Abstract
The immediate-early 2 (IE2) protein of human herpesvirus 6 is a potent transactivator of cellular and viral promoters. To better understand the biology of IE2, we generated a LexA-IE2 fusion protein and screened, using the yeast two-hybrid system, a Jurkat T-cell cDNA library for proteins that could interact with IE2. The most frequently isolated IE2-interacting protein was the human ubiquitin-conjugating enzyme 9 (Ubc9), a protein involved in the small ubiquitin-like modifier (SUMO) conjugation pathway. Using deletion mutants of IE2, we mapped the IE2-Ubc9-interacting region to residues 989 to 1037 of IE2. The interaction was found to be of functional significance to IE2, as Ubc9 overexpression significantly repressed promoter activation by IE2. The C93S Ubc9 mutant exhibited a similar effect on IE2, indicating that the E2 SUMO-conjugating function of Ubc9 is not required for its repressive action on IE2. No consensus sumoylation sites or evidence of IE2 conjugation to SUMO could be demonstrated under in vivo or in vitro conditions. Moreover, expression levels and nuclear localization of IE2 were not altered by Ubc9 overexpression, suggesting that Ubc9's repressive function likely occurs at the transcriptional complex level. Overall, our results indicate that Ubc9 influences IE2's function and provide new information on the complex interactions that occur between herpesviruses and the sumoylation pathway.
Human herpesvirus 6 (HHV-6) is a betaherpesvirus initially isolated from human immunodeficiency virus (HIV)-infected individuals and from patients suffering from lymphoproliferative disorders (66). HHV-6 is the etiologic agent of the childhood disease roseola or exanthem subitum (76). Links between HHV-6 infection and other pathologies, such as meningoencephalitis (33), organ transplant rejection (10, 79), and AIDS (40, 63), have also been suggested.
Molecular, biological, and immunological analysis between various isolates has led to the subdivision into HHV-6 variants A and B (1). Primary HHV-6B infection is associated with exanthem subitum (76), but a pathological role for HHV-6A is still unclear. Although HHV-6A and HHV-6B are closely related, there is no genetic gradient between them, and recombinant viruses have never been detected (16, 32). Interestingly, the most variable region between variants of HHV-6 is observed within the immediate-early A (IE-A) locus and could account for the diverse biological properties between these viruses (16, 32). The HHV-6 IE-A locus includes two genetic units termed IE1 and IE2, corresponding to open reading frames U90/U89 and U90/U86, respectively. We have previously characterized the IE1 variant B (26) and IE2 variant A (27) proteins translated from spliced transcripts of the IE-A locus.
It has been reported that the HHV-6 IE2 protein is a potent transcriptional activator of heterologous promoters (19, 27). Indeed, the HHV-6 open reading frame U86 gene product was found to transactivate the human CD4 promoter (19). Moreover, cotransfection experiments in T cells indicated that IE2 variant A can induce the transcription of complex promoters, such as the one present in the HIV long terminal repeat (LTR), as well as simpler promoters whose expression is driven by a unique set of responsive elements (cyclic AMP-responsive element, NF-AT, and NF-κB) (17, 27). Finally, the C-terminal domain encompassing the final 436 residues of IE2A was shown to bind a DNA fragment containing the transcription initiation site, TATA box, and upstream sequence of the IE-A promoter (57).
Clues pertaining to the nature of HHV-6 IE2 functional domains can be derived from the numerous studies of human cytomegalovirus (HCMV) immediate-early protein IE2. HCMV, like HHV-6, is a betaherpesvirus and shares amino acid sequence similarities, immunological cross-reactivity, and overall gene organization with HHV-6 (45, 53, 78). HCMV gene UL122, coding for protein IE2, is a positional homologue of HHV-6 open reading frame U86 (55), which codes for the C-terminal portion of HHV-6 IE2. The similarity between HCMV IE2 and the carboxy-terminal region of HHV-6 IE2 is 45% (55). HCMV IE2 is an 86-kDa protein whose biological functions are well defined and include transactivation of heterologous promoters (60), repression of its own promoter (28), association with the viral DNA replication compartment (3), blocking of cell cycle progression (75), and modulation of apoptosis (80).
Viruses have evolved numerous mechanisms to overcome host defenses and to use the host biological pathways to the virus's advantage. One type of virus-host interaction that is well established and widespread is regulation of viral protein function by posttranslational modification systems, such as phosphorylation, glycosylation, and ubiquitinylation. Sumoylation is another host cell posttranslational modification system that has been characterized to a greater depth in recent years. While not yet completely understood in terms of functional effects, sumoylation appears to control protein activity and/or intracellular location.
Sumoylation is the process of covalently attaching a small ubiquitin-related modifier (SUMO) moiety to a target protein. In humans, at least four SUMO proteins can be found, with 47% amino acid identity between SUMO-1 and SUMO-2 and 95% amino acid identity between the closely related SUMO-2 and SUMO-3. Although there seem to be significant differences between SUMO-1 and SUMO-2/3 distribution, availability, and substrates, there are still few studies so far on SUMO-2/3. SUMO-1 is itself a 101-amino-acid polypeptide with sequence relatedness to ubiquitin (44). The mechanism of conjugation of SUMO-1 to various targets is somewhat analogous to that of ubiquitination. It involves an activating E1 enzymatic heterodimer (Uba2/Aos1) (13, 23, 56), a conjugating E2 enzyme (Ubc9) (14, 22, 36, 68), and one of the many SUMO-1 E3 ligases, such as PIASy or RanBP2 (58, 62). In contrast to ubiquitination, however, SUMO-1 conjugation does not appear to target proteins to proteasomal degradation; rather, it affects the ability of modified proteins to interact with other cellular factors. Effects of SUMO-1 conjugation on target proteins include targeting to specific subcellular compartments, alteration of functional activity, and regulation of ubiquitin-mediated proteolysis (29).
Besides the many known cellular substrates of sumoylation, such as RanGAP1 (48), IκBα (12), primary biliary cirrhosis autoantigen Sp100 (72), promyelocytic leukemia protein (17), tumor suppressor p53 protein (24, 61), and homeodomain-interacting protein kinase 2 (39), there are more and more viral protein targets of SUMO-1 currently being identified. The first known viral targets were HCMV IE1 and IE2 proteins (30, 52). Promoter transactivation by HCMV IE2 is strongly reduced in SUMO-1 conjugation-defective mutants, suggesting a functional relevance of the SUMO-1 conjugation pathway in the biological activity of IE2 protein (30). However, it was subsequently demonstrated that sumoylation of HCMV IE2 is not essential for virus growth in cultured HF cells (46). The consequence of sumoylation for HCMV IE1 is not yet clearly understood; however, it was shown that a sumoylation-defective IE1 mutant has reduced levels of the HCMV IE2 transcript and impaired viral replication (54). Sumoylation of Epstein-Barr virus immediate-early protein Zta, encoded by the BZLF1 gene, leads to reduced activity of this transactivator on specific promoters (2). However, another immediate-early Epstein-Barr virus transactivator, Rta (encoded by the BRLF1 gene), shows increased transactivating activity upon SUMO-1 conjugation (11).
During the ongoing work to characterize the immediate-early proteins of HHV-6, our laboratory reported that HHV-6B IE1 is also a target of SUMO-1 conjugation (26), a finding subsequently confirmed for HHV-6A IE1 in another laboratory (71). SUMO-1 overexpression leads to increased levels of IE1 expression through an undefined mechanism (25, 71).
Because of the growing numbers of immediate-early herpesviral transactivators being shown to be subject to sumoylation, we investigated the possibilities of SUMO conjugation to HHV-6 IE2. This hypothesis was further supported by the homology between IE2 and a known sumoylation target, HCMV IE2. SUMO-1 conjugation to target proteins involves the carboxy-terminal glycine residue (G97) of SUMO-1, the Ubc9 conjugating enzyme, and a lysine (K) residue on the target protein. The sumoylation consensus motif is generally identified as a lysine residue immediately preceded by a hydrophobic amino acid (ψ) and followed by a variable residue (X) and a glutamic acid residue (E) (17, 37, 39). This widely accepted consensus motif (ψKXE) is not, however, inclusive of all described SUMO-1 conjugation sites (9, 35, 39). HCMV IE2 is SUMO-1 modified on lysine residues 175 and 180, both described by the conjugation consensus motif ψKXE (4, 30). Computer analysis of the HHV-6 IE2 sequence revealed no typical consensus sumoylation sites.
Using a yeast two-hybrid system, we screened a human Jurkat T-cell line cDNA expression library for proteins that could interact with HHV-6 IE2. We report that HHV-6 IE2 and Ubc9 physically interact together. Previous research indicates that a SUMO-1 or Ubc9 interaction in the two-hybrid assay is often indicative of SUMO-1 conjugation to the interacting protein (50). Our results suggest that Ubc9 represses IE2's transactivating activity in a SUMO-independent fashion. We also determined that Ubc9 does not alter the levels of IE2 expression or the nuclear localization of IE2. These results suggest a new way by which cells or viruses can regulate gene transcription as well as further contribute to our knowledge of the complex relation that exists between herpesviruses and the sumoylation pathway.
