Abstract
Hendra virus (HeV) is a recently identified paramyxovirus that is fatal in humans and could be used as an agent of bioterrorism. The HeV receptor-binding protein (G) is required in order for the fusion protein (F) to mediate fusion, and analysis of the triggering/activation of HeV F by G should lead to strategies for interfering with this key step in viral entry. HeV F, once triggered by the receptor-bound G, by analogy with other paramyxovirus F proteins, undergoes multistep conformational changes leading to a six-helix bundle (6HB) structure that accomplishes fusion of the viral and cellular membranes. The ectodomain of paramyxovirus F proteins contains two conserved heptad repeat regions (HRN and HRC) near the fusion peptide and the transmembrane domains, respectively. Peptides derived from the HRN and HRC regions of F are proposed to inhibit fusion by preventing F, after the initial triggering step, from forming the 6HB structure that is required for fusion. HeV peptides have previously been found to be effective at inhibiting HeV fusion. However, we found that a human parainfluenza virus 3 F-peptide is more effective at inhibiting HeV fusion than the comparable HeV-derived peptide.
Hendra virus (HeV) is a zoonotic paramyxovirus that emerged in Australia, causing fatalities in both horses and humans. It is closely related to Nipah virus (NiV), which infects pigs and has caused outbreaks of severe encephalitis in humans in Singapore, Malaysia, and Bangladesh. Together, these two viruses make up a new genus within the Paramyxovirinae, called Henipavirus (45, 46). The study of these viruses has been designated as a priority of the NIAID Biodefense Research Agenda, based on their virulence and transmissibility and their potential for use as agents of bioterrorism.
At the onset of infection, the HeV virion binds to the target cell via interaction of the viral receptor-binding molecule with receptor molecules on the cell surface. G, a type II membrane glycoprotein, serves the dual purpose of binding to the recently identified receptor, Ephrin-B2 (3, 32), and activating the viral fusion protein (F), leading to merger of the virus and host cell membranes. The viral nucleocapsid that is released into the cytoplasm after fusion contains the genome RNA in association with the viral nucleocapsid protein (NP). This RNA/protein complex is the template both for transcription and for replication of the genome RNA that is packaged into progeny virions. The six viral genes encode the two surface glycoproteins G and F, the matrix protein which is involved in assembly and budding, the RNA polymerase proteins (L and P), the nucleocapsid protein (NP) and, through alternative reading frames and RNA editing, one or more proteins that are expressed only in the infected cell (16).
The identification of Ephrin B2 as a cellular receptor for both HeV and NiV (3, 32), as well as the recent finding that Ephrin B3 can serve as an alternate NiV receptor (33), has shed light on several of the pathological features of the diseases caused by these viruses. Ephrins are ligands for the Eph family of receptor tyrosine kinases, and the signaling mediated by the Eph-Ephrin interaction is critical to a series of developmental pathways, including angiogenesis and axonal guidance, as well as to tumorigenesis. Ephrin B2 is expressed specifically on endothelial cells, neurons, and the smooth muscle cells surrounding arterioles, a distribution pattern that parallels the tropism of NiV and HeV diseases. Ephrin B3 is not expressed in the endothelium but rather in the central nervous system, notably in some locations where Ephrin B2 is lacking but NiV disease is manifested. Interaction of NiV and HeV glycoproteins with the Ephrin receptors provides a key target for antiviral development.
The HeV F glycoprotein, like the F from all paramyxoviruses, mediates fusion between the viral and host cell membranes during infection (21, 37). The paramyxovirus F protein forms a trimer during synthesis; for HeV, once F reaches the cell surface it is again internalized and then cleaved by cathepsin L, yielding a membrane-distal and a membrane-anchored subunit (27, 35). The carboxyl terminal of the membrane-anchored subunit of paramyxovirus F proteins is anchored to the viral membrane, while the newly exposed amino terminal contains the hydrophobic residues, termed the “fusion peptide,” that insert into target membranes during fusion, which occurs at neutral pH (reviewed in reference 15). Initially, the paramyxovirus fusion peptide lies deep within the hydrophobic core of the F protein. In order for the virion to come into close proximity with the target membrane, F must undergo an activation step exposing the fusion peptide. This general mechanism appears to apply to HeV, but details of this process, as well as the conformational changes that F must undergo, need to be further scrutinized.
Paramyxovirus fusion proteins belong to the group of “class I” fusion proteins (reviewed in reference 9) that also includes the influenza virus hemagglutinin protein, the human immunodeficiency virus (HIV) gp120 fusion protein, and the Ebola virus fusion protein. The trigger that initiates the series of conformational changes in F, leading to membrane merger, differs depending on the pathway that the virus uses to enter the cell and thus whether fusion occurs at the surface at neutral pH or in the endosome. The fusion process for paramyxoviruses occurs at the surface of the target cell, at neutral pH, as it does for HIV, for which the trigger is gp120's binding to receptor. We found that for the paramyxovirus human parainfluenza virus 3 (HPIV3), the F protein is activated when the adjacent receptor-binding protein (hemagglutinin-neuraminidase [HN]) binds to a sialic acid-containing receptor, permitting fusion to occur. Upon receptor binding, HN triggers F to fuse (29, 39). For this reason, both the receptor-binding and fusion proteins of HPIV3 must be present, and the receptor-binding protein must interact with its respective receptor, in order for fusion to occur (21, 30, 31, 39, 40). This has been shown to be true in general for paramyxoviruses (9), with some exceptions where F can undergo triggering without the respective receptor-binding protein, although the receptor-binding protein nonetheless serves to facilitate the process (24, 42, 43). Indeed, the requirement for the receptor-binding protein (G) to trigger F has been found to be true for HeV as well (4, 5).
It has been predicted that this process of paramyxovirus F activation after receptor binding requires a conformational change in the fusion protein, which would propel the fusion peptide from the core to the surface of F, generating a transient but necessary intermediate and allowing the fusion peptide to become inserted in the host cell membrane. The ectodomain of the membrane-anchored subunit of the F protein contains two hydrophobic domains, the fusion peptide, which inserts into the cellular target membrane during fusion, and the transmembrane-spanning domain. Each of these domains is adjacent to one of two conserved heptad repeat (HR) regions: the fusion peptide is adjacent to the N-terminal heptad repeat (HRN), and the transmembrane domain is adjacent to the C-terminal heptad repeat (HRC). These HR domains are α-helical and can oligomerize into coiled-coils composed of several α-helices. The transient intermediate of F that is anchored to both viral and cell membranes is thought to refold and assemble into a fusogenic six-helix bundle (6HB) structure as the HRN and HRC associate into a tight complex with N and C peptides aligned in an antiparallel arrangement. The refolding is thought to relocate the fusion peptide and TM anchor to the same side, pulling the viral and cell membranes into close proximity. The formation of a 6HB during this step would generate the free energy for the membranes to bend towards each other and serve as the driving force for membrane fusion (42, 52).
Peptides derived from the HRN and HRC regions of the F protein that interfere with fusion intermediates of F (22, 41, 51) are prime candidates for interfering with viral entry. The ability of HR peptides to interfere with the class I fusion process for HIV led to a clinically effective peptide inhibitor of HIV infection (T-20, or enfuvirtide) (12, 47, 48). The HIV gp160 attaches to cellular receptors via its gp120 subunit and mediates fusion via its gp41 subunit. HIV peptides corresponding to the HRC domain of gp41 block viral entry and are effective for treatment of HIV in humans; T-20 was the first synthetic HRC peptide approved for HIV treatment (20). Peptides derived from the HRN or HRC regions of paramyxovirus F proteins can also interfere with fusion intermediates of paramyxovirus F proteins (2, 22, 41, 51). The HRC peptide regions of a number of paramyxoviruses, including Sendai virus, measles virus (MeV), Newcastle disease virus (NDV), respiratory syncytial virus (RSV), and simian virus 5, can inhibit viral infectivity in vitro (17, 22, 41, 49, 51, 54, 55). It has been proposed that this inhibition occurs because the peptides bind to their complementary HR region and thereby prevent HRN and HRC from refolding into the 6HB stable structure required for fusion (2, 9, 42). Inhibition of the fusion process, by peptides that interact with the HR regions of the activated F protein, is a highly promising area for development of antiviral therapies.
