited from the original C. arenosa ancestor. We suspect that many of these AFLP products represent activated genes and are not polymorphisms because induction of new bands was also observed in azadC-treated Arabidopsis, which has no polymorphisms. Further, we used untreated sibling allotetraploids to compare expression patterns of the putatively azadC-activated genes with the untreated controls, which allowed us to further decrease the number of false positives.
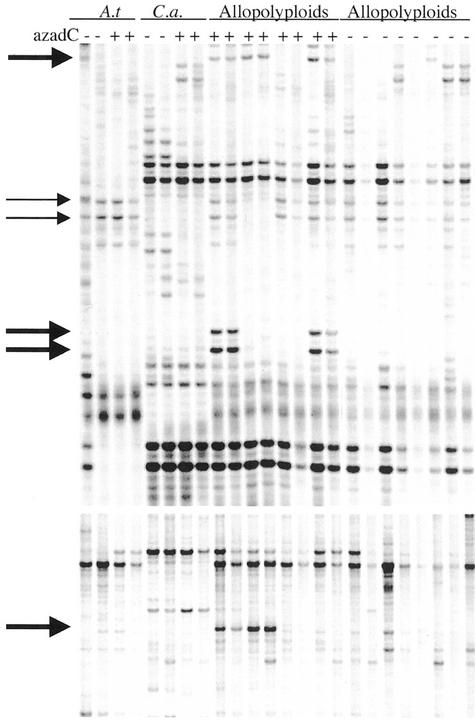
Figure 5.
Example of cDNA-amplified fragment length polymorphism (AFLP) analysis. Selections of autoradiographs from cDNA-AFLP polyacrylamide gels. A.t., Arabidopsis; C.a., C. arenosa; + or − indicate the presence or absence of azadC during seed germination. Bold arrows indicate products that were activated in azadC-treated allotetraploids. Thin arrows indicate Arabidopsis products that were differentially silenced azadC-treated allotetraploids. All samples were run in duplicate representing separate RNA preparations from comparable tissues.
Table V.
Transcriptional changes caused by azadC in parents and F3 allotetraploids
| Compared Lines and
Treatments
|
Totala | % | P | Scoredb | |||
|---|---|---|---|---|---|---|---|
| Line 1 | azadC | Line(s) 2c | azadC | ||||
| Arabidopsisd | + | Arabidopsis | – | 13 /1,500 | 0.9 | – | Any change |
| Arabidopsis | – | Allo | – | 7 /1,500 | 0.5 | – | If Arabidopsis ≠ any Alloe |
| Arabidopsis | + | Allo | + | 45 /1,500 | 3.0 | 8E-9 | If Arabidopsis ≠ any Allo |
| Arabidopsis | – | Allo | + | 41 /1,500 | 2.7 | 5E-7 | If Arabidopsis ≠ any Allo |
Changed AFLP-cDNA bands/no. of cDNA bands scored.
Bands were scored if they were absent in one of the two lines compared or, in the case of the allotetraploids, if any of the four lines sampled differed from Arabidopsis. Bands present in C. arenosa but absent in allotetraploids were not scored because they could be caused by polymorphisms in the parent. Bands present in allotetraploids and not in the parents were scored (see “Results” for further comments).
Four allotetraploid individuals were sampled.
Allo, F3 allotetraploids of Arabidopsis and C. arenosa.
Two-tailed probability of the null hypothesis (no difference) from Fisher exact test of independence in which row 2 data are compared with rows 3 or 4.