MATERIALS AND METHODS
Virus and cell lines.
The HSB-2 and Molt-3 leukemia human T-cell lines were cultured in MegaCell RPMI 1640 medium supplemented with 10% fetal bovine serum (Sigma-Aldrich Canada, Oakville, Ontario, Canada) and M-plasmocin (InvivoGen, San Diego, CA) to prevent mycoplasma contamination. The HEK293T human epithelial kidney cell line was cultured in MegaCell Dulbecco's modified Eagle's medium (Sigma-Aldrich Canada) supplemented with 3% fetal bovine serum and M-plasmocin and passaged every 2 days. HHV-6A (GS strain) was propagated in HSB-2 cells as previously described (18). HHV-6B (Z29 strain) was propagated in Molt-3 cells as described previously (26).
Plasmid generation.
To generate the LexA-IE2 N-terminal fusion construct (pHybLex-IE2 1-720, encoding amino acids 1 to 720 of IE2), mRNA from HHV-6-infected HSB-2 cells was reverse transcription-PCR amplified with primers matching the N-terminal region of IE2 (forward primer, 5′-taggtacccacc ATG GAG CCA GCA AAA C-3′; reverse primer, 5′-tctgactcgag AGC CAT GGT GCA ACT TT-3′; adapters not homologous with the viral sequence are indicated by lowercase script, and KpnI and XhoI restriction sites are underlined) and ligated in frame into KpnI/XhoI-digested pHybLex/Zeo vector (Invitrogen, Carlsbad, CA). Similarly, to generate the LexA-IE2 C-terminal fusion construct (pHybLex-IE2 925-1466), infected-cell mRNA was reverse transcription-PCR amplified with primers matching the C-terminal region of IE2 (forward primer, 5′-gagctc AAA GCC TCC AGC AGA GCC TCC AG-3′; reverse primer, 5′-gtcgacag TTA ACA TTT TGA AAG TGT AC-3′; adapters not homologous with the viral sequence are indicated by lowercase script, and SacI and SalI restriction sites are underlined), ligated in pCR3.1 cloning vector (Invitrogen), and then subcloned in frame into SacI/SalI-digested pHybLex/Zeo vector. Two different deletion mutants of the C-terminal portion of IE2 were generated by digestion of pHybLex-IE2 925-1466 with the restriction enzymes PstI or AccIII/SalI (Promega, Madison, WI), yielding, respectively, plasmids pHybLex-IE2 925-988 and pHybLex-IE2 925-1233. In order to further map the interaction region between IE2 and Ubc9, the following constructs were generated by single-base mutagenesis following the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) instructions. Plasmid pHybLex-IE2 925-1466 was mutated to pHybLex-IE2 925-1176 using the stop codon mutagenic primer 5′-GAA CAA TCC AAT CAT TCC TAA AAC GCC ATT GAT GAA G-3′ and its complementary oligonucleotide (the mutated base is underlined). Constructs pHybLex-IE2 925-1131, pHybLex-IE2 925-1083, and pHybLex-IE2 925-1037 were generated using, respectively, primers 5′-CA GAT TCA AAA CAC TAA ACT ACA AAC ATG TCT TCA G-3′, 5′-ACA CAA TTT TAC TAT TAG TCT TCC AGA ACT AGA TC-3′, and 5′-TCT CAC TGT CGA AAT TAA CCA GAT TCA CTG ACC-3′ (the mutated base is underlined) and their complementary oligonucleotides.
Library-isolated two-hybrid prey plasmid pYESTrp-Ubc9 was mutated into non-SUMO-1-conjugating pYESTrp-Ubc9 C93S by using mutagenic primer 5′-CG GGG ACA GTG AGC CTG TCC ATC TTA G-3′ and its complementary oligonucleotide (the mutated base is underlined). Furthermore, SUMO-1 was extracted from pcDNA3-6His-SUMO-1 (kind gift of R. T. Hay) by BamHI digestion and subcloned into pYESTrp (Invitrogen) to yield pYESTrp-SUMO-1. Lastly, plasmid pHybLex-Ubc9 was generated by in-frame cloning of Ubc9 cDNA from pYESTrp-Ubc9 into the EcoRI restriction site of pHybLex.
For the transactivation assays, complete wild-type HHV-6A IE2 was PCR amplified from pBK-IE2A (27) (forward primer, 5′-aggtacc GGA GCC AGC AAA ACC-3′; reverse primer, 5′-cctcgagg TTA ACA TTT TGA AAG TGT AC-3′; adapters not homologous with the viral sequence are indicated by lowercase script, and KpnI and XhoI restriction sites are underlined), ligated in pCR3.1 cloning vector, and then subcloned in frame into KpnI/XhoI-digested pcDNA4/HisMaxA vector (Invitrogen) to yield pcDNA4-IE2A. Ubc9 interaction mutant pcDNA-IE2Δ993-1037 was generated by directed mutagenesis of pcDNA-IE2A using mutagenic primer 5′-TGC AGC TCG ACT CCA GAG CTC ACC AGA TTC ACT GAC C-3′ and its complementary oligonucleotide. Ubc9 cDNA isolated from the Jurkat library was subcloned in the EcoRI restriction site of pcDNA3.1 (Invitrogen). Construct pcDNA3.1-Ubc9 was subsequently mutated into non-SUMO-1-conjugating pcDNA3.1-Ubc9 C93S by using primer 5′-CG GGG ACA GTG AGC CTG TCC ATC TTA G-3′ and its complementary oligonucleotide (the mutated base is underlined).
Finally, full-length HIV-1 Tat cDNA was subcloned from pREP-Tat (kindly provided by M. Tremblay) into the BamHI restriction site of pcDNA4/HisMaxA. Construct pcDNA3-Myc-RanGAP1 was kindly provided by S. Muller. Hemagglutinin (HA)-tagged construct pCMV-SUMO-1 was cloned as previously described (25).
Yeast two-hybrid screen.
pHybLex-IE2 925-1466 plasmid was introduced into the L40 strain of Saccharomyces cerevisiae by lithium acetate transformation (15). Zeocin-resistant L40 cells were subsequently transformed with a human Jurkat cell cDNA library cloned into the pYESTrp plasmid. Protein-protein interaction led to the activation of two yeast reporter genes, LacZ and HIS3. Expression of HIS3 allowed yeast cells to grow on His-negative media, while LacZ led to β-galactosidase expression. The screening was performed following the Invitrogen Hybrid Hunter protocol. Interactions were first selected on His-negative plates, followed by a second screen for β-galactosidase expression. For Western blot analysis, cells were subjected to mechanical lysis with glass beads in urea-containing cracking buffer (8 M urea, 5% sodium dodecyl sulfate [SDS], 40 mM Tris-HCl, pH 6.8, 0.1 mM EDTA, 1% β-mercaptoethanol). Expression of the bait and prey proteins was assessed by Western blotting using an anti-LexA rabbit polyclonal antibody and an anti-V5 mouse monoclonal antibody, respectively (both from Invitrogen). Plasmids coding for proteins interacting with IE2 were recovered from yeast cells by mechanical lysis with glass beads, sodium acetate precipitation, and transformation into Escherichia coli XL2-Blue ultracompetent bacteria (Stratagene). Sequencing of plasmids was performed by automated sequence analysis using the dye termination reaction in an ABI (Weiterstadt, Germany) sequencer.
Mapping of IE2 domain interacting with Ubc9.
S. cerevisiae strain L40 cells were transformed with pYESTrp-Ubc9, pYESTrp-Ubc9 C93S, or pYESTrp-SUMO-1 plasmids and either pHybLex-IE2 1-720, pHybLex-IE2 925-1466, pHybLex-IE2 925-1233, pHybLex-IE2 925-1176, pHybLex-IE2 925-1131, pHybLex-IE2 925-1083, pHybLex-IE2 925-1037, or pHybLex-IE2 925-988. Cells transformed with both bait and prey plasmids were selected on Trp-negative (His-positive) zeocin media plates. Extracts were prepared following Applied Biosystems technical notes. In brief, each yeast colony was resuspended in 60 μl of lysis buffer (50 μl Tropix Galacto-Light lysis solution [Applied Biosystems, Bedford, MA], 2.5 μl 0.1% SDS, 7.5 μl chloroform) and vortexed for 10 seconds. Subsequently, 5 μl of extract was added to 67 μl of diluted Galacton (Applied Biosystems) and incubated at room temperature for 60 min. Chemiluminescence indicating β-galactosidase activity was assayed by adding 100 μl of accelerator solution using an MLX microtiter plate luminometer (Dynex Technologies, Chantilly, VA). The chemiluminescent signal was normalized for the amount of protein (colony size) in each sample. At least eight colonies were assayed for each different transformation, and the average signal was compared to that of negative control cells cotransformed with pYESTrp-Ubc9 and pHybLex.
Immunofluorescence.