In the case of HIV, the fusion-inhibitory effect of the T-20 peptide has been recently shown to derive from interacting with multiple targets, not only preventing 6HB formation but also binding to gp120 in the region of coreceptor interaction (25). In order to design the best inhibitors, we must explore whether effective HR peptides interact directly with HRN sequences, with HRC sequences, or with other regions of F and which interaction determines the antientry effectiveness. Surprisingly, we have found that a heterologous HRC peptide is highly effective, more so than the homologous peptide, at inhibiting HeV fusion and entry.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were maintained in Eagle's minimal essential medium supplemented with 10% fetal bovine serum and antibiotics in 5% CO2. 293T (human kidney epithelial) and Vero (African green monkey kidney) cells were grown in Dulbecco's modified Eagle's medium (Cellgro, Mediatech) supplemented with 10% fetal bovine serum and antibiotics in 5% CO2. For NDV infections, a recombinant green fluorescent protein (GFP)-expressing NDV B1 (vaccine strain) virus (36) was obtained from Peter Palese. For quantitation of the effects of peptides on NDV viral entry, Vero cell monolayers grown in 24-well plates were incubated for 90 min with a multiplicity of infection (MOI) of 0.5 of GFP-expressing NDV B1 virus in medium containing various concentrations of inhibitors, as we have performed previously (38). The dishes were incubated at 37°C for 24 h, and fluorescent cells in the control and experimental wells were counted under a fluorescence microscope (Nikon Eclipse TE-2000-U) and photographed using a Photometrics CoolSnap CF camera. The effect of peptides on HPIV3 plaque number was assessed by a plaque reduction test performed as described previously (23). Briefly, Vero cell monolayers were inoculated with an MOI of 6 × 10−4 of HPIV3 in the presence of various concentrations of peptides. After 120 min, 2× minimal essential medium containing 10% fetal bovine serum was mixed with 1% agarose and added to the plates, which were then incubated at 37°C for 24 h. After removing the agarose overlay, the cells were immunostained for plaque detection. The numbers of plaques in the control (no peptide, scrambled peptide) and experimental wells were counted under a dissecting stereoscope.
Plasmids and reagents.
HeV wild-type (wt) G and wt F in pCAGGS were a gift from Lin-Fa Wang. HIV Tat in the pSV2 vector was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and was used to transfect target Vero cells for fusion assays. To generate the shortened cytoplasmic tail variant of HeV G (HeV G-CT32), an internal primer containing an EcoR1 site and initiating at position 32 of the open reading frame was used for nested PCR. The primer sequence was 5′ GGAATTCGGCACAATGGACATCAAG 3′. Soluble Fc-fusion ephrin-B2 protein was purchased from Sigma and was dissolved in 1× phosphate-buffered saline (PBS; Mediatech, Cellgro) to a concentration of 0.5 μg/μl. Stock solutions were stored at −20°C. Anti-HeV G antibodies were a gift from Benhur Lee.
Transient expression of G, F, luciferase, and tat genes.
Transfections were performed according to the Lipofectamine 2000 reagent manufacturer's protocols (Invitrogen).
Luminescence fusion assay.
A previously described luciferase reporter gene assay for cell fusion (43) was adapted for quantifying cell fusion promoted by HeV envelope proteins. Twenty-four-well plates of HeLa cells were transfected with LTR-luciferase plus HeV F and wt G or CT32 G. Vero cells transfected with pSV2-TAT (NIH AIDS Research and Reference Reagent Program) were added to the plates of Vero cells, with or without HRC peptides, and incubated at 37°C for 24 h. The cells were then washed, lysed (using luciferase lysis buffer [Promega]), and luciferase activity resulting from fusion of the two cell types was quantified using luciferase assay substrate (Promega) and a Spectromax M5 (Molecular Devices) luminescence microplate reader.
HR peptides.
Peptides were synthesized on a Symphony peptide synthesizer (Protein Technologies Inc., Massachusetts) by standard Fmoc/2-(1 H-benzo-triazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate methods, purified to homogeneity by high-performance liquid chromatography (Shimadzu Corp., Kyoto, Japan), and characterized with a matrix-assisted laser desorption ionization-time of flight mass spectrometer (Voyager DE; Applied Biosystems, Foster City, CA). Peptides were weighed and then completely dissolved in dimethyl sulfoxide to a final concentration of 5 mM, based on the molecular weight provided by the synthesizer. The sequences of the scrambled peptides that were used as controls were as follows: HPIV3 HRC 45, N terminal-RSIDLIWKVDATLELKISEASNKGPDNIKINLESQSLDNIEDNRS-C terminal; HeV HRC 42, N terminal-SDMIPASKLEVSIDTIPQYNLKVQNQSIQDPVSQSDYSKLTK-C terminal.
Pseudotyped virus infection assay.
VSV-ΔG-RFP is a recombinant vesicular stomatitis virus (VSV) derived from the cDNA of VSV Indiana, in which the G gene is replaced with the red fluorescent protein (RFP) gene. We obtained VSV-ΔG-RFP complemented with VSV-G from Michael Whitt (University of Tennessee Health Science Center and GTX Inc.). Pseudotypes with HeV F and G were generated as described elsewhere (32, 44). Briefly, 293T cells were transfected with either VSV-G (gift from M. Whitt), HeV-G CT32/F, HeV-G CT32, or HeV-F. At 24 h posttransfection, the dishes were washed and infected (MOI of 1) with VSV-ΔG-RFP complemented with VSV-G. Supernatant fluid containing pseudotyped virus (HeV F/CT32-G or VSV-G) was collected 18 h postinfection and stored at −80°C. For infection assays, HeV F/CT32-G or VSV-G pseudotypes were used at an MOI of 0.25 to infect Vero cells in the absence of serum. Peptides (HRN or HRC peptides derived from either HeV F or HPIV3 F), Ephrin B2, or anti-HeV G antibodies were added at various concentrations. RFP production at 36 h was analyzed by fluorescence microscopy (38) and fluorescence-activated cell sorter (FACS; Becton Dickinson FACSCalibur) analysis.
Computational methods.
Our computational methodology consists of model building and hydropathic analysis. The available crystallographic structure of the HPIV3 F protein in its postfusion conformation (pdb 1ZTM, resolution of 3.05 Å) (53) and that of the Hendra virus fusion core (pdb 1WP8, resolution of 2.2 Å) (50) were used to build models for the interactions between the HRC and HRN regions of the fusion proteins. The Sybyl molecular modeling suite of programs (version 7.1; www.tripos.com) was used for model building and optimization and for visual inspection. The N and C termini were added and modeled as charged; hydrogen atoms, generally not present in the pdb files, were added and minimized. The case of HPIV3 F presents several modeling limitations, i.e., 100 residues have side chains with missing atoms. These were fixed with the Biopolymer menu in Sybyl, and the side chains of the whole protein were allowed to relax with energy minimization to an energy gradient of 0.005 kcal mol−1 Å−1. The default parameters were used, except that the distance-dependent dielectric was set to 4 due to the solvent accessibility of the protein. The mutants were built from the optimized crystallographic wild-type structure, and the regions around the mutation (“hot” radius of 4 Å) were allowed to relax with energy minimization. The models for the interaction between HRC from HPIV3 and HRN from Hendra virus were built from the available crystallographic structures, superimposing the HRC peptide from HPIV3 on HRC from Hendra virus. The HPIV3 HRC peptide was then manually docked, and the resulting HRC peptide-HRN complex was energy minimized with the same methods and parameters used earlier.