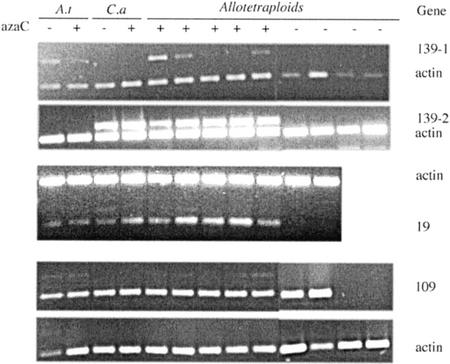
For ultimate confirmation of transcriptional changes, several differentially displayed AFLP bands were excised and eluted from the gel, re-amplified, sequenced, and the sequence compared with the GenBank database. Most of the sequences originated from genes, but a few were also from intergenic regions (data not shown). In a few cases, we verified differential expression of the sequences by RT-PCR using cDNA from the original plants as template. Figure 6 shows examples of differentially regulated genes. Where possible, we designed species-specific primers to amplify exclusively the transcript from either the Arabidopsis genome or the C. arenosa genome (139-1 and 139-2). Gene 139 was well expressed in the untreated parents. Expression of both the C. arenosa homeolog and the Arabidopsis homeolog appeared strongly reduced in the untreated allotetraploids. AzadC treatment diminished gene expression in Arabidopsis but not in the C. arenosa parent. In the allotetraploids, azadC restored expression of the C. arenosa homeolog with high efficiency and of the Arabidopsis homeolog with partial efficiency. In cases where species-specific primers could not be made (genes 19 and 109, Fig. 6), RT-PCR merely revealed differential expression between treated and untreated allotetraploids. For gene 19, only two untreated allotetraploids were tested and found to show no transcription, whereas azadC-treated allotetraploids displayed vigorous transcription. Gene 109 showed a similar pattern: All tested azadC-treated allotetraploids displayed transcription, whereas transcription was turned off in two of the four tested untreated allotetraploids. The sequences of genes 19, 109, and 139 were compared with sequences in the GenBank database. The GenBank annotation states similarity of gene 19's product to a dehydrogenase from Arabidopsis, containing an Myb-like DNA binding domain (GenBank accession no. AAF81310.1). Gene 109's product displays similarity to a regulator of chromosome condensation (GenBank accession no. BAB10107.1), and gene 139's product resembles a putative DNA binding protein (GenBank accession no. AAK64037.1).
Figure 6.
Reverse transcriptase (RT)-PCR verification of differential cDNA-AFLP products. Three bands that displayed differential transcription patterns in the AFLP-cDNA analysis were chosen and subjected to RT-PCR verification. Primers were made either specifically to the parental species (no. 139) or, if that was not possible, to the consensus sequence (nos. 19 and 109). A.t, Arabidopsis; C.a, C. arenosa. Actin was used as a control gene and either amplified in the same PCR reaction (multiplex, no. 19, and no. 139) or, if competition between bands did not allow multiplexing, amplified in separate reaction tubes (no. 109). A, 139-1 (Arabidopsis specific) and 139-2 (C. arenosa specific) are not transcribed in the azadC-treated allotetraploids. B, 19 is not transcribed in two untreated allotetraploids but highly active in the azadC-treated allotetraploids. C, 109 is transcribed in all azadC-treated, but only two of four untreated, allotetraploids. The c-DNA used for the RT-PCR analysis of all three genes was prepared from the original RNA used for c-DNA-AFLP analysis in Figure 5.
DISCUSSION
The allotetraploid offspring from crosses of Arabidopsis and C. arenosa display phenotypic instability, high embryo lethality, and frequent gene silencing (Comai et al., 2000). The mechanisms underlying this phenomenon, however, are poorly understood. To test the hypothesis that allotetraploidization results in partial loss of epigenetic gene regulation affecting genome methylation and gene expression, we investigated methylation patterns in newly formed allotetraploids and explored the effect of the demethylating agent azadC on phenotypic and transcriptional changes in these plants.
We set out by investigating overall CG DNA methylation at TaqI restriction sites in F3 allotetraploids and found no gross changes between parents and the allotetraploids. To sample defined sequence sites for methylation changes we employed MSAP analysis, which revealed frequent changes in F4 allotetraploids, involving both increases and decreases in methylation but no overall hyper- or hypomethylation (Fig. 2; Tables I and II). Interestingly, many of the changes in methylation state were observed in different individual siblings, indicating that at least some methylation changes were either inherited or were locus specific. Close to one-half of the sequenced differential MSAP DNA fragments did not have a match in the sequence databases: They could represent heterochromatic regions that either have not been sequenced or that are unique to C. arenosa.