HSB-2 cells were infected for 48 h with HHV-6A, fixed with cold (−20°C) acetone, and incubated with various antibodies described hereafter. IE2 expression was detected with a mouse anti-IE2 monoclonal (P6H8) antibody raised against a glutathione S-transferase-IE2 fusion protein (7) and coupled to the Alexa 488 green fluorescent dye (Molecular Probes, Eugene, OR). Ubc9 expression was analyzed with a mouse anti-Ubc9 antibody (BD Biosciences, Mississauga, Ontario, Canada) followed by a rabbit anti-mouse antibody coupled to the Alexa 568 red dye (Molecular Probes). SUMO-1 was detected with a rabbit anti-SUMO-1 antibody raised against glutathione S-transferase-SUMO-1 and Alexa 568 coupled. Cells were examined on a Leitz Aristoplan epifluorescence microscope (Leica Microsystems Canada, Richmond Hill, Ontario, Canada) with 488-nm (green dye, IE2) and 568-nm (red dye, Ubc9 and SUMO-1) filters. Representative cell fields were captured using a black-and-white digital camera (Dage-MTI, Michigan City, IN) and Bioquant NOVA software (Bioquant-R&M Biometrics, Nashville, TN). The same technique was used for immunofluorescence analysis of transfected HEK293T cells.
Sumoylation assay in infected cells.
HSB-2 cells were infected for 72 h with HHV-6A. Infected cells (1 × 107) were pelleted, lysed, and sonicated in a 1:3 dilution of buffer I and II containing 5 mM N-ethylmaleimide, as described previously (12). Clarified supernatants were incubated overnight with anti-IE1 (26) or anti-IE2 (7) antibodies and protein A-Sepharose beads (Pierce, Rockford, IL), followed by three washes with lysis buffer. Beads were resuspended in Laemmli buffer and boiled for 5 min. Immunoprecipitated proteins were electrophoresed, and Western blotting was carried out using anti-IE1, anti-IE2, or anti-SUMO-1 antibodies as described below.
Western blotting.
For Western blot analysis, cells were washed in phosphate-buffered saline, lysed in an appropriate volume of Laemmli buffer, and boiled. Samples were electrophoresed through an SDS-polyacrylamide gel, transferred to polyvinylidene fluoride membranes, and blotted for 1 h at room temperature. After three 10-min washes with Tris-buffered saline-Tween, the blots were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h. The blots were then washed with Tris-buffered saline-Tween, and the proteins were visualized with enhanced chemiluminescence (Perkin-Elmer, Boston, MA) using a PhosphorImager system (Fuji Medical Systems, Stamford, CT). The following primary antibodies were used: P6H8 mouse anti-HHV-6A IE2, rabbit anti-HHV-6B IE1, and rabbit anti-SUMO-1 antibodies as previously described (7, 25); mouse anti-Ubc9 (BD Biosciences); mouse anti-His (Amersham Biosciences, Baie d'Urfé, Quebec, Canada); 12CA5 mouse anti-HA; and 9E10 mouse anti-Myc.
Transfections and luciferase assays.
Transfections were performed using the calcium phosphate precipitation procedures. HEK293T cells were plated at 200,000 cells/well (six-well plate) the day prior to transfection. Cells were transfected with 500 ng of reporter plasmid and up to 6.5 μg of expression vectors per well and brought to a total of 7 μg of DNA per well for each condition with the pcDNA4 control plasmid. Cells were lysed 48 h after transfection. Transactivation was evaluated using pLTR-Luc (kindly provided by M. Tremblay) and p2-1900 (kindly provided by M. Fresno; described in reference 31) reporter constructs coding for the luciferase gene driven by HIV LTR and human COX-2 promoters, respectively. Luciferase activity was measured on an MLX microtiter plate luminometer (Dynex Technologies). The values obtained are means of three distinct experiments performed in duplicate and are normalized for protein concentration in each sample, as determined by a bicinchoninic acid colorimetric assay (Pierce).
Molt-3 cells were transfected by electroporation using a Gene Pulser apparatus and capacitance extender (Bio-Rad Laboratories, Hercules, CA). Briefly, 18 μg total DNA was added to 107 cells in 400 μl RPMI medium. Cells were pulsed at 0.25 kV with a 960-μF capacitance in a 0.4-cm-gap electroporation cuvette (Bio-Rad Laboratories). Cells were transferred into 10 ml of culture medium and lysed after 48 h of growth. Transactivation of reporter constructs was assayed as described above.
Cell fractionation.
Transfected HEK293T cells (4 × 105) were harvested 48 h after transfection, washed in phosphate-buffered saline, and incubated for 5 min on ice in 250 μl of F buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 5 mM MgCl2, and 0.5% NP-40) to which Complete protease inhibitor cocktail was added (Roche Diagnostics, Laval, Quebec, Canada). Cells were centrifuged for 5 min at 500 × g and 4°C, upon which supernatant (cytoplasmic fraction) was set aside. The remaining pellet was resuspended in 250 μl of F buffer (nuclear fraction). To each fraction was added Laemmli buffer, and samples were boiled for 5 min before SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Statistical analysis.
Statistical analysis of the data, which were normally distributed, was performed using an unpaired two-tailed Student's t test. Differences between means were considered significant at a P value of <0.05. Data analysis was performed with Prism software version 4.03 (GraphPad Software, San Diego, CA).
RESULTS
Identification of Ubc9 as an IE2-interacting protein.
To identify cellular proteins interacting with HHV-6A IE2, we made use of the yeast two-hybrid system with the IE2 protein as a bait and a Jurkat cDNA expression library as preys. Initial tests indicated that the N-terminal portion of IE2 (residues 1 to 720) strongly transactivates the yeast β-galactosidase reporter gene without the need for an interaction partner. Consequently, we used for our library screen the C-terminal portion (amino acids 925 to 1466), which demonstrated no nonspecific transactivation. Screening of 37 million clones resulted in the isolation of 7 individual yeast colonies showing strong and specific interaction with IE2 925-1466. Sequence analysis of the various clones identified four out of seven clones as coding for the human Ubc9 protein (GenBank accession number X96427). The human ubc9 gene is 1.3 kb long and codes for an 18-kDa enzyme (74, 77) essential for the posttranslational conjugation of small ubiquitin-related modifiers to various cellular proteins.
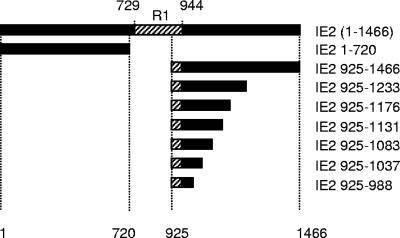
To map the interaction domain between IE2 and Ubc9, we generated six deletion or stop codon mutants of pHybLex-IE2 925-1466 (Fig. 1) and subjected them to a two-hybrid experiment with pYESTrp-Ubc9 formerly isolated from the Jurkat cDNA library. Double transformant colonies that grew on selective media (His negative) were assayed for β-galactosidase activity using a chemiluminescence liquid assay. Table 1 shows positive interaction, as determined by both growth on minimal medium (His negative) and β-galactosidase activity, for all mutants except IE2 925-988. This suggests that residues 989 to 1037 include a minimal sequence required for Ubc9 interaction.
FIG. 1.
Mapping of the IE2-Ubc9 interaction domain. Deletion mutants of IE2 were generated by restriction digest or mutagenesis of wild-type IE2. R1 is the SSRA/SSRD amino acid repeat region of IE2.
TABLE 1.
Mapping of the IE2-Ubc9 interaction domain by yeast two-hybrid screening
| Bait (pHybLex fusion) | Prey (pYESTrp fusion)
|
|||
|---|---|---|---|---|
| Ubc9
|
Ubc9 C93S
|
|||
| His-negative growtha | β-Gal activityb | His-negative growtha | β-Gal activityb | |
| Control (empty vector)c | − | 1.00 ± 0.38 | − | 1.02 ± 0.17 |
| IE2 1-720 | + | 35.6 ± 7.62* | + | 38.8 ± 7.30* |
| IE2 925-1466 | + | 9.40 ± 2.89* | + | 14.6 ± 2.31* |
| IE2 925-1233 | + | 7.73 ± 2.41* | + | 10.0 ± 1.92* |
| IE2 925-1176 | + | 4.23 ± 1.48* | + | 6.00 ± 1.19* |
| IE2 925-1131 | + | 4.09 ± 1.74* | + | 6.81 ± 1.34* |
| IE2 925-1083 | + | 7.46 ± 2.25* | + | 7.89 ± 1.64* |
| IE2 925-1037 | + | 14.4 ± 2.83* | + | 20.9 ± 4.50* |
| IE2 925-988 | − | 1.45 ± 0.51 | − | 1.52 ± 0.40 |
A + indicates growth, while a − indicates that there was no growth.
Results are expressed as means ± standard deviations from three distinct experiments. Reference activity was that of control cells cotransformed with pYESTrp-Ubc9 and pHybLex. Asterisks indicate that P was <0.05 compared with reference activity. β-Gal, β-galactosidase.
Control cotransformations with empty prey vector pYESTrp and each of the bait constructs were all negative for His-negative growth except for pHybLex-IE2 1-720 (β-Gal activity = 41.0 ± 10.8; P < 0.05). Cotransformations with prey pYESTrp-SUMO-1 and each of the bait constructs were all negative for His-negative growth, except for pHybLex-IE2 1-720 (β-Gal activity = 39.5 ± 11.1; P < 0.05) and pHybLex-Ubc9 (12.1 ± 2.05; P < 0.05).