The hydropathic analysis, i.e., log P calculation and evaluation of the binding energies, was performed with the HINT (hydropathic interactions) software (www.tripos.com). In this work, HINT version 3.10S, which incorporates new local modifications, was used (1, 19). This software, which is based on experimentally measured solvent partitioning coefficients to calculate free energies of binding (10, 14, 18), has been used to evaluate the interaction between the HRC and HRN regions of the fusion protein. For the hydrogens, the “essential” option, which only considers explicitly polar hydrogens, was chosen, while the solvent-accessible surface area for protein backbone nitrogens was corrected with the “+20” option. In previous work (7, 10, 14, 18), we have shown that about −500 HINT score units correlates with 1.0 kcal mol−1. This relationship was used in reporting relative free energies in this work.
RESULTS
Peptides corresponding to the HRC region of HeV and HPIV3 F inhibit HeV G/F-mediated cell fusion.
In order to predict the location of the two coiled-coil regions on HeV and HPIV3 F, HRC, and HRN, we used the program Learncoil along with information from previous reports (4, 13, 22). These domains are found on opposite ends of the F protein and are separated by almost 300 amino acids (aa). Using the predicted coiled-coil regions, we synthesized corresponding peptides, whose sequences are presented in Table 1.
TABLE 1.
Peptide sequences used in this studya
Derived from the HRC regions of HeV F and HPIV3 F. The heptad repeat domains are shown in green. Mutations are shown in red in the left column and are also indicated in red in the peptide sequence. The HPIV3-derived peptides are ordered according to their IC50 for HeV-pseudotyped virion entry (shown in blue).
For experiments assessing the potency of HR peptides at inhibiting HeV fusion, we generated a mutated HeV G, with a shortened cytoplasmic tail, that is highly effective at activating F (a manuscript in preparation details the properties of this mutant). The pairing of this mutated G with HeV F results in enhanced fusion and thereby allows for amplification of differences in potency between inhibitory compounds. The complete cytoplasmic tail (CT) of HeV G, including the transmembrane domain, consists of 46 amino acids and contains three methionine residues, each of which is a potential initiation site. The CT of the mutant G that we generated initiates at the third methionine, so that 32 amino acids are lost and only 14 remain, with the resulting “G-CT32” activating F to mediate fusion much more effectively than HeV G-wt. We hypothesized that differences in potency of inhibitors would be more evident in the attempt to block F-mediated fusion in cells expressing G-CT32 than in cells expressing G-wt.
The efficacy of the HR peptides was first tested using a luciferase reporter-gene fusion assay (43) that we adapted for quantifying fusion mediated by the envelope proteins of HeV. HeLa cells coexpressing HeV F with G-wt or G-CT32 were transfected with a vector that contains the luciferase gene under the control of an HIV-LTR promoter and were overlaid with cells that express HIV-Tat. Thus, luciferase expression only occurs when cells containing Tat fuse with those transfected with luciferase. Fusion in the presence or absence of synthetic peptides, added together with the overlay cells, was measured after incubation at 37°C for 24 h. Addition of peptides together with the overlay cells ensured that the peptide was present during the entire process and might interrupt either triggering of the fusion protein or fusion itself.
Figure 1 shows the quantitation of fusion mediated by HeV F coexpressed with HeV G-wt (Fig. 1A), HeV G-CT32 (Fig. 1B), HPIV3 F coexpressed with HPIV3 HN (Fig. 1C), or NDV-AV F coexpressed with NDV-AV HN (Fig. 1D) at 24 h, in the presence of increasing concentrations of HPIV3 HRC peptide or HeV HRC peptide. Fusion is quantitated between viral envelope glycoprotein (HeV G/F, HPIV3 HN/F, or NDV-AV HN/F)/HIV-LTR-luciferase expressing cells and HIV-Tat-expressing cells, and the results are expressed as percent inhibition of fusion at 24 h (y axis) (compared to control cells) by different peptide concentrations (x axis). It may be seen from Fig. 1A that fusion mediated by the HeV glycoproteins F and G-wt was decreased by HeV-HRC in a dose-dependent manner; significant inhibition was caused by 0.01 μM peptide, with about 0.1 μM required for 50% inhibition. Surprisingly, HPIV3-HRC inhibited fusion more effectively than the homotypic HeV HRC (Fig. 1A). The concentrations of the heterotypic HPIV3 HRC that inhibited fusion by 75% and by 100% were 0.1 and 1.0 μM, while 100% inhibition by HeV HRC required a 10 μM concentration of peptide.
FIG. 1.
Inhibition of cell fusion mediated by HeV wt G/F, HeV CT32/F, HPIV3 HN/F, and NDV AV HN/F by HRC peptides derived from HeV F and HPIV3 F. The reporter gene assay quantitates fusion between target Vero cells that express HIV Tat (which activates LTR-luciferase production) and effector HeLa cells transfected with luciferase and either HeV wt G/F (A), HeV CT32/F (B), HPIV3 HN/F (C), or NDV AV HN/F (D). The target and effector cells were combined along with different concentrations of the HeV F- or HPIV3 F-derived HRC peptides. Luminescence was measured after 24 h at 37°C. The x axis shows the concentration of HRC peptide derived from HeV F or HPIV3 F. The results are expressed as percent inhibition of fusion compared to cells that were not treated with peptide (y axis). The values are means (± standard deviations) of results from at least three experiments.
Figure 1B shows that cell fusion mediated by HeV F coexpressed with the variant G-CT32 was also inhibited by both HRC peptides and that, at each concentration, the HPIV3 HRC peptide was more inhibitory than the homotypic HeV peptide. The difference in effectiveness is apparent at each concentration of the HeV or HPIV3 HRC peptide. A 0.1 μM concentration of the HPIV3 peptide completely inhibited fusion in cells expressing the variant G-CT32 protein, while the HeV peptide only achieved 50% inhibition at this concentration. The differences between effectiveness of the HPIV3 and HeV peptides are apparent at the 0.01 μM and 0.1 μM concentrations. Figure 1C shows that only the HPIV3 HRC peptide inhibits fusion mediated by the HPIV3 glycoproteins, although at higher concentrations than those required for inhibition of HeV. Specificity is indicated by the lack of inhibitory activity versus NDV; as seen in Fig. 1D, fusion mediated by NDV-AV glycoproteins is not affected by either HRC peptide. In this and every subsequent experiment, a scrambled peptide (see Materials and Methods) was used as a negative control and was ineffective (data not shown).
These experiments demonstrated that the HPIV3 HRC peptide not only inhibited HeV G/F-mediated cell fusion but also was far more effective than the homotypic peptide derived from HeV F itself. The differences between the two peptides in inhibition of fusion in cells coexpressing F with wt G (Fig. 1A) or G-CT32 (Fig. 1B) confirm our suggestion that the advantage of the stronger inhibitor is more evident when HeV F is coexpressed with the highly fusion-activating G-CT32 variant than with wt G. The result is specific because NDV fusion is inhibited by neither HPIV3 nor HeV peptides, and HPIV3 fusion is inhibited only by the homotypic peptide. These findings led us to further explore the potential of HPIV3 HRC peptides as inhibitors and to investigate the properties of these peptides that make them effective against HeV.
Shorter peptides are effective for inhibition of HeV F-mediated cell fusion.