To investigate if large-scale methylation changes occur in heterochromatic regions of the genome, we compared cytosine methylation of the 180-bp centromeric repeats, which represent 1% to 2% of the genome (Martinez-Zapater et al., 1986; Round et al., 1997) and are almost completely methylated in wild-type Arabidopsis (Vongs et al., 1993). We confirmed this finding and also showed, using species-specific probes, that the repeats in C. arenosa, A. suecica, and the allotetraploids are highly methylated (Fig. 4A), suggesting that the observed phenotypic variability is not a result of gross differences in heterochromatic methylation. Treatment with azadC led to demethylation in all tested genotypes (Fig. 4A). In one of the C. arenosa individuals, however, demethylation was not as pronounced as in the other individuals tested (Fig. 4A). When we tested several allotetraploid individuals for their centromeric repeat methylation state, we noticed that one of the three individuals in Figure 4B showed demethylation of only one of its genomes (compare lane 3 with 7). This might indicate preferential methylation of one of the two genomes and is reminiscent of nucleolar dominance in which one set of rRNA genes in A. suecica is preferentially silenced (Chen et al., 1998). We also found an instance of differential methylation in two branches of the same allotetraploid plant differing in morphology (Fig. 4C).
The frequent gene silencing and phenotypic abnormalities observed previously in synthetic F2 allotetraploids could be explained by the simple assumption that many loci in these plants became methylated and thus silenced. We hypothesized, therefore, that a demethylating treatment might relieve many of the observed abnormalities. Contrary to this expectation, treatment with azadC caused exaggerated phenotypic instability specific to the synthetic allotetraploids, which showed frequent morphological abnormalities (Fig. 3), whereas azadC-treated parents remained unaffected in the case of Arabidopsis or lightly affected in the case of C. arenosa. Remarkably, the natural allotetraploid A. suecica was also unaffected by azadC. Although untreated allotetraploids also display instability in the first few generations after the allotetraploidization event, the phenotypic changes in untreated allotetraploids were in some cases similar but much less intense than those observed in the azadC-treated allotetraploids. For example, light fasciation as shown in Figure 3A is common in newly constructed allotetraploids and mostly affects inflorescence stems. Allotetraploids treated with azadC, however, often displayed entire sectors with vastly exaggerated fasciation (Fig. 3B). In some cases, flowers were affected and displayed various levels of homeotic abnormalities (Fig. 3, C–K). Therefore, it appears as if demethylation of the genome does not restore the normal phenotype in allotetraploids but further exaggerates the abnormalities.
To further test our original hypothesis that demethylation would reactivate genes that had become methylated and thus silenced in response to allotetraploidization, we used cDNA-AFLPs to quantify the level of transcriptional changes in the azadC-treated plants as compared with the untreated cohort. If the original hypothesis was correct, allotetraploids would display azadC-induced changes comparable with those displayed by the azadC-treated parents and fewer than the changes displayed by the untreated allotetraploids. However, analogous to our phenotypic observations, the results from the cDNA-AFLP analysis (Fig. 5; Table V) showed that allotetraploids treated with azadC displayed more than three times the number of transcriptional changes observed in parental Arabidopsis treated with azadC. This result suggests that demethylation with azadC leads to greater transcriptional instability in newly formed allotetraploids than in their parents. Therefore, demethylating treatment of allotetraploids increases both phenotypic and transcriptional abnormalities as if an already disturbed chromatin structure might be more susceptible than the corresponding structure of the parents.
To verify the transcriptional changes in azadC-treated allotetraploids, we performed RT-PCR analysis on a number of differentially regulated genes (Fig. 6). In the documented cases, genes were chosen that were active in parent plants and had become partially or fully silenced in individual untreated allotetraploids. In some cases, these genes were reactivated by azadC treatment (19 and 109). In other cases (139-1), reactivation varied from weak to complete but differed in pattern between the two different sets of homeologs.
Why did the azadC-treated allotetraploids show such increased phenotypic variability? Are the silenced or (re-) activated genes directly responsible for the observed phenotypic changes? It is not possible to assign a cause-and-effect relationship between the putative functions of the differentially expressed genes and the observed phenotypes in our experiments. We note, nevertheless, that the genes shown in Figure 6 contained DNA-binding domains, as did silenced genes described in previous studies (Comai et al., 2000; Lee and Chen, 2001). This category of genes may be more susceptible to allotetraploidization and to perturbed chromatin regulation. For example, the silenced gene encoding the putative transcription factor RAP2.1 (an APETALA2-class gene) contains repeated elements in its 5′ promoter region and is naturally methylated in Arabidopsis, yet actively transcribed (Comai et al., 2000). Interestingly, in the methylation-deficient cmt3 background, RAP2.1 was shown to be hypomethylated (Tompa, et al., 2002), suggesting epigenetic regulation of this gene. Further, it has been suggested that genes encoding transcription factors are more likely than other genes to gain novel regulatory patterns through insertion of transposable elements in their promoter regions (White et al., 1994). If this is the case, genes encoding transcription factors may also be more susceptible to epigenetic regulation (Martienssen, 1998) and could become selectively unstable under compromised heterochromatin suppression. Destabilization of this class of genes may be responsible for the phenotypic effects we observed in the allotetraploids.