Previous research indicates that a Ubc9 or SUMO-1 interaction in the two-hybrid assay is often indicative of SUMO-1 conjugation to the interacting protein (50). A pYESTrp-SUMO-1 hybrid was therefore tested for interaction with all of our pHybLex-IE2 constructs. No significant interaction between SUMO-1 and any of the IE2 forms was detected (Table 1). In order to validate the pYESTrp-SUMO-1 construct, we tested it for interaction with a pHybLex-Ubc9 plasmid. As expected (22), a strong Ubc9-SUMO-1 reporter activity was detected, indicative that an interaction between Ubc9 and SUMO-1 occurred in yeast cells.
Next, to determine whether the SUMO-1-conjugating activity of Ubc9 is required for Ubc9 interaction with IE2, we mutated cysteine 93 of pYESTrp-Ubc9 to serine in order to remove its catalytic SUMO-1 binding residue (20, 73). The pYESTrp-Ubc9 C93S construct was assayed in the two-hybrid interaction test described above. Interaction with all previously described IE2 constructs was not significantly altered (Table 1), indicating that the sumoylating activity of Ubc9 is not required for interaction with IE2. Thus, although IE2 segment 989 to 1037 interacts with Ubc9 and Ubc9 C93S, no SUMO-1 interaction was detected with any of the IE2 constructs.
IE2-Ubc9 colocalization in infected and transfected cells.
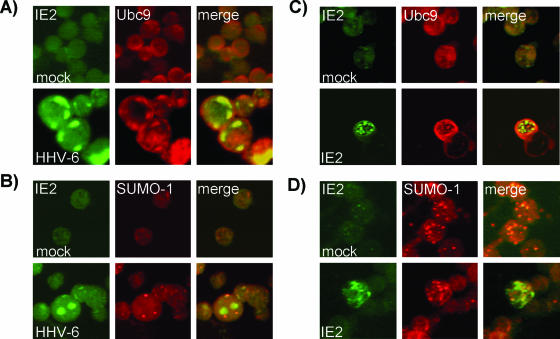
In order to confirm that the observed interaction between IE2 and Ubc9 is physiologically relevant, we examined HHV-6-infected and IE2-transfected cells by immunofluorescence for colocalization of IE2, Ubc9, and/or SUMO-1. Figure 2A shows that in HHV-6-infected HSB-2 cells, Ubc9 has a pericellular and somewhat diffuse pattern, while IE2 forms patches that were previously shown to be nuclear (27). Ubc9 colocalizes with IE2 in some of the patches. In order to confirm that IE2 is not sumoylated, we also analyzed the cellular distribution of SUMO-1. In mock-infected HSB-2 cells, the SUMO-1 signal is barely detectable. In infected cells, however, SUMO-1 forms strong nuclear dots that do not colocalize with IE2 (Fig. 2B). It was shown previously that in infected cells, SUMO-1 dots colocalize with PML nuclear bodies and HHV-6 IE1 (26). Because the possibility of other viral proteins acting as a scaffold between IE2 and Ubc9 could not be excluded, we also transfected HEK293T human epithelial cells with a wild-type IE2 construct. In transfected cells, IE2 shows a punctuate pattern that colocalizes with some of the Ubc9 patches (Fig. 2C), while IE2 and SUMO-1 do not colocalize (Fig. 2D). These data suggest that the interaction between IE2 and Ubc9 does not involve other viral proteins or SUMO-1 conjugation.
FIG. 2.
(A) HHV-6A IE2 protein relative localization with Ubc9 in infected cells. HSB-2 cells were infected with HHV-6A and processed for immunofluorescence as described in Materials and Methods. Left panels represent cells reacted with anti-IE2 antibody (IE2 in green), and middle panels show the same fields colored with anti-Ubc9 antibody (Ubc9 in red). Colocalizing proteins are represented in the merged panels by the yellow color. (B) HHV-6A IE2 protein relative localization with SUMO-1 in infected cells. Left panels represent cells reacted with anti-IE2 antibody (IE2 in green), and middle panels show the same fields colored with anti-SUMO-1 antibody (SUMO-1 in red). (C) HHV-6A IE2 protein relative localization with Ubc9 in transfected cells. HEK293T cells were transfected with the pcDNA4-IE2 construct and processed for immunofluorescence as described in Materials and Methods. Left panels represent cells reacted with anti-IE2 antibody (IE2 in green), and middle panels show the same fields colored with anti-Ubc9 antibody (Ubc9 in red). (D) HHV-6A IE2 protein relative localization with SUMO-1 in transfected cells. Left panels represent cells reacted with anti-IE2 antibody (IE2 in green), and middle panels show the same fields colored with anti-SUMO-1 antibody (SUMO-1 in red).
Sumoylation tests.
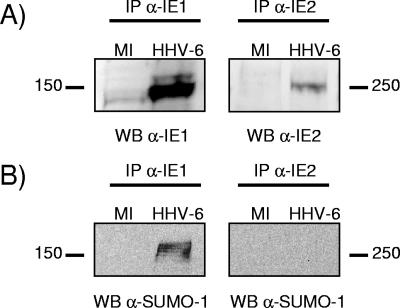
HHV-6A-infected HSB-2 cells and HHV-6B-infected Molt-3 cells were lysed in a buffer designed for preservation of sumoylated forms, as described previously (12). After immunoprecipitation of IE1B or IE2A, samples were analyzed by electrophoresis and Western blotted with anti-IE1, anti-IE2, or anti-SUMO-1 antibodies. While sumoylated forms of IE1 could be detected in HHV-6-infected cells, as previously described (26), no slow-migrating forms indicative of SUMO-1 conjugation to IE2 could be detected in the same cells (Fig. 3).
FIG. 3.
HHV-6 IE1 but not IE2 is sumoylated in vivo. HHV-6A-infected HSB-2 cells and HHV-6B-infected Molt-3 cells were lysed as described in Materials and Methods. After immunoprecipitation (IP) of IE1B or IE2A, samples were analyzed by electrophoresis and Western blotted (WB) with anti-IE1, anti-IE2, or anti-SUMO-1 antibodies. (A) Immunoprecipitated samples were analyzed by SDS-PAGE and probed for IE1B or IE2A using appropriate antibodies. MI, mock-infected cells. (B) The same samples were probed using an anti-SUMO-1 antibody.
To rule out the possibility of IE2 as a target of sumoylation, an in vitro SUMO-1 conjugation assay developed by Desterro et al. (12) was used. Briefly, in vitro [35S]Met-transcribed/translated IE2 proteins (the N-terminal or C-terminal portions were tested separately) were incubated with a HeLa cell extract exhibiting SUMO-1 E1 activity, recombinant Ubc9, and SUMO-1 proteins in a reaction buffer including an ATP-regenerating system. An HHV-6 IE1 protein control was used to test conjugation efficacy. While IE1 was efficiently modified by SUMO-1, no conjugation could be detected for either the N-terminal or C-terminal portions of IE2 (data not shown). Given the in vitro and in vivo conjugation assays, the immunofluorescence results, and the two-hybrid interaction tests, we conclude that IE2 is not likely to undergo sumoylation.
Promoter transactivation activity.
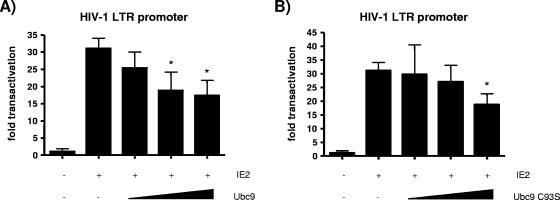
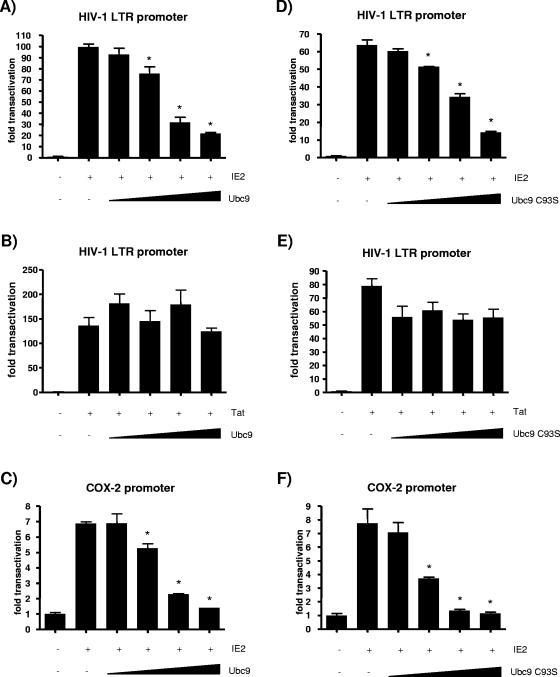
The currently known function of HHV-6 IE2 is to promiscuously promote transcriptional activation (27). Knowing that IE2 interacts with Ubc9, we next proceeded to study the effects of Ubc9 expression on promoter transactivation by IE2. We first performed transfection in Molt-3 T cells, a cell line efficiently infected by HHV-6. Cells were transfected with pLTR-Luc, a luciferase reporter plasmid driven by the HIV-1 LTR promoter, and wild-type IE2 in the absence or presence of Ubc9. IE2 is known to transactivate the LTR promoter efficiently (27, 47). The luciferase activity measured the effect of overexpressed Ubc9 on the transactivation of the promoter by IE2. Wild-type IE2 strongly transactivated the HIV-1 LTR promoter construct, while coexpression of Ubc9 partly inhibited IE2 transactivation in a dose-dependent manner (Fig. 4A). Because the sumoylation activity of Ubc9 is not required for interaction with IE2, we needed to ascertain whether the catalytic C93 residue of Ubc9 played a role in the downregulation of IE2 activity. Molt-3 cells were transfected with pLTR-Luc, wild-type IE2, and increasing quantities of the Ubc9 C93S construct. As Fig. 4B illustrates, the same dose-dependent inhibition of IE2 activity is observed when sumoylation-defective Ubc9 is overexpressed and when wild-type Ubc9 is overexpressed.