Since both the HeV and HPIV3 HRC peptides inhibited HeV fusion, we hypothesized that their action might be attributable to homologous segments and that the nonhomologous segments might be eliminated without loss of function. Shorter inhibitory peptides may be better potential therapeutic candidates, as well as providing useful tools for studying the mechanisms that regulate conformational changes in the activated F protein. While peptides of 45 aa in length would be unwieldy and expensive as a treatment tactic, shorter but equally effective peptides could be therapeutic agents. In order to determine which sections of the peptides are critical for inhibition, we aligned the sequences of the HPIV3 HRC 45-mer and HeV HRC 42-mer peptides using SIM (http://us.expasy.org) to detect homologous regions. The best alignment obtained using the BLOSUM62 matrix was a 36-amino-acid segment in the two peptides showing 30.6% sequence identity. Using the SIM prediction, the two HRC 36-aa peptides were then synthesized (for sequences, see Table 1) and compared with the full-length HRCs.
Inhibition of fusion by cells coexpressing HeV F and G-CT32 was used to evaluate the HeV and HPIV3 HRC 36-mers alongside their longer counterparts. The use of the variant G in these experiments allowed for amplification of differences in potency between individual peptides.
Figure 2 contains the results of experiments using HeV and HPIV3 peptides to inhibit fusion mediated by HeV glycoproteins (Fig. 2A). The effects of the different peptide concentrations (x axis; log scale) are expressed as the percentage of inhibition of cell fusion compared to untreated control cells (y axis). For inhibition of HeV glycoprotein-mediated fusion, both 36-aa peptides retained inhibitory powers equal to that of their longer counterparts. Any differences between HPIV3 HRC 45 and 36 and HeV HRC 42 and 36 were insignificant at all concentrations. The difference between the HPIV3- and HeV-derived peptides in inhibiting HeV fusion, however, is clear along the entire concentration range. HPIV3 HRC peptides inhibit much more effectively than HeV HRC peptides; this is most apparent at the lower concentrations of peptide, for example, at 0.005 μM (5 nM), where inhibition by HPIV3 peptides is over 40% and HeV peptides inhibit fusion by only about 12%. Figure 2B shows that the shorter HPIV3 HRC peptide is equally effective at inhibiting fusion mediated by HPIV3 glycoproteins and confirms that the HeV peptides have no heterotypic inhibitory effect on HPIV3.
FIG. 2.
Fusion inhibition by shortened HeV F- and HPIV3 F-derived HRC peptides. The effect of HRC peptides derived from HeV F or HPIV3 F on fusion by luciferase and HeV CT32/F-expressing cells (A) or HPIV3 HN/F-expressing cells (B) was determined as described for Fig. 1. The results are shown as percent inhibition of fusion compared to cells not treated with peptide (y axis) as a function of the concentration (x axis, log scale) of full-length peptides (black symbols) or 36-residue peptides (white symbols) present during the 24-h incubation. The values are means (± standard deviations of the means) of results from at least three experiments.
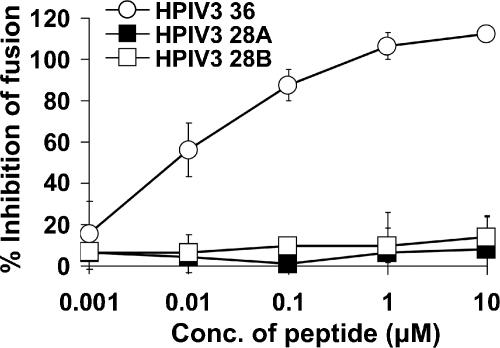
The more effective HPIV3 36-aa HRC peptide was then used to examine the consequences of removing another 8 amino acids (Table 1). Truncation at the C terminus of this HRC peptide yielded peptide 28A, and the product of truncation at the N terminus of the molecule was called peptide 28B. The two 28-mers were tested for their effects on the fusion of cells transfected with HeV or HPIV3 envelope proteins. Figure 3 contains the results of experiments using HPIV3 peptides to inhibit fusion mediated by HeV CT32 G/F. The effects of the different peptide concentrations (x axis; log scale) are expressed as the percentage of inhibition of cell fusion compared to untreated control cells (y axis). Neither of the two truncated HPIV3 HRC 28-aa peptides significantly inhibited fusion by HeV G-CT32/F-coexpressing cells (Fig. 3) or by HPIV3 HN/F-coexpressing cells (data not shown), whereas the corresponding HPIV3 36-aa HRC peptide prevented fusion of cells expressing either set of proteins. The 50% inhibitory concentration (IC50) values are also shown in Table 1. These results suggest that both 8-amino-acid segments, or parts of them, that were removed from HPIV3 HRC-36 contain residues that are essential for fusion-inhibitory action.
FIG. 3.
Fusion inhibition by 36-residue and 28-residue HPIV3 HRC peptides. The effect of HRC peptides derived from HPIV3 F on fusion by HeV CT32/F/luciferase-expressing cells was determined as described for Fig. 1. The results are shown as percent inhibition of fusion compared to cells not treated with peptide (y axis) as a function of the concentration (x axis, log scale) of HPIV3 36-residue peptide, HPIV3 28-residue peptide A, or HPIV3 28-residue peptide B present during the 24-h incubation. The values are means (± standard deviations of the means) of results from at least three experiments.
Mutation of the effective HPIV3 HRC peptide at residues predicted to increase interaction with the HPIV3 HRN domain sequences results in loss of inhibitory activity versus HeV.
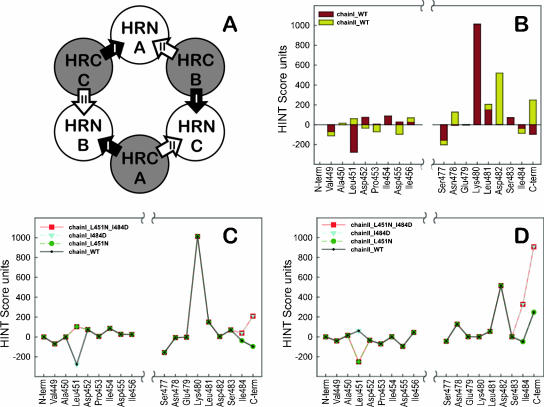
We undertook to determine whether rational modeling of the interaction between the HPIV3 fusion protein and its derived peptide could be used to design more effective peptide inhibitors. Starting with the crystal structure of the uncleaved paramyxovirus (HPIV3) fusion protein (pdb code 1ztm) (53), we analyzed the interactions between the HRN and HRC domains. Each of the three HRCs has two different “faces” with respect to HRNs. We called these “chain I” and “chain II” (Fig. 4A). Although the trimer is itself symmetric, because this crystal structure is not required by its crystallographic space group to be symmetric, there were noticeable structural differences, including different missing atoms among the three instances of each HRC domain (and their chain I and chain II interactions). Thus, each of our calculations was an average over all three HRCs. Figure 4B illustrates the interactions for the “required segments” (residues 449 to 456 and 477 to 484) of an “average” HRC with its chain I and chain II neighbors, on a residue-by-residue basis. These interactions were scored with the HINT program (18), a software tool that analyzes with equal efficacy polar and apolar interactions and calculates scores that can be correlated with free energies of binding (10, 14, 18). Within the required segment ends of the peptide we selected Leu451, which has a quite unfavorable interaction with chain I, and Ile484, which has unfavorable interactions with both chain I and chain II, as possible sites for mutation.
FIG. 4.