An important and still largely unanswered question is the nature of the signals that induce epigenetic regulation. Given that both parents and the derived allopolyploids are tetraploids, the observed effects most likely result from hybridization rather than from a change in ploidy. Therefore, incompatibilities between loci located on different genomes may play a major role in the establishment of allotetraploids. Previously described epigenetic phenomena suggest further possibilities. In some cases, such as the PAI gene system or in posttranscriptional silencing of transgenes (Luff et al., 1999), one gene organized as an inverted tandem duplication can trigger its own transcriptional silencing and methylation as well as that of unlinked homologous loci, perhaps through formation of dsRNAs (Muskens et al., 2000; Wassenegger, 2000). In other cases, such as the formation of SUPERMAN epialleles, global genome demethylation triggers hypermethylation of certain loci (Jacobsen and Meyerowitz, 1997). It is not known whether a sequence-specific property of SUPERMAN or the location of this gene within the genome context causes the observed methylation changes in the clark kent alleles (Jacobsen and Meyerowitz, 1997). Similar mechanisms to those discussed above could contribute to epigenetic remodeling of allopolyploids.
CONCLUSION
We have shown that phenotypic instability in newly formed allotetraploids is accompanied by nonrandom changes in the methylation state of the combined genomes and by remodeled transcription involving both gene silencing and gene activation. Hypersensitivity of the allotetraploids to azadC is consistent with compromised chromatin regulation, resulting in transcriptional changes at sensitive loci. We suggest that this syndrome is triggered by incompatibilities between the two genomes. The same mechanisms may play a role in phenomena that occur when divergent genomic combinations are made such as in wide hybridizations, crosses resulting in hybrid vigor or heterosis, or when hybrids display unexpected behavior during the introgression of genes from wild accessions into established cultivars. Thus, remodeling of chromatin in wide crosses could have economic and evolutionary implications by affecting changes in phenotype that may be either disruptive or adaptive.
MATERIALS AND METHODS
Plant Lines, Growth, and Allotetraploidization
Allotetraploid plants were generated as described elsewhere (Comai et al., 2000). In brief, Arabidopsis tetraploids (ecotype Ler, LC612; 4x = 2n = 20) were used as the female recipient of pollen from Cardaminopsis arenosa (Washington University no. 9509; 4x = 2n = 32). Four separate F1s were produced (49-2A, 49-2B, 605A, and 605B) using pollen from one C. arenosa individual. Inbreeding lines were subsequently advanced by self-pollination and were called α1, α2, α3, and α4, respectively. A. suecica Sue-1 (2n = 26) was obtained from Fumiaki Katagiri (Massachusetts Institute of Technology, Cambridge). Plants were grown in a soil-less peat mix (Sunshine no. 5) in a growth room at 22°C ± 3°C under 16 h of artificial daylight (TL80 fluorescent bulbs, Philips, Eindhoven, The Netherlands) and 8 h of darkness. Some plants (see below) were grown in a greenhouse at 21°C ± 4°C., with supplemental light to provide 16-h days.
For azadC treatment, F2 or F3 allotetraploid seed and seed from each parental line and from several Arabidopsis ecotypes (Metz, Niederzenz, No-0, Columbia, and Ler) was surface sterilized, cold-treated for 2 d, and plated on one-half-strength Murashige and Skoog salts (Invitrogen, Rockville, MD) supplemented with 40 μg mL−1 of azadC (10 μm). Controls were plated on medium lacking azadC. Seedlings were transplanted to potting soil when their roots reached 2 cm in length and were subsequently grown in growth rooms or greenhouse as described above. Plants were scored at the juvenile and adult stage on a subjective scale of abnormalities, from normal (5) to severely abnormal (1). The observations were converted to numerical values (normal versus affected) in the following way: azadC-treated individuals were scored as affected if they ranked lower than any individual in the control. In one experiment, five azadC-treated C. arenosa individuals were scored “4” and one was scored “3.” All controls scored “5.” Thus, six azadC-treated plants were scored as affected. It should be noted that none of these plants displayed the severity of symptoms of treated synthetic allopolyploids, which were often scored as “1” through “3.” The data in Table IV were subjected to Fisher Exact Test for independence (http://home.clara.net/sisa/fisher.htm) by pair-wise comparison with the C. arenosa data in a 2 × 2 table format. The two-tailed probability was computed using the “sum of small P values” method.