FIG. 4.
Inhibitory effect of Ubc9 on HIV-1 LTR promoter transactivation by HHV-6A IE2. Molt-3 cells were transfected as described in Materials and Methods with 2 μg pLTR-Luc reporter construct, 4 μg pcDNA4-IE2, and 0, 4, 8, or 12 μg of either pcDNA3.1-Ubc9 (A) or pcDNA3.1-Ubc9 C93S (B). Cells were lysed 48 h after electroporation. Transactivation of the pLTR-Luc reporter was quantified by measurement of luciferase activity, expressed as transactivation (n-fold) relative to control (pcDNA-transfected cells). Results are means ± standard deviations (SD) from four distinct experiments and were normalized for protein content in each sample. *, P < 0.05 compared with IE2-transfected cells.
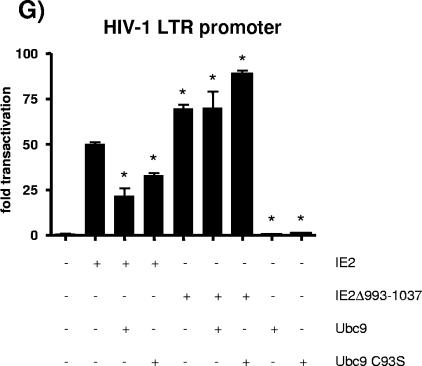
Because transfection efficiency is relatively low in T-cell lines, it was difficult to obtain consistent results with some of the reporters and conditions tested. For these reasons, we switched to HEK293T cells, which can be transfected more efficiently. Our results (Fig. 5A) indicate that transactivation of the HIV-1 LTR promoter by wild-type IE2 in the absence or in the presence of Ubc9 follows closely what was observed in Molt-3 T cells. These results suggest that IE2 behaves similarly in both cell lines. Figure 5A shows that in HEK293T cells, Ubc9 can downregulate IE2 activity, in a dose-dependent manner, by as much as fivefold. We also cotransfected IE2 and SUMO-1, SUMO-2, or SUMO-3 to verify whether those modifiers would have an effect on IE2 transactivation. None of the SUMO proteins could modulate IE2 activation of the LTR promoter (data not shown). HIV-1 protein Tat transactivation of the LTR promoter was not significantly altered by Ubc9 overexpression (Fig. 5B), suggesting that the Ubc9 effect is somewhat specific to IE2 and not the result of a general promoter-repressive activity. IE2 has previously been reported to transactivate the human COX-2 promoter (34). To determine whether this effect was specific to the LTR promoter, we investigated whether Ubc9 could influence the COX-2 promoter as well. As shown in Fig. 5C, Ubc9 had a dose-dependent inhibitory effect on the transactivation of a COX-2 promoter luciferase reporter plasmid, indicative that the effect is not specific for any given promoter.
FIG.5.
(A to F) Inhibitory effect of Ubc9 on promoter transactivation by HHV-6A IE2. HEK293T cells were transfected as described in Materials and Methods with 1 μg reporter construct (pLTR-Luc encoding the HIV-1 LTR promoter or p2-1900 encoding the human COX-2 promoter), 0.1 μg transactivator (pcDNA4-IE2 or control pcDNA4-Tat), and 0, 0.12, 0.25, 0.5, or 1 μg of either pcDNA3.1-Ubc9 or pcDNA3.1-Ubc9 C93S. Cells were lysed 48 h after transfection. Transactivation of the reporters was quantified by measurement of luciferase activity expressed as transactivation (n-fold) relative to control (pcDNA-transfected cells). Results are means ± SD from three distinct experiments of duplicate transfections and were normalized for protein content in each sample. *, P < 0.05 compared with IE2- or Tat-transfected cells. (G) Effect of the deletion of the Ubc9-interacting domain of HHV-6A IE2 on HIV-1 LTR promoter transactivation. HEK293T cells were transfected with 1 μg pLTR-Luc reporter construct, 0.1 μg pcDNA4-IE2 or pcDNA4-IE2Δ993-1037, and 1 μg pcDNA3.1-Ubc9 or pcDNA3.1-Ubc9 C93S. Luciferase activity measurement is described in Materials and Methods. *, P < 0.05 compared with IE2-transfected cells.
The inhibitory activity of Ubc9 C93S was also studied in HEK293T cells. As Fig. 5D illustrates, a dose-dependent inhibition of IE2 activity is observed when sumoylation-defective Ubc9 is overexpressed. Tat transactivation of the LTR promoter was not downregulated by Ubc9 C93S (Fig. 5E), while IE2 transactivation of a heterologous promoter, that of human COX-2, was strongly inhibited (Fig. 5F).
We also generated an IE2 mutant lacking the Ubc9 interaction site (993 to 1037 region), as identified in the yeast two-hybrid assay. Transactivation of the LTR promoter by IE2Δ993-1037 is stronger than that by wild-type IE2 (Fig. 5G), which was expected, since the absence of a Ubc9-interacting region makes IE2Δ993-1037 impervious to downregulation by endogenous Ubc9. While IE2 transactivation is downregulated by Ubc9 or Ubc9 C93S coexpression, IE2Δ993-1037 activity is not negatively influenced by them, which demonstrates that the deleted region participates in the inhibition of IE2 function by Ubc9. These observations suggest that Ubc9 substantially inhibits heterologous promoter transactivation by IE2 and that SUMO-1-conjugating activity is not essential for this effect. Taken together with the aforementioned IE2-Ubc9 interaction, our results suggest a mechanism by which Ubc9 represses IE2 activity without involving IE2 sumoylation.
Expression and nuclear localization of IE2.
In an attempt to better define the mechanism through which Ubc9 alters IE2 function, we studied the expression and localization of IE2 when coexpressed with Ubc9. Transfected HEK293T cells were fractionated into cytoplasmic and nuclear fractions and analyzed by Western blotting for IE2 and Ubc9 expression. The same samples were assayed for transactivation activity of IE2 as described previously. Figure 6 shows that although Ubc9 is overexpressed and IE2 activity is partly inhibited, IE2 levels are roughly stable and the viral protein is detected only in the nuclear fractions. Thus, IE2-Ubc9 interaction seems to be a nuclear event that does not downregulate IE2 transactivation by IE2 exclusion from the nucleus or by modulating IE2 expression at a transcriptional level.
FIG. 6.
Nuclear localization of IE2 in transfected cells is not altered by overexpression of Ubc9. HEK293T cells were transfected as described in Materials and Methods with 1 μg pLTR-Luc reporter construct, 2 μg pcDNA4-IE2 and 0, 1, 2, or 3 μg pcDNA3.1-Ubc9. Cells were harvested 48 h after transfection and split for the following assays. (A) Transactivation of the pLTR-Luc reporter was quantified by measurement of luciferase activity, expressed as transactivation (n-fold) relative to control (pcDNA-transfected cells). Results (±SD) represent one typical experiment out of three and were normalized for protein content in each sample. (B) The HEK293T-transfected cells were also processed for cellular fractionation into cytoplasmic (C) and nuclear (N) protein fractions as described in Materials and Methods. Samples were analyzed by SDS-PAGE and probed using anti-IE2 or anti-Ubc9 antibodies.
Impact of IE2 on the sumoylation pathway.