Selection of mutation sites for the HPIV3 HRC-derived peptide. (A) Definition of chain I and chain II with respect to HRN. Since each HRC presents two different faces to neighboring HRN chains in the six-helix bundle, we define the clockwise neighbor as chain I and the counterclockwise neighbor as chain II. (B) The HINT score interactions for each residue of the required segments of the peptide with both chain I and chain II HRNs (average of each of the three HRCs in the 1ztm crystal structure as they interact with the HRN chains [I and II]). Favorable interactions have positive HINT scores, while unfavorable interactions have negative scores. From this analysis, mutations at two sites, Leu451 and Ile484, were chosen for further investigation. (C) Comparison of residue-by-residue HINT scores for the wild-type (solid line, purple diamonds), I484D mutant (blue triangles), L451N mutant (green circles), and double mutant (red squares) peptides interacting with chain I. (D) Comparison of residue-by-residue HINT scores for the wild-type (solid line, purple diamonds), I484D mutant (blue triangles), L451N mutant (green circles), and double mutant (red squares) peptides interacting with chain II.
In silico calculations of these two residues using a palette of putative mutations, again using HINT scoring, suggested that the I484D and L451N mutated peptides, and also the double mutant, may have the best profiles for improved binding with chain I and chain II. The resulting score profiles of these mutants are illustrated in Fig. 4C for chain I and in Fig. 4D for chain II. It should be noted that the structure optimization procedure (Sybyl, using the Tripos force field [8]) used to relax the molecular models after mutation overrelaxes the C-terminal end of the HRC, which is manifested as unrealistically favorable interactions between the terminal residue and chain I and chain II. This tends to obscure the actual effects of the mutation at 484.
These “best-case” mutations would be expected to bind somewhat better to HPIV3 F, but not substantially so. The calculations indicated an improvement of only 0.2 to 0.5 kcal/mol in binding energy for the mutants versus wild-type peptides. This was confirmed in that the mutants were similar or slightly better in inhibiting HPIV3 fusion (see data in Fig. 5). However, the mutants have significantly different properties in that both mutations were replacements of hydrophobic residues with polar residues within the required segments. Thus, they are excellent probes for examining the unique phenomenon of HPIV3-derived peptides inhibiting Hendra virus.
FIG. 5.
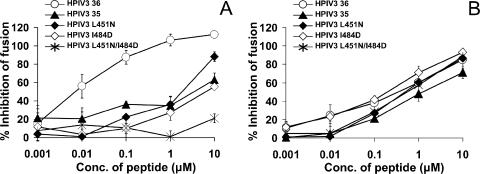
Fusion inhibition by HPIV3 HRC peptides mutated at residues predicted to increase interaction with the homotypic HRN domain. The effect of HRC peptides derived from HPIV3 HRC on fusion by luciferase and HeV CT32/F-expressing cells (A) or HPIV3 HN/F-expressing cells (B) was determined as described for Fig. 1. The results are shown as percent inhibition of fusion compared to cells not treated with peptide (y axis) as a function of the concentration (x axis, log scale) of HPIV3 HRC 36-wt, HPIV3 HRC 36-L451N, HPIV3 HRC 36-I484D, double mutant HPIV3 HRC 36-L451N/I484D, or HPIV3 HRC 35 (22) present during the 24-h incubation. The values are means (± standard deviations of the means) of results from at least three experiments.
In the experiments shown in Fig. 5, we tested these mutated peptides for inhibition of HeV glycoprotein-mediated fusion (Fig. 5A) and HPIV3 glycoprotein-mediated fusion (Fig. 5B). We also tested for inhibition of NDV-mediated fusion (data not shown). Also included in the panel of peptides was a previously described 35-residue HPIV3 peptide (22). This peptide (Table 1) is shifted towards the C terminus and lacks the first 5 residues of our 36-residue HPIV3 HRC peptide. For HeV-mediated fusion (Fig. 5A), the single-residue mutations decrease activity and the double mutation abolishes inhibitory activity. The 35-residue peptide lacking the N terminus of our peptide has a markedly decreased inhibitory activity. In the case of HPIV3 glycoprotein-mediated fusion (Fig. 5B), the mutated peptides inhibit with similar efficiency compared to the original peptides. Specificity is indicated by the complete lack of activity against NDV glycoprotein-mediated fusion (data not shown). Thus, the mutations at the N and C termini of the HRC inhibitory peptide abolish heterotypic inhibition of HeV glycoprotein-mediated fusion by HPIV3 peptides, while homotypic inhibition is maintained.
HRC peptides inhibit entry of VSV pseudotyped with HeV envelope proteins and inhibit entry of HPIV3 virions.
In order to test the effectiveness of each peptide in an assay that more closely mimicked the conditions of viral infection, we set up a virion-based infection assay for HeV that allowed use of the G-CT32 protein. HeV glycoproteins were pseudotyped onto a recombinant VSV that expresses RFP but lacks VSV-G. The resulting pseudotyped virus (VSV-ΔG-RFP-HeV G-CT32/F) contains the binding and fusion proteins from HeV. Entry of the pseudotyped virus into the target cells in the absence and presence of the HRC peptides was quantified by assessing red fluorescence through FACS analysis (Becton Dickinson FACSCalibur). For comparison, we tested infection with HPIV3 virions.
The results of these experiments are shown in Fig. 6. At the same time that cells were infected with the various pseudotyped viruses or viruses, peptide inhibitors were added at different concentrations and then incubated for 36 h. The effects of the indicated agents on viral entry are expressed as percent inhibition of viral entry compared to cells infected in the absence of peptide (y axis). HRC peptides derived from both viruses avidly inhibited entry of HeV pseudotypes (Fig. 6A). At a 1 μM concentration of all the peptides, viral entry was reduced by about 95% for all four peptides. However, at the lower concentrations, a difference can be seen between the HeV HRC peptides and HPIV3 HRC peptides. The difference is clear at a concentration of 0.1 μM, where the HeV peptides only inhibit viral entry by about 60% while the HPIV3 peptides caused about 90% inhibition. The IC50 of the HPIV3 peptides—approximately 0.02 μM (20 nM)—is more than three times lower than that of the HeV peptides. In agreement with Fig. 2, the 36-residue peptides inhibit viral entry at the same rate as their longer counterparts; no inhibitory action is lost with the shortening of these peptides. This encouraging result suggests that the HPIV3 HRC peptide might be effective in inhibiting viral infection at nanomolar concentrations. In order to verify that peptide inhibition occurs via a mechanism involving the HeV surface glycoproteins, the experiment was repeated using VSV-G-RFP, which contains the surface glycoproteins from VSV itself (data not shown). These control experiments showed that the infectivity of VSV-G-RFP was unaffected by HPIV3 or HeV HRC peptides and suggested that inhibition of entry of the HeV-pseudotyped virus, VSV-ΔG-RFP-HeV G-CT32/F, is attributable to interference with HeV envelope protein function. Since VSV-G requires endocytosis for infection, we performed experiments with other viruses whose fusion proteins mediate fusion at neutral pH to confirm the specificity of the observed HeV inhibition. Figure 6B shows that while HRC peptides derived from HPIV3 inhibit entry of HPIV3 virions, the peptides derived from HeV do not inhibit HPIV3 entry. In addition, infection with NDV-B1-GFP was not affected by addition of any of the peptides (data not shown). Anti-HeV G antiserum and soluble EphB2, which competes with the cell membrane-bound EphB2 receptor to bind G, inhibited viral entry as expected (data not shown).
FIG. 6.
Inhibition by HPIV3 HRC peptides of infection with HeV G-CT32/F-pseudotyped VSV or infection with HPIV3 virions. (A) Vero cells were infected with pseudotyped VSV-ΔG-RFP-HeV G-CT32/F virus at an MOI of 0.25 in the presence of peptides derived from HeV F or HPIV3 F. At 36 h after infection, the number of fluorescent (infected) cells was determined using FACS analysis. (B) Vero cells were infected with HPIV3 at an MOI of 6 × 10−4 in the presence of these peptides, overlaid with agarose at 120 min, and stained at 24 h postinfection. The results (y axis) are shown as percent inhibition of viral entry (compared to control cells that were infected without inhibitor) as a function of the concentration (x axis, log scale) of full-length (black symbols) or 36-residue (white symbols) peptides. Values are means (± standard deviations) of results from five (HeV) or three (HPIV3) experiments.