Taq Site Methylation Analysis
Taq site analysis was performed as described (Cedar et al., 1979). In brief, genomic DNA (0.5–2.0 μg) was treated with RNAse A and RNAseT1 and cleaved with TaqI. The terminal cytosine was labeled with [γ-32P] ATP. The labeled and digested DNA was separated by thin-layer chromatography and exposed to a PhosphorImager screen. Quantification was performed using ImageQuant software (IQMac version 1.2, Molecular Dynamics, Sunnyvale, CA). The 5-methylcytosine levels were calculated by the formula 5-methyl deoxycitidine monophosphate/(5-methyl deoxycitidine monophosphate + deoxycitidine monophosphate).
DNA Analysis
DNA was prepared as described elsewhere (Comai et al., 2000). To assess the methylation status of the 180-bp repeats, 0.25 μg of HpaII-digested (with 10 units of enzyme μg−1 of DNA) genomic DNA were separated on a 0.8% (w/v) agarose gel, blotted on Biodyne-B nylon membrane, and hybridized to species-specific centromeric probes. To verify the completeness of HpaII digestion of the genomic DNA, we examined the appearance of stereotypical banding in the ethidium-bromide-stained gels of the digested DNA. To prepare the Arabidopsis centromeric probe, we first amplified the insert of the pUC derivative plasmid pARR20 (a gift of Eric Richards, Washington University, St. Louis) with forward and reverse M13 primers. To prepare the C. arenosa centromeric probe, we amplified the 180-bp repeat from C. arenosa genomic DNA using primers CAcenL (AGCTTCTTATTGCTCTCAACGG) and CAcenR (TTAGAAGCTCCAAAACCGAAAA). For the PCR, we used the time-release enzyme AmpliTaq Gold (Perkin-Elmer Applied Biosystems, Foster City, CA) with the following cycle conditions: 9 min at 94°C, followed by 35 cycles (20 s at 94°C, 30 s at 57°C, and 40 s at 72°C). Radiolabeled probes were prepared by single-stranded PCR of the above PCR products using the M13 reverse primer and the CAcenL primer, respectively. Hybridized blots were washed in 0.2× SSC at 60°C and visualized using a PhosphorImager.
MSAP Analysis
For MSAP analysis, DNA was isolated from four α1 F3 sibling plants derived from the same F2 plant (631), which in turn was the offspring of the original F1 allotetraploid (49-2A; Comai et al., 2000). Two of the siblings were grown in a growth room, and two in a greenhouse (see above). For the analysis, we followed a modified AFLP protocol developed to assay DNA methylation. This adaptation incorporates the use of methylation-sensitive frequent-cutter restriction enzymes, such as the isoschizomers HpaII and MspI, with a rare cutter such as EcoRI. The adapters for EcoRI were the same as those used in the AFLP protocol (Bachem et al., 1996). The adapters for HpaII-MspI digest fragments were designed according to Xiong and colleagues (1999).