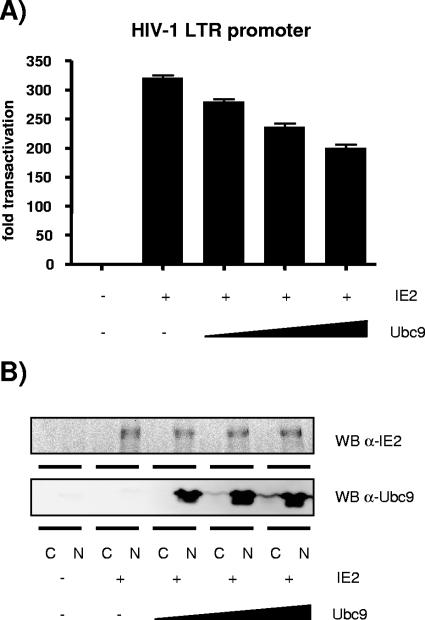
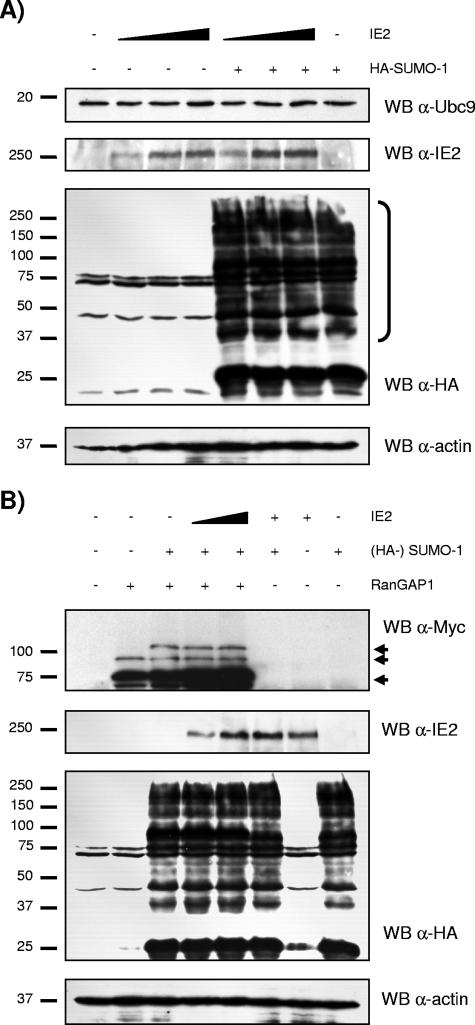
Since interaction with Ubc9 does not lead to sumoylation of IE2 and the sumoylating activity of Ubc9 is not required for the interaction, we next determined whether the IE2-Ubc9 interaction had an impact on the sumoylation pathway in the host cell. For doing so, we first verified whether IE2 could alter the expression of Ubc9. HEK293T cells were transfected with increasing doses of wild-type IE2 and analyzed by Western blotting for endogenous Ubc9 expression. Figure 7A (upper panel) shows that Ubc9 expression remained steady regardless of exogenous IE2 overexpression. Next, to verify whether transfected IE2 could alter the sumoylation pattern of cellular proteins, we cotransfected HEK293T cells with SUMO-1 together with increasing concentrations of IE2. Because the pool of free SUMO-1 available for conjugation in the cell at any given moment is very small (64), we needed to overexpress SUMO-1 in this experiment. Figure 7A (third panel) shows that the sumoylation patterns of control and IE2-transfected cells are very similar, leading us to conclude that the IE2-Ubc9 interaction has no major impact on the cell sumoylation pathway. We also tested a known sumoylation target, RanGAP1, by cotransfecting RanGAP1, HA-tagged SUMO-1, and IE2 constructs. Western blot analysis revealed no significant difference in the levels of sumoylation of RanGAP1 in the presence or absence of exogenous IE2 (Fig. 7B). Therefore, we conclude that the IE2-Ubc9 interaction, rather than the Ubc9 sumoylation activity, modulates the function of IE2.
FIG. 7.
(A) Overexpression of HHV-6A IE2 does not alter Ubc9 expression or the sumoylation pattern of total cellular proteins. HEK293T cells were transfected with 2, 4, or 6 μg pcDNA4-IE2 and 2 μg HA-tagged construct pCMV-SUMO-1. Cells were harvested 48 h after transfection. Total lysates were analyzed by SDS-PAGE and probed for Ubc9, IE2, HA-tagged sumoylated proteins (designated by a bracket), and actin. (B) Overexpression of HHV-6A IE2 does not alter the sumoylation pattern of RanGAP1. HEK293T cells were transfected with 2 or 4 μg pcDNA4-IE2, 2 μg pCMV-SUMO-1, and 2 μg pcDNA3-Myc-RanGAP1. Total lysates were analyzed by SDS-PAGE and probed for RanGAP1 using an anti-Myc antibody. Arrows indicate (from top to bottom) HA-SUMO-1-modified Myc-RanGAP1, endogenous SUMO-1-modified Myc-RanGAP1, and unconjugated Myc-RanGAP1. HA-tagged SUMO-1 is 26 kDa, while endogenous SUMO-1 is 18 kDa.
DISCUSSION
Since many herpesvirus immediate-early proteins, including HHV-6B IE1, have been shown to undergo sumoylation, we were expecting a similar in vivo modification for IE2. To isolate cellular proteins interacting with IE2, we screened, using the yeast two-hybrid system, an expression cDNA library from Jurkat cells. More than half of the interactors identified match human Ubc9, an enzyme involved in the conjugation of SUMO-1 to target proteins (14, 22, 36, 68). SUMO-1 (reviewed in reference 65) is a small protein covalently linked to many cellular and viral targets. Our laboratory previously reported that HHV-6 IE1 is also a substrate of SUMO-1 conjugation (26). The IE2 protein of HHV-6 is able to functionally transactivate multiple promoters (27, 47), and interestingly, promoter transactivation by the moderately similar IE2 protein of HCMV is strongly affected by SUMO-1 conjugation mutants, suggesting a functional relevance for the SUMO-1 conjugation pathway in the biological activity of IE2 (30).
The IE2-Ubc9 interaction site has been mapped in between residues 989 and 1037 of IE2. Analysis of a hydropathicity plot of IE2 indicates that this region is strongly hydrophilic, suggesting an external surface position after protein folding and thus a likely protein interaction site. Although many Ubc9-interacting proteins have been described already, especially assuming that all SUMO-conjugated proteins do interact with Ubc9, no simple motif has been defined for the interacting region. Even though a SUMO-interacting motif has been brought to light (70), it cannot account for sumoylation-independent interactions—with the Ubc9 C93S mutant, for instance.
The in vitro and in vivo SUMO-1 conjugation assays, the lack of IE2-SUMO-1 colocalization in immunofluorescence assays, and the absence of IE2-SUMO-1 interaction in the yeast two-hybrid system suggest that IE2 is not a suitable substrate for sumoylation. No typical ψKXE SUMO-1 acceptor site could be identified in IE2, reinforcing our conclusion that unlike HCMV IE1 and IE2, HHV-6 IE1, and other immediate-early herpesvirus proteins, HHV-6 IE2 does not undergo sumoylation.
Although SUMO-1 conjugation is the principal known function of Ubc9, it was also reported to interact with proteins without leading to SUMO-1 modification. For example, Ubc9 interaction with tumor necrosis factor alpha receptor 1 and kinase MEKK1 leads to upregulation of signal transduction to NF-κB (67). In both cases, no SUMO-1 conjugation was detected, suggesting that Ubc9 is involved in ways other than through SUMO-1 association. Interaction between Ubc9 and importin 13, an importin β-related receptor primarily involved in nuclear import, suggests an involvement for Ubc9 in nuclear translocation of cytoplasmic targets (51). Ubc9 has equally been shown to interact with a nuclear localization signal sequence of homeobox protein Vsx-1, thus mediating Vsx-1 nuclear localization (43). Vsx-1 does not appear to be sumoylated, and a C93S mutant of Ubc9 is still able to restore Vsx-1 nuclear localization in a cell line with a low level of endogenous Ubc9 (43). Ubc9 functions as a corepressor (41) or a coactivator (42) of chicken ovalbumin upstream promoter-transcription factor I as tested on two different promoters, always in a manner distinct from its SUMO-1-conjugating activity. Finally, Ubc9 interaction also suppresses the dinucleoside polyphosphate hydrolase activity of the antitumoral protein Fhit irrespective of the addition of SUMO-1 in the assays (21).
Moreover, in some cases the sumoylating catalytic activity of Ubc9 is dispensable for functional regulation of the target protein, even if the target can be sumoylated. For instance, RNA helicase A is a sumoylation target whose transactivating activity is enhanced by the interaction with Ubc9 independently of the SUMO-1-conjugating activity of Ubc9 (6). Ubc9 also binds and modulates the induction properties of glucocorticoid receptors irrespective of its ability to transfer SUMO-1 to the receptor (38). These examples demonstrate that Ubc9 activities range far beyond its role in sumoylation and suggest that regulation of transactivation is an emerging important function for this cellular protein.
Past publications show that the HCMV IE2 protein is SUMO-1 modified on lysine residues 175 and 180, both described by conjugation consensus motif ψKXE (4, 30). Mutation of both residues (but not either alone) led to a dramatic decrease in transactivation of promoters, including the HIV LTR and HCMV IE1/2 promoters (30), but only a slight decrease in transactivation of Pol and cyclin E promoters (4). Moreover, transactivating activity of wild-type but not conjugation-deficient HCMV IE2 was upregulated by Ubc9 and SUMO-1 cotransfection (4). We describe here a decrease in LTR promoter-transactivating activity of HHV-6 IE2 following interaction with Ubc9. In this case, SUMO-1 conjugation does not appear to be directly involved, and exogenous SUMO-1 does not potentiate promoter transactivation by IE2.