Mutation of the effective HPIV3 HRC peptide at residues predicted to increase interaction with the HPIV3 HRN domain sequences results in loss of inhibitory activity versus HeV-pseudotyped viral entry.
In the experiments shown in Fig. 7, we tested the mutated peptides that were described in Fig. 5 (see also Table 1) for their effects on entry of the pseudotyped virus (VSV-ΔG-RFP-HeV G-CT32/F) that contains the binding and fusion proteins from HeV (Fig. 7A). These experiments were performed as for those in Fig. 6, and entry into the target cells in the absence and presence of the original and mutated HRC peptides was quantified by assessing red fluorescence through FACS analysis (Becton Dickinson FACSCalibur). The results are in accord with the findings on inhibition of HeV glycoprotein-mediated fusion (Fig. 5) and indicate the importance of those residues for heterotypic interaction. Figure 7B shows that, in contrast, the mutations in the HPIV3 peptides do not alter their effectiveness at inhibiting HPIV3 entry. For HeV inhibition, mutation at both ends of the peptide abolished inhibitory effectiveness at the lower concentrations tested; at higher concentrations this doubly mutated peptide appears to somewhat enhance entry of the HeV pseudotypes, and this phenomenon is being explored.
FIG. 7.
Inhibition of infection with HeV G-CT32/F-pseudotyped VSV or infection with HPIV3 by HPIV3 F HRC peptides mutated at residues predicted to increase interaction with the homotypic HRN domain. Vero cells were infected with either pseudotyped VSV-ΔG-RFP-HeV G-CT32/F virus (A) or HPIV3 (B) as described for Fig. 6, in the presence of peptides derived from HPIV3 F. The results (y axis) are shown as percent inhibition of viral entry (compared to control cells that were infected without inhibitor) as a function of the concentration (x axis, log scale) of HPIV3 HRC 36-wt, HPIV3 HRC 36-L451N, HPIV3 HRC 36-I484D, double mutant HPIV3 HRC 36-L451N/I484D, or HPIV3 HRC 35 (22). The values are means (± standard deviations of the means) of results from five (HeV) or three (HPIV3) experiments.
Modeling and analysis of the effective HPIV3 36-residue HRC peptide compared to ineffective mutated peptides in the context of the HeV HRC structure.
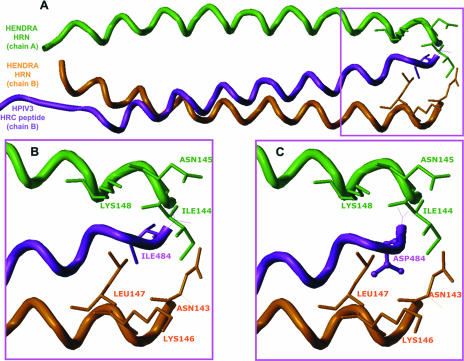
Without experimental structural data on how the HPIV3-derived peptide might bind to the Hendra virus fusion protein, it is difficult to develop a comprehensive mechanism for this inhibition process. There are many ways in which the HPIV3 HRC peptide might bind to the Hendra virus fusion protein and interrupt its fusion, and it is likely to be a combination of more than one contributing pathway that yields the overall mechanism. Nevertheless, by analogy to the HPIV3 HRC peptide inhibiting HPIV3 fusion, we developed molecular models of the HPIV3 HRC peptide as it might bind to the Hendra virus fusion protein and inhibit its association with other monomers. Figure 8 illustrates one such model, in which the HPIV3 HRC peptide was aligned with the Hendra virus HRN chains by assuming that the C terminus of the peptide is approximately in register with the N termini of the HRN chains. Figure 8A shows the overall alignment of the peptide and two of the Hendra virus HRN chains; Fig. 8B shows the region of Ile484, illustrating a close and favorable hydrophobic interaction with the Ile144 of HeV; mutation of Ile484 to Asp484 (Fig. 8C) disrupts the favorable interaction by placing the mutant polar Asp side chain in a hydrophobic region. Our calculations indicate that this mutant HeV HRN complex would be about 1 kcal/mol less stable than the wt HeV complex. Similar loss of effectiveness at the second mutation site, as seen with the double mutant peptide, would yield an almost-2-orders-of-magnitude loss of binding efficacy. Since both of the site mutations from hydrophobic to polar residues were coupled with loss of inhibition efficacy, this suggests that the original interactions between the peptide and the HeV chains are tight. Mutations of other types, e.g., acid for base, hydrophobic for polar, etc., could be assumed to be associated with loss of hydrogen bonds and display concomitantly dramatic effects.
FIG. 8.
Possible molecular model depicting the interaction of HPIV3 HRC-derived peptides interacting with the HeV HRN chain. This model was built by assuming that the C terminus of the HPIV3 HRC-derived peptide would be in register with the N terminus of the complementary HeV HRN chains of the F monomer when bound. (A) Overall chain trace of the peptide (purple) and protein (green and orange) backbones. Chain A and chain B are as described in the legend for Fig. 4. (B) The peptide C-terminal region, paying particular attention to the peptide Ile484 residue. This residue makes a favorable hydrophobic contact with the Ile144 residue of chain A (green) or the Leu147 of chain B (orange). (C) The peptide C-terminal region, after mutation of Ile484 to Asp. The favorable hydrophobic interactions seen between the wild-type peptide and the HeV HRN chains are lost and replaced by repulsive hydrophobic polar interactions.
The peptide might, in fact, interact with the HRC chains of Hendra virus or bridge between two chains and act like a “strut” to hold them apart and inhibit fusion. Various other mechanistic/structural possibilities cannot yet be ruled out. Further experimental data will be required before definitive molecular models can be made regarding this mixed-species fusion inhibition.
DISCUSSION
The possibility of inhibiting paramyxovirus fusion with HRC peptides has been raised (22), and various peptides have been proposed as candidate therapeutic agents. HRC peptides have generally been the peptides discussed in this context, since they are more potent than HRN peptides because they acquire less coiled-coil structure in solution and therefore aggregate less (22, 26, 47). Our experiments yielded the result that 45-residue or 36-residue peptides derived from the HRC region of HPIV3 F are highly effective inhibitors of HeV infection and far more effective than the published HeV peptide inhibitor (4, 5). A recent report (6) showed that a shorter (36-mer) modified NiV peptide inhibited HeV fusion at similar concentrations to those we report here for the 36-mer HeV peptide; however, the period during which fusion inhibition was assessed was 10-fold shorter (2.5 h compared to 24 h). We suggest that our assay methods—a 24-h incubation in medium that contains serum—provide a test of effectiveness that may be more reflective of potential clinical utility than the shorter fusion assays used in other studies; such fusion assays with a shorter overlay time may tend to favor peptides that might not be the best clinical choices. Most importantly, under identical conditions, the HPIV3-derived peptide reported here is more effective, with a lower IC50 when tested under identical conditions in our assays, than previously reported peptides. The HPIV3 peptide we report here is more effective based upon several criteria: it is more effective than the HeV peptide at inhibiting fusion mediated by HeV F in concert with the fusogenic variant G-CT32, and it is more effective at inhibiting entry of pseudotyped virus with G-CT32. The HPIV3 peptide also retains effectiveness for at least a 48-h incubation period.