All the primers designed for the EcoRI fragments had the same core and enzyme-specific sequence (5′-GACTGCGTACCAATTC-3′) and the following combinations of three selective nucleotides were added to the basic sequence: ACA, AGA, ACC, AAA, and AAC. The EcoRI primers were used in combination with two HpaII-MspI primers that bear four selective nucleotides: 5′-CATGAGTCCTGCTCGGTCAA-3′ and 5′-CATGAGTCCTGCTCGGTCCA-3. The genomic DNA (1 μg) was digested with 20 units of EcoRI (New England Biolabs, Beverly, MA) in a final volume of 40 μL of the appropriate buffer for 3 h at 37°C. For the second digestion, 20 units of HpaII (Life Technologies/Gibco-BRL, Cleveland) or MspI (New England Biolabs) were used. The digested fragments were ligated to the adapters in a buffer containing 0.5 mm dithiothreitol, 1 mm ATP, and 20 units of T4 DNA ligase (New England Biolabs), and incubated at 37°C for 2 h. The pre-amplification was performed by using 1 μL of the ligation products and 0.2 μm of the EcoRI and HpaII-MspI primers, without the selective nucleotides, in a final volume of 50 μL containing 1× PCR buffer, 0.1 mm dNTP, and 1 unit of Taq polymerase. The PCR reactions were performed with the following program: 30 s at 72°C, 3 min at 94°C, and 30 cycles consisting of 1 min at 94°C, 1 min at 56°C, 2 min at 72°C, and a final extension step of 5 min at 72°C. The pre-amplification products were diluted 1:10 (v/v) and 1 μL was used in the selective amplification reaction with the EcoRI and HpaII-MspI primers, end labeled with [γ-32P]ATP, in a final volume of 20 μL. The other components were the same as the pre-amplification reactions. The PCR program was the same as in the original AFLP protocol.
The specificities of HpaII and MspI are described in the REBASE database of restriction enzymes (Roberts and Macelis, 2001). In summary, when methylation affects both DNA strands, HpaII is blocked by methylation of either the internal or external cytosine, whereas MspI is blocked only by methylation of the external cytosine. Hemimethylation of the outside C blocks MspI but not HpaII. To follow changes in methylation, MSAP analysis for each genotype was performed in duplicate, displaying the HpaII-EcoRI and MspI-EcoRI products, respectively (referred to as HpaII and MspI lanes). A product appearing in both lanes with the same relative intensity indicates that the corresponding CCGG site is either un-methylated or only partially methylated. Partial methylation, due to differences in methylation state between copies of the same locus, results in changes in product intensity between genotypes. A difference in product intensity between the HpaII and the MspI lanes of the same genotype indicates differential methylation between the internal and external cytosine. A stronger MspI product is caused by hypomethylation of the external relative to the internal cytosine residue. A stronger HpaII fragment is due to hemimethylation of the external cytosine.
Isolation and Direct Sequencing of the MSAP Fragments
The MSAP fragments were eluted by rehydrating the gel in boiling water for 5 min and re-amplified with the appropriate primers. The eluted DNA was diluted 1:1,000 (v/v) and 1 μL was used for PCR amplification in a final volume of 50 μL, the reaction and cycling conditions were the same as the selective amplification described above. The PCR products were checked by agarose gel electrophoresis and 1 μL was used as a template for direct sequencing according to standard Big Dye terminator protocols (ABI, Sunnyvale, CA). The analysis of DNA similarity of the sequences obtained was performed by using the Advanced BLASTN program at the National Center for Biotechnology Information site.
cDNA-AFLP Analysis and RT-PCR
RNA was prepared from the inflorescence tips (buds, flowers, petioles, and connected stem) of four azadC-treated plants and four comparable controls. The four azadC-treated plants had the following genotype and phenotype: plant 1, F2 α1 line, semidwarf, thick stems, small non-serrated leaves, and small flowers; plant 2, F2 α3 line, semidwarf, regular stems, small serrated leaves, and small flowers often displaying fasciated carpels; plant 3, F2 α3 line, normal size, and flowers and leaves but heavily fasciated; and plant 4, F2 α4 line, dainty dwarf, thin stems, and small flowers with many in terminal position on inflorescence. The four control plants had typical phenotypes with larger bodies, serrated leaves, and larger, usually normal, flowers. The cDNA-AFLP analysis was performed as described (Bachem et al., 1996; Ditt et al., 2001), except that the amplification primers contained three base extensions. Verification of AFLP patterns by RT-PCR was essentially performed as described (Ditt et al., 2001). All samples were run in duplicate and variations were scored only if they were clearly apparent in both replicates. As in the MSAP analysis, we disregarded differential bands involving C. arenosa products if they could also be interpreted as polymorphisms that were not inherited in the tested allotetraploids. In addition, we disregarded subtle changes in band intensity (see Fig. 5 for examples). Isolation and sequencing of cDNA-AFLP products was performed as described for MSAP fragments above. The data in Table V were subjected to Fisher Exact Test for independence (http://home.clara.net/sisa/fisher.htm) by pair-wise comparison in a 2 × 2 table format. The two-tailed probability was computed using the “sum of small P values” method.