Ubc9 and sumoylation also play a role in nucleocytoplasmic transport (reviewed in reference 59). It is believed that Ubc9 can influence the localization of proteins independently of its enzymatic function, since Ubc9 is carried inside the nucleus by transport receptor importin 13 and could act as an adapter between this receptor and diverse binding partners (51). In the absence of IE2 sumoylation, we hypothesized that the interactions between Ubc9 and IE2 may lead to impaired nuclear transport of the transactivating viral protein. However, our experiments demonstrated that nuclear levels of IE2 remain constant when Ubc9 is overexpressed, ruling out nuclear exclusion as an explanation for the reduced activity of IE2.
Alternatively, IE2 could block conjugation of SUMO-1 to cellular proteins by antagonizing Ubc9 activity, thus affecting cellular pathways where SUMO-1 modification is required for normal operation, such as NF-κB- or RanGAP1-related pathways. Our results do not provide evidence that IE2 alters sumoylation patterns in transfected cells, and sumoylation of one major SUMO-1 target, RanGAP1, was not affected.
Recently, two other cellular protein interaction partners for IE2 have been identified. The N-terminal region of HHV-6B IE2 was found to interact in yeast two-hybrid, pull-down, and coimmunoprecipitation assays with heterogenous nuclear riboprotein K and the beta subunit of casein kinase 2 (69). A potential function of heterogenous nuclear riboprotein K is in pre-mRNA processing (49), and it has been hypothesized that it created a docking platform to facilitate communication among molecules involved in gene expression and signal transduction (8). Casein kinase 2 is a multifunctional, second messenger-independent serine/threonine kinase present in the nucleus and cytoplasm of all eukaryotic cells and composed of catalytic (α) and regulatory (β) subunits (5). The importance of both protein partners in regard to the functionality of IE2 has not been established, so it remains to be seen whether the transactivating activity of IE2 is regulated by interactions within its N-terminal domain.
Since our results show that Ubc9-conjugating activity is not required for the observed repression of IE2-transactivating activity and that IE2 is not conjugated itself, we propose that the IE2-Ubc9 interaction facilitates recruitment of corepressor molecules to the transcription complex formed by IE2 or, alternatively, that Ubc9 itself acts as a repressor to the activity of IE2. This may be a way for the virus or the cell to negatively regulate its immediate-early transactivators once initiation of viral replication is well on its way. Thus, interaction with Ubc9 could represent a new mechanism by which HHV-6 uses cellular pathways, and this study provides valuable knowledge of the biology of this virus. It also sheds light on the importance of the SUMO conjugation pathway to the biology of herpesviruses in general and provides insights on how these viruses utilize the cellular pathways to control the infectious processes.
Acknowledgments
We thank Ronald T. Hay, Stefan Muller, Michel Tremblay, and Manuel Fresno for generously providing DNA constructs and Serge Desnoyers and Sylvain Picard for the use of their microscopy and imaging systems.
This work was supported by grant 14437 from the Canadian Institutes of Health Research (CIHR) to Louis Flamand. Louis Flamand is a senior scholar from the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Ablashi, D. V., N. Balachandran, S. F. Josephs, C. L. Hung, G. R. Krueger, B. Kramarsky, S. Z. Salahuddin, and R. C. Gallo. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184:545-552. [DOI] [PubMed] [Google Scholar]
- 2.Adamson, A. L. 2005. Effects of SUMO-1 upon Epstein-Barr virus BZLF1 function and BMRF1 expression. Biochem. Biophys. Res. Commun. 336:22-28. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, J.-H., W.-J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn, J.-H., Y. Xu, W.-J. Jang, M. J. Matunis, and G. S. Hayward. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75:3859-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allende, J. E., and C. C. Allende. 1995. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9:313-323. [DOI] [PubMed] [Google Scholar]
- 6.Argasinska, J., K. Zhou, R. J. Donnelly, R. T. Hay, and C. G. Lee. 2004. A functional interaction between RHA and Ubc9, an E2-like enzyme specific for Sumo-1. J. Mol. Biol. 341:15-25. [DOI] [PubMed] [Google Scholar]
- 7.Arsenault, S., A. Gravel, J. Gosselin, and L. Flamand. 2003. Generation and characterization of a monoclonal antibody specific for human herpesvirus 6 variant A immediate-early 2 protein. J. Clin. Virol. 28:284-290. [DOI] [PubMed] [Google Scholar]
- 8.Bomsztyk, K., I. Van Seuningen, H. Suzuki, O. Denisenko, and J. Ostrowski. 1997. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 403:113-115. [DOI] [PubMed] [Google Scholar]
- 9.Buschmann, T., S. Y. Fuchs, C. G. Lee, Z. Q. Pan, and Z. Ronai. 2000. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell 101:753-762. [DOI] [PubMed] [Google Scholar]
- 10.Carrigan, D. R., W. R. Drobyski, S. K. Russler, M. A. Tapper, K. K. Knox, and R. C. Ash. 1991. Interstitial pneumonitis associated with human herpesvirus-6 infection after marrow transplantation. Lancet 338:147-149. [DOI] [PubMed] [Google Scholar]
- 11.Chang, L. K., Y. H. Lee, T. S. Cheng, Y. R. Hong, P. J. Lu, J. J. Wang, W. H. Wang, C. W. Kuo, S. S. Li, and S. T. Liu. 2004. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 279:38803-38812. [DOI] [PubMed] [Google Scholar]
- 12.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 13.Desterro, J. M., M. S. Rodriguez, G. D. Kemp, and R. T. Hay. 1999. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274:10618-10624. [DOI] [PubMed] [Google Scholar]
- 14.Desterro, J. M., J. Thomson, and R. T. Hay. 1997. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417:297-300. [DOI] [PubMed] [Google Scholar]
- 15.Dohmen, R. J., A. W. Strasser, C. B. Honer, and C. P. Hollenberg. 1991. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7:691-692. [DOI] [PubMed] [Google Scholar]
- 16.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 18.Flamand, L., J. Gosselin, M. D'Addario, J. Hiscott, D. V. Ablashi, R. C. Gallo, and J. Menezes. 1991. Human herpesvirus 6 induces interleukin-1β and tumor necrosis factor alpha, but not interleukin-6, in peripheral blood mononuclear cell cultures. J. Virol. 65:5105-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flamand, L., F. Romerio, M. S. Reitz, and R. C. Gallo. 1998. CD4 promoter transactivation by human herpesvirus 6. J. Virol. 72:8797-8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giraud, M. F., J. M. Desterro, and J. H. Naismith. 1998. Structure of ubiquitin-conjugating enzyme 9 displays significant differences with other ubiquitin-conjugating enzymes which may reflect its specificity for sumo rather than ubiquitin. Acta Crystallogr. D 54:891-898. [DOI] [PubMed] [Google Scholar]
- 21.Golebiowski, F., A. Szulc, A. Szutowicz, and T. Pawelczyk. 2004. Ubc9-induced inhibition of diadenosine triphosphate hydrolase activity of the putative tumor suppressor protein Fhit. Arch. Biochem. Biophys. 428:160-164. [DOI] [PubMed] [Google Scholar]
- 22.Gong, L., T. Kamitani, K. Fujise, L. S. Caskey, and E. T. Yeh. 1997. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272:28198-28201. [DOI] [PubMed] [Google Scholar]
- 23.Gong, L., B. Li, S. Millas, and E. T. Yeh. 1999. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 448:185-189. [DOI] [PubMed] [Google Scholar]
- 24.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gravel, A., V. Dion, N. Cloutier, J. Gosselin, and L. Flamand. 2004. Characterization of human herpesvirus 6 variant B immediate-early 1 protein modifications by small ubiquitin-related modifiers. J. Gen. Virol. 85:1319-1328. [DOI] [PubMed] [Google Scholar]
- 26.Gravel, A., J. Gosselin, and L. Flamand. 2002. Human herpesvirus 6 immediate-early 1 protein is a sumoylated nuclear phosphoprotein colocalizing with promyelocytic leukemia protein-associated nuclear bodies. J. Biol. Chem. 277:19679-19687. [DOI] [PubMed] [Google Scholar]
- 27.Gravel, A., A. Tomoiu, N. Cloutier, J. Gosselin, and L. Flamand. 2003. Characterization of the immediate-early 2 protein of human herpesvirus 6, a promiscuous transcriptional activator. Virology 308:340-353. [DOI] [PubMed] [Google Scholar]
- 28.Hermiston, T. W., C. L. Malone, and M. F. Stinski. 1990. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J. Virol. 64:3532-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodges, M., C. Tissot, and P. S. Freemont. 1998. Protein regulation: tag wrestling with relatives of ubiquitin. Curr. Biol. 8:R749-R752. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iniguez, M. A., S. Martinez-Martinez, C. Punzon, J. M. Redondo, and M. Fresno. 2000. An essential role of the nuclear factor of activated T cells in the regulation of the expression of the cyclooxygenase-2 gene in human T lymphocytes. J. Biol. Chem. 275:23627-23635. [DOI] [PubMed] [Google Scholar]