Removal of 8 residues from either terminus of the effective HPIV3 36-residue peptide results in loss of activity, as shown in Fig. 3. While this may suggest that length is critical, we propose instead that specific important interactions occur at these termini. While it has been proposed that HRC peptides work by preventing the F protein from forming the 6HB (HRN-HRC interaction) required for fusion, other mechanisms, for example, peptide interaction with another region of F, have not been ruled out. HRC peptides have been generally proposed to act by interacting with the corresponding HRN domain (13, 17, 28), and in fact the interaction of HRN and HRC domains has been used to map the sequential stages of fusion (42). In the case of HIV, the fusion-inhibitory effect of the T-20 peptide has recently been shown to derive from interaction with multiple targets, not only preventing 6HB formation but also binding to gp120 in the region of coreceptor interaction (25).
Our experimental results, combined with computational modeling of the possible interactions of the HPIV3 peptide with the 6HB region of the HeV F protein, suggest a distinct balance of structural features on the HPIV3 HRC peptide that render it so effective in inhibiting Hendra virus fusion. As predicted by modeling, the two fairly minor mutations on this peptide had a relatively small effect on the peptide's ability to inhibit HPIV3 fusion. This concordance gives us some confidence that the mechanism relying on the HRC peptides binding to the fusion protein HRN chains has at least partial validity and that it can be modeled when enough experimental structural data are available. While the fact that the two ends (8-mers) are so critical in maintaining efficacy of the peptides as inhibitors may suggest that length is important, a more intriguing hypothesis is that they are positioning and holding in place the middle of the peptide sequence. Figure 9 illustrates these concepts in terms of a simple model, with a proposed mechanism for the inhibition of HeV fusion protein by the native HPIV3 HRC-derived 36-mer peptide. Figure 9A shows fusion inhibition for HPIV3 with the HPIV3 HRC-derived peptide. The structural/functional features of the residue side chains, e.g., hydrogen bond donor, hydrogen bond acceptor, and hydrophobic, are indicated as shapes. In this model, shape complementarity yields stronger interactions between the peptide and the HRN chain. In general, the shape match is good, particularly within the required segments, but suggests that mutations of the peptide might improve several properties. Figure 9B shows inhibition of the HeV F protein with the HPIV3 HRC peptide, indicating how that peptide may bind even more strongly to chains of HeV, particularly within the required segments. The shape match between the peptide and the HeV HRN (or possibly HRC) is very good within the required segments, allowing tighter binding of the peptide ends to a chain of the HeV F. This complex does not allow subsequent binding of another monomer, thus blocking fusion. A mutation to this peptide that would improve or not affect its binding to HPIV3 might, in contrast, significantly compromise its binding to HeV.
FIG. 9.
Proposed mechanism for the inhibition of HeV fusion protein by the native HPIV3 HRC-derived 36-mer peptide. (A) Inhibition of HPIV3 fusion with the HPIV3 HRC-derived peptide (green; required segments of eight residues on either end of the peptide are indicated) as it interacts with the F monomer (HRC in red, HRN in blue). The structural/functional features of the residue side chains, e.g., hydrogen bond donor, hydrogen bond acceptor, hydrophobicity, etc., are indicated as shapes. Shape complementarity yields stronger interactions between the peptide and the HRN chain. (B) Inhibition of the HeV F protein with the HPIV3 HRC peptide. The shape match between the peptide and the HeV HRN (or possibly HRC) is very good within the required segments, allowing tighter binding (yellow boxes) of the peptide ends to a chain of the HeV F. This complex does not allow subsequent binding of another monomer, thus blocking fusion.
Although the HR sequences of F proteins are somewhat conserved among the paramyxoviruses, fusion inhibition by the corresponding HR peptides has generally been found to be virus specific. For example, peptides corresponding to the HRC domains of HPIV3 or HPIV2 prevented cell fusion mediated only by the same F protein; each peptide was completely virus specific (51). HRC peptides derived from either of the two henipavirus F proteins, HeV or NiV, were found to equally inhibit the other henipavirus but to have no effect on fusion mediated by either MeV F or canine distemper virus F (4, 5). In an earlier study (22), peptides corresponding to the HRC F domains of HPIV3, MeV, RSV, or HIV-1 were found to inhibit fusion mediated by the virus of origin, and there was little if any cross-inhibition between measles virus, respiratory syncytial virus, or HIV-1. However, while HPIV3 fusion was not inhibited by the heterotypic HRC peptides, the HPIV3 HRC peptide inhibited fusion mediated by both the morbillivirus MeV and the pneumovirus RSV. This potential for interaction of the HPIV3 F's HRC with paramyxoviruses of two different genera stimulated our hypothesis that HPIV3 peptides might inhibit HeV fusion and might suggest a new inhibitory mechanism. Our results not only support the original hypothesis and the findings of Lambert et al. but, surprisingly, show that the HPIV3 HRC used in our study was far more effective than the homotypic (HeV) HRC peptide presently proposed as a candidate antiviral molecule (6). This particular HPIV3 peptide was highly effective both in inhibiting fusion mediated by HeV G and F and in preventing infection with particles pseudotyped with G and F, even those with a highly fusion-promoting G protein.
Several possibilities are under consideration to explain the finding that the HPIV3 HRC peptide inhibits HeV glycoprotein-mediated fusion and entry more effectively than it inhibits HPIV3-mediated fusion or entry. Fusion in cell culture systems occurs more rapidly for the HPIV3 proteins than for the HeV proteins (data not shown), and we are currently investigating whether the HPIV3 HN is more effective than the HeV G at triggering its corresponding F protein to fusion readiness, thus limiting the time window for action of the inhibitory peptides. A longer time lag to fusion activation for HeV might offer more opportunity for inhibitory peptide action. Alternatively, it is possible that the target for the HPIV3 HRC inhibitory peptide is different in the two viruses, and this is an area of investigation.
Future studies will further explore the mechanism of HPIV3 HRC inhibition of HeV fusion. We will assess the timing of interaction of HPIV3 peptide with HeV and determine whether this interaction occurs when F is expressed alone or when G actively interacts with F. This information is critical for the development of potent viral inhibitors, which must target the virus at the most effective moment during infection. Taking advantage of the information presented here, together with the crystal structures of the HPIV3 F protein and additional molecular modeling of the protein-peptide interactions, new peptides will be designed and tested for their effectiveness at reducing infection. With an understanding of the determinants of peptide effectiveness, we believe that it will be possible to design shorter and more effective peptides as superior candidate antivirals.
HR peptides derived from F protein can only inhibit fusion during a transient phase of F's conformational transition. HR peptides that interact with F's HRC domain do not inhibit infection unless F has been triggered (11, 34, 42) and act only during the period of conformational shift. On the other hand, receptor-mimicking molecules that act by blocking binding can act only before F has been triggered. We propose that combining the two classes of potential antiviral agents may be advantageous, and future studies will test this concept. While we have shown that specific non-HeV paramyxovirus HRC peptides are highly effective at inhibiting HeV fusion, far more effective than the published HeV peptide inhibitor (6), we contend that any approach that works only after F triggering and insertion occurs carries inherent risks and would benefit from combination with the strategy of inactivating or blocking virus before it can reach the target cell.
Acknowledgments
This work was supported by Public Health Service grant AI056185 to A.M. from the National Institutes of Health (NIH), by the NIH (NIAID) Northeast Center of Excellence for Bio-defense and Emerging Infections Disease Research (U54AI057158 to W. I. Lipkin; P.I. of the Developmental Research Grant is A.M.), and by Public Health Service grant GM71894 to G.E.K. from the NIH.