ACKNOWLEDGMENTS
We thank Yvonne Stevens for help with the azadC treatment and Nicole Riddle for providing her methylcytosine thin-layer chromatography analysis protocol. We thank our collaborators researching the functional genomics of plant polyploids, and J. Chris Pires for stimulating discussions and comments.
Footnotes
This work was supported in part by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant to L.C.) and by the National Science Foundation Plant Genome Research Program (grant no. NSF 0077774).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003095.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bachem CWB, van der Hoefen RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RGF. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 1996;9:745–753. doi: 10.1046/j.1365-313x.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- Cedar H, Solage A, Glaser G, Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6:2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Comai L, Pikaard CS. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsisallopolyploids. Proc Natl Acad Sci USA. 1998;95:14891–14896. doi: 10.1073/pnas.95.25.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. Genetic and epigenetic interactions in allopolyploid plants. Plant Mol Biol. 2000;43:387–399. doi: 10.1023/a:1006480722854. [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B. Phenotypic instability and rapid gene silencing in newly formed Arabidopsisallotetraploids. Plant Cell. 2000;12:1551–1568. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditt RF, Nester EW, Comai L. Plant gene expression response to Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2001;98:10954–10959. doi: 10.1073/pnas.191383498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Meyerowitz EM. Hypermethylated SUPERMANepigenetic alleles in Arabidopsis. Science. 1997;277:1100–1103. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- Lee HS, Chen ZJ. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci USA. 2001;98:6753–6758. doi: 10.1073/pnas.121064698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff B, Pawlowski L, Bender J. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol Cell. 1999;3:505–511. doi: 10.1016/s1097-2765(00)80478-5. [DOI] [PubMed] [Google Scholar]
- Martienssen R. Transposons, DNA methylation and gene control. Trends Genet. 1998;14:263–264. doi: 10.1016/s0168-9525(98)01518-2. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Estelle MA, Somerville CR. A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet. 1986;204:417–423. [Google Scholar]
- Muskens MW, Vissers AP, Mol JN, Kooter JM. Role of inverted DNA repeats in transcriptional and posttranscriptional gene silencing. Plant Mol Biol. 2000;43:243–260. doi: 10.1023/a:1006491613768. [DOI] [PubMed] [Google Scholar]
- Ozkan H, Levy A, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS. Nucleolar dominance and silencing of transcription. Trends Plant Sci. 1999;4:478–483. doi: 10.1016/s1360-1385(99)01501-0. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Macelis D. REBASE-restriction enzymes and methylases. Nucleic Acids Res. 2001;29:268–269. doi: 10.1093/nar/29.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round EK, Flowers SK, Richards EJ. Arabidopsis thalianacentromere regions: genetic map positions and repetitive DNA structure. Genome Res. 1997;7:1045–1053. doi: 10.1101/gr.7.11.1045. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Osborn TC. Novel flowering time variation in resynthesized polyploid Brassica napus. J Hered. 2000;91:242–246. doi: 10.1093/jhered/91.3.242. [DOI] [PubMed] [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell. 2001;13:1749–1759. doi: 10.1105/TPC.010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. The dynamic nature of polyploid genomes. Proc Natl Acad Sci USA. 1995;92:8089–8091. doi: 10.1073/pnas.92.18.8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa R, McCallum CM, Delrow J, Henikoff JG, van Steensel B, Henikoff S. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr Biol. 2002;12:65–68. doi: 10.1016/s0960-9822(01)00622-4. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thalianaDNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wassenegger M. RNA-directed DNA methylation. Plant Mol Biol. 2000;43:203–220. doi: 10.1023/a:1006479327881. [DOI] [PubMed] [Google Scholar]
- White SE, Habera LF, Wessler SR. Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proc Natl Acad Sci USA. 1994;91:11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong LZ, Xu CG, Saghai Maroof MA, Zhang Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet. 1999;261:439–446. doi: 10.1007/s004380050986. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Magill CW, Magill JM, Newton RJ. An apparent case of nonsymmetrical and sustained strand-specific hemimethylation in the Dc8 gene of carrot. Genome. 1998;41:23–33. doi: 10.1139/g97-100. [DOI] [PubMed] [Google Scholar]