- 32.Isegawa, Y., T. Mukai, K. Nakano, M. Kagawa, J. Chen, Y. Mori, T. Sunagawa, K. Kawanishi, J. Sashihara, A. Hata, P. Zou, H. Kosuge, and K. Yamanishi. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 73:8053-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishiguro, N., S. Yamada, T. Takahashi, Y. Takahashi, T. Togashi, T. Okuno, and K. Yamanishi. 1990. Meningo-encephalitis associated with HHV-6 related exanthem subitum. Acta Paediatr. Scand. 79:987-989. [DOI] [PubMed] [Google Scholar]
- 34.Janelle, M. E., A. Gravel, J. Gosselin, M. J. Tremblay, and L. Flamand. 2002. Activation of monocyte cyclooxygenase-2 gene expression by human herpesvirus 6. Role for cyclic adenosine monophosphate responsive element binding protein and activator protein-1. J. Biol. Chem. 277:30665-30674. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, E. S., and G. Blobel. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147:981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson, E. S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272:26799-26802. [DOI] [PubMed] [Google Scholar]
- 37.Kamitani, T., K. Kito, H. P. Nguyen, H. Wada, T. Fukuda-Kamitani, and E. T. Yeh. 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273:26675-26682. [DOI] [PubMed] [Google Scholar]
- 38.Kaul, S., J. A. Blackford, Jr., S. Cho, and S. S. Simons, Jr. 2002. Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J. Biol. Chem. 277:12541-12549. [DOI] [PubMed] [Google Scholar]
- 39.Kim, Y. H., C. Y. Choi, and Y. Kim. 1999. Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc. Natl. Acad. Sci. USA 96:12350-12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knox, K. K., and D. R. Carrigan. 1995. Active human herpesvirus (HHV-6) infection of the central nervous system in patients with AIDS. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:69-73. [PubMed] [Google Scholar]
- 41.Kobayashi, S., H. Shibata, I. Kurihara, K. Yokota, N. Suda, I. Saito, and T. Saruta. 2004. Ubc9 interacts with chicken ovalbumin upstream promoter-transcription factor I and represses receptor-dependent transcription. J. Mol. Endocrinol. 32:69-86. [DOI] [PubMed] [Google Scholar]
- 42.Kurihara, I., H. Shibata, S. Kobayashi, N. Suda, Y. Ikeda, K. Yokota, A. Murai, I. Saito, W. E. Rainey, and T. Saruta. 2005. Ubc9 and protein inhibitor of activated STAT 1 activate chicken ovalbumin upstream promoter-transcription factor I-mediated human CYP11B2 gene transcription. J. Biol. Chem. 280:6721-6730. [DOI] [PubMed] [Google Scholar]
- 43.Kurtzman, A. L., and N. Schechter. 2001. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc. Natl. Acad. Sci. USA 98:5602-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapenta, V., P. Chiurazzi, P. van der Spek, A. Pizzuti, F. Hanaoka, and C. Brahe. 1997. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics 40:362-366. [DOI] [PubMed] [Google Scholar]
- 45.Lawrence, G. L., M. Chee, M. A. Craxton, U. A. Gompels, R. W. Honess, and B. G. Barrell. 1990. Human herpesvirus 6 is closely related to human cytomegalovirus. J. Virol. 64:287-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee, H. R., and J. H. Ahn. 2004. Sumoylation of the major immediate-early IE2 protein of human cytomegalovirus Towne strain is not required for virus growth in cultured human fibroblasts. J. Gen. Virol. 85:2149-2154. [DOI] [PubMed] [Google Scholar]
- 47.Martin, M. E., J. Nicholas, B. J. Thomson, C. Newman, and R. W. Honess. 1991. Identification of a transactivating function mapping to the putative immediate-early locus of human herpesvirus 6. J. Virol. 65:5381-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matunis, M. J., W. M. Michael, and G. Dreyfuss. 1992. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 12:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melchior, F. 2000. Sumo nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 51.Mingot, J. M., S. Kostka, R. Kraft, E. Hartmann, and D. Gorlich. 2001. Importin 13: a novel mediator of nuclear import and export. EMBO J. 20:3685-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neipel, F., K. Ellinger, and B. Fleckenstein. 1991. The unique region of the human herpesvirus 6 genome is essentially collinear with the UL segment of human cytomegalovirus. J. Gen. Virol. 72:2293-2297. [DOI] [PubMed] [Google Scholar]
- 54.Nevels, M., W. Brune, and T. Shenk. 2004. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 78:7803-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicholas, J. 1994. Nucleotide sequence analysis of a 21-kbp region of the genome of human herpesvirus-6 containing homologues of human cytomegalovirus major immediate-early and replication genes. Virology 204:738-750. [DOI] [PubMed] [Google Scholar]
- 56.Okuma, T., R. Honda, G. Ichikawa, N. Tsumagari, and H. Yasuda. 1999. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem. Biophys. Res. Commun. 254:693-698. [DOI] [PubMed] [Google Scholar]
- 57.Papanikolaou, E., V. Kouvatsis, G. Dimitriadis, N. Inoue, and M. Arsenakis. 2002. Identification and characterization of the gene products of open reading frame U86/87 of human herpesvirus 6. Virus Res. 89:89-101. [DOI] [PubMed] [Google Scholar]
- 58.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 59.Pichler, A., and F. Melchior. 2002. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381-387. [DOI] [PubMed] [Google Scholar]
- 60.Pizzorno, M. C., P. O'Hare, L. Sha, R. L. LaFemina, and G. S. Hayward. 1988. trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez, M. S., J. M. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saito, Y., L. R. Sharer, S. Dewhurst, B. M. Blumberg, C. B. Hall, and L. G. Epstein. 1995. Cellular localization of human herpesvirus-6 in the brains of children with AIDS encephalopathy. J. Neurovirol. 1:30-39. [DOI] [PubMed] [Google Scholar]
- 64.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 65.Saitoh, H., R. T. Pu, and M. Dasso. 1997. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem. Sci. 22:374-376. [DOI] [PubMed] [Google Scholar]
- 66.Salahuddin, S. Z., D. V. Ablashi, P. D. Markham, S. F. Josephs, S. Sturzenegger, M. Kaplan, G. Halligan, P. Biberfeld, F. Wong-Staal, B. Kramarsky, and R. C. Gallo. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596-601. [DOI] [PubMed] [Google Scholar]
- 67.Saltzman, A., G. Searfoss, C. Marcireau, M. Stone, R. Ressner, R. Munro, C. Franks, J. D'Alonzo, B. Tocque, M. Jaye, and Y. Ivashchenko. 1998. hUBC9 associates with MEKK1 and type I TNF-alpha receptor and stimulates NFkappaB activity. FEBS Lett. 425:431-435. [DOI] [PubMed] [Google Scholar]
- 68.Schwarz, S. E., K. Matuschewski, D. Liakopoulos, M. Scheffner, and S. Jentsch. 1998. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA 95:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shimada, K., K. Kondo, and K. Yamanishi. 2004. Human herpesvirus 6 immediate-early 2 protein interacts with heterogeneous ribonucleoprotein K and casein kinase 2. Microbiol. Immunol. 48:205-210. [DOI] [PubMed] [Google Scholar]
- 70.Song, J., L. K. Durrin, T. A. Wilkinson, T. G. Krontiris, and Y. Chen. 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA 101:14373-14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stanton, R., J. D. Fox, R. Caswell, E. Sherratt, and G. W. Wilkinson. 2002. Analysis of the human herpesvirus-6 immediate-early 1 protein. J. Gen. Virol. 83:2811-2820. [DOI] [PubMed] [Google Scholar]
- 72.Sternsdorf, T., K. Jensen, and H. Will. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139:1621-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tong, H., G. Hateboer, A. Perrakis, R. Bernards, and T. K. Sixma. 1997. Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system. J. Biol. Chem. 272:21381-21387. [DOI] [PubMed] [Google Scholar]
- 74.Wang, Z. Y., Q. Q. Qiu, W. Seufert, T. Taguchi, J. R. Testa, S. A. Whitmore, D. F. Callen, D. Welsh, T. Shenk, and T. F. Deuel. 1996. Molecular cloning of the cDNA and chromosome localization of the gene for human ubiquitin-conjugating enzyme 9. J. Biol. Chem. 271:24811-24816. [DOI] [PubMed] [Google Scholar]
- 75.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamanishi, K., T. Okuno, K. Shiraki, M. Takahashi, T. Kondo, Y. Asano, and T. Kurata. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet 1:1065-1067. [DOI] [PubMed] [Google Scholar]
- 77.Yasugi, T., and P. M. Howley. 1996. Identification of the structural and functional human homolog of the yeast ubiquitin conjugating enzyme UBC9. Nucleic Acids Res. 24:2005-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yasukawa, M., Y. Yakushijin, M. Furukawa, and S. Fujita. 1993. Specificity analysis of human CD4+ T-cell clones directed against human herpesvirus 6 (HHV-6), HHV-7, and human cytomegalovirus. J. Virol. 67:6259-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshikawa, T., S. Suga, Y. Asano, T. Nakashima, T. Yazaki, R. Sobue, M. Hirano, M. Fukuda, S. Kojima, and T. Matsuyama. 1991. Human herpesvirus-6 infection in bone marrow transplantation. Blood 78:1381-1384. [PubMed] [Google Scholar]
- 80.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]