We acknowledge the Northeast Center of Excellence for Bio-defense and Emerging Infections Disease Research's Proteomics Core for peptide synthesis and purification. Reagents were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We thank Michael Whitt (University of Tennessee Health Science Center and GTx) and GTx, Inc., for the kind gift of VSV-ΔG-RFP VSV-G and Ernest Yakob and Ernest Levroney for helpful discussions and thoughtful editing of the manuscript.
REFERENCES
- 1.Amadasi, A., F. Spyrakis, P. Cozzini, D. J. Abraham, G. E. Kellogg, and A. Mozzarelli. 2006. Mapping the energetics of water-protein and water-ligand interactions with the “natural” HINT forcefield: predictive tools for characterizing the roles of water in biomolecules. J. Mol. Biol. 358:289-309. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Bonaparte, M. I., A. S. Dimitrov, K. N. Bossart, G. Crameri, B. A. Mungall, K. A. Bishop, V. Choudhry, D. S. Dimitrov, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 102:10652-10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossart, K. N., L. F. Wang, B. T. Eaton, and C. C. Broder. 2001. Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology. 290:121-135. [DOI] [PubMed] [Google Scholar]
- 5.Bossart, K., L. Wang, M. Flora, K. Chua, S. Lam, B. Eaton, and C. Broder. 2002. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 76:11186-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossart, K. N., B. A. Mungall, G. Crameri, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Inhibition of Henipavirus fusion and infection by heptad-derived peptides of the Nipah virus fusion glycoprotein. Virol. J. 2:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnett, J. C., P. Botti, D. Abraham, and G. Kellogg. 2001. Computationally accessible method for estimating free energy changes resulting from site-specific mutations of biomolecules: systematic model building and structural/hydropathic analysis of deoxy and oxy hemoglobins. Proteins Struct. Funct. Genet. 42:355-377. [DOI] [PubMed] [Google Scholar]
- 8.Clark, M., R., D. I. Cramer, and N. van Opdenhosch. 1989. Validation of the general purpose Tripos 5.2 force field. J. Comp. Chem. 10:982-1012. [Google Scholar]
- 9.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 10.Cozzini, P., M. Fornabaio, A. Marabotti, D. J. Abraham, G. E. Kellogg, and A. Mozzarelli. 2002. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J. Med. Chem. 45:2469-2483. [DOI] [PubMed] [Google Scholar]
- 11.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. USA 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 14.Fornabaio, M., F. Spyrakis, A. Mozzarelli, P. Cozzini, D. J. Abraham, and G. E. Kellogg. 2004. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 3. The free energy contribution of structural water molecules in HIV-1 protease complexes. J. Med. Chem. 47:4507-4516. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 16.Horvath, C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 271:4621-4628. [DOI] [PubMed] [Google Scholar]
- 17.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 18.Kellogg, G. E., and D. J. Abraham. 2000. Hydrophobicity. Is logPo/w more than the sum of its parts? Eur. J. Med. Chem. 35:651-661. [DOI] [PubMed] [Google Scholar]
- 19.Kellogg, G. E., M. Fornabaio, D. Chen, D. Abraham, F. Spyrakis, P. Cozzini, and A. Mozzarelli. 2006. Tools for building a comprehensive modeling system for virtual screening under real biological conditions: the computational titration algorithm. J. Mol. Graph. Model. 24:434-439. [DOI] [PubMed] [Google Scholar]
- 20.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 21.Lamb, R. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin-Perlman, S., M. Jordan, R. Brossmer, O. Greengard, and A. Moscona. 1999. The use of a quantitative fusion assay to evaluate HN-receptor interaction for human parainfluenza virus type 3. Virology. 265:57-65. [DOI] [PubMed] [Google Scholar]
- 24.Li, J., V. R. Melanson, A. M. Mirza, and R. M. Iorio. 2005. Decreased dependence on receptor recognition for the fusion promotion activity of L289A-mutated Newcastle disease virus fusion protein correlates with a monoclonal antibody-detected conformational change. J. Virol. 79:1180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, S., H. Lu, J. Niu, Y. Xu, S. Wu, and S. Jiang. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 280:11259-11273. [DOI] [PubMed] [Google Scholar]
- 26.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 27.Meulendyke, K. A., M. A. Wurth, R. O. McCann, and R. E. Dutch. 2005. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12643-12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, J. P., and R. W. Doms. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. USA 100:10598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moscona, A. 2005. Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J. Clin. Investig. 115:1688-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moscona, A., and R. W. Peluso. 1991. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J. Virol. 65:2773-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moscona, A., and R. W. Peluso. 1993. Relative affinity of the human parainfluenza virus 3 hemagglutinin-neuraminidase for sialic acid correlates with virus-induced fusion activity. J. Virol. 67:6463-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negrete, O. A., E. L. Levroney, H. C. Aguilar, A. Bertolotti-Ciarlet, R. Nazarian, S. Tajyar, and B. Lee. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401-405. [DOI] [PubMed] [Google Scholar]
- 33.Negrete, O. A., M. C. Wolf, H. C. Aguilar, S. Enterlein, W. Wang, E. Muhlberger, S. V. Su, A. Bertolotti-Ciarlet, R. Flick, and B. Lee. 2006. Two key residues in Ephrin B3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Netter, R. C., S. M. Amberg, J. W. Balliet, M. J. Biscone, A. Vermeulen, L. J. Earp, J. M. White, and P. Bates. 2004. Heptad repeat 2-based peptides inhibit avian sarcoma and leukosis virus subgroup a infection and identify a fusion intermediate. J. Virol. 78:13430-13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pager, C. T., and R. E. Dutch. 2005. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12714-12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plemper, R. K., A. S. Lakdawala, K. M. Gernert, J. P. Snyder, and R. W. Compans. 2003. Structural features of paramyxovirus F protein required for fusion initiation. Biochemistry 42:6645-6655. [DOI] [PubMed] [Google Scholar]
- 38.Porotto, M., M. Murrell, O. Greengard, M. Lawrence, J. McKimm-Breschkin, and A. Moscona. 2004. Inhibition of parainfluenza type 3 and Newcastle disease virus hemagglutinin-neuraminidase receptor binding: effect of receptor avidity and steric hindrance at the inhibitor binding sites. J. Virol. 78:13911-13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN): an HN mutation diminishing the rate of F activation and fusion. J. Virol. 77:3647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porotto, M., M. Murrell, O. Greengard, L. Doctor, and A. Moscona. 2005. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 79:2383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rapaport, D., M. Ovadia, and Y. Shai. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 14:5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2004. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J. Virol. 78:13727-13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 94:14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3:279-287. [DOI] [PubMed] [Google Scholar]
- 46.Wang, L. F., M. Yu, E. Hansson, L. I. Pritchard, B. Shiell, W. P. Michalski, and B. T. Eaton. 2000. The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J. Virol. 74:9972-9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wild, T. F., and R. Buckland. 1997. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J. Gen. Virol. 78:107-111. [DOI] [PubMed] [Google Scholar]
- 50.Xu, Y., S. Gao, D. K. Cole, J. Zhu, N. Su, H. Wang, G. F. Gao, and Z. Rao. 2004. Basis for fusion inhibition by peptides: analysis of the heptad repeat regions of the fusion proteins from Nipah and Hendra viruses, newly emergent zoonotic paramyxoviruses. Biochem. Biophys. Res. Commun. 315:664-670. [DOI] [PubMed] [Google Scholar]
- 51.Yao, Q., and R. W. Compans. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223:103-112. [DOI] [PubMed] [Google Scholar]
- 52.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, J. K., R. P. Hicks, G. E. Wright, and T. G. Morrison. 1997. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology 238:291-304. [DOI] [PubMed] [Google Scholar]
- 55.Young, J. K., D. Li, M. C. Abramowitz, and T. G. Morrison. 1999. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J. Virol. 73:5945-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]