Abstract
If we conduct the same experiment in two laboratories or repeat a classical study many years later, will we obtain the same results? Recent research with mice in neural and behavioral genetics yielded different results in different laboratories for certain phenotypes, and these findings suggested to some researchers that behavior may be too unstable for fine-scale genetic analysis. Here we expand the range of data on this question to additional laboratories and phenotypes, and, for the first time in this field, we formally compare recent data with experiments conducted 30–50 years ago. For ethanol preference and locomotor activity, strain differences have been highly stable over a period of 40–50 years, and most strain correlations are in the range of r = 0.85–0.98, as high as or higher than for brain weight. For anxiety-related behavior on the elevated plus maze, on the other hand, strain means often differ dramatically across laboratories or even when the same laboratory is moved to another site within a university. When a wide range of phenotypes is considered, no inbred strain appears to be exceptionally stable or labile across laboratories in any general sense, and there is no tendency to observe higher correlations among studies done more recently. Phenotypic drift over decades for most of the behaviors examined appears to be minimal.
Keywords: agonistic behavior, anxiety, ethanol preference, gene–environment interaction, locomotor activity
If we conduct the same experiment in two laboratories or repeat a classical study many years later, will we obtain the same results? Recent research with mice in neural and behavioral genetics yielded somewhat different results in different laboratories for certain phenotypes (1–4), and these findings evoked what Pfaff (5) termed an “old stereotype,” i.e., that behavior may be too unstable for fine-scale genetic analysis. Here we expand the range of data on this question to additional laboratories and phenotypes, and, for the first time in this field, we formally compare recent data with experiments conducted 30–50 years ago. We find that several kinds of behavior are genetically quite stable.
It is now common practice to backcross targeted mutations in mice onto the same standard, inbred strain background in different laboratories. If the background genotype on which mutations are placed does not behave consistently across laboratories, our understanding of the effects of gene manipulations on behaviors may be compromised. Comprehensive information on a wide range of inbred strain characteristics is now being compiled in the Mouse Phenome Database (6) in a manner that facilitates comparisons among different phenotypes studied in different laboratories. Many investigators are now using gene expression information for brain tissue of recombinant inbred strains from the Gene Network (www.genenetwork.org) to compare phenotypes across strains (7). What constitutes a high between-trait strain correlation depends critically on the magnitude of the strain correlation for the same trait measured in different laboratories or at different times.
To repeat a genetic experiment, the genotype of the animals must be replicable. Brother-by-sister mating can achieve genetic purity in an inbred strain after 60 generations (8). Derivation of standard strains of mice commenced with the DBA strain in 1909, BALB in 1913, and C57BL in 1921 (9). By 1950, many strains maintained by The Jackson Laboratory (Bar Harbor, ME) had undergone >40 generations of full sibling mating. Today, when two laboratories obtain samples of the same strain, they can be confident that the genotypes are almost identical, but genetic contamination can occur when rigorous inbreeding is not practiced (10–12). Consequently, in this analysis we compare only data reported for the same substrains from one supplier, the The Jackson Laboratory (laboratory code J), in almost all cases.
Only simultaneous study of identical tests given to identical mouse strains can evaluate replicability in the strict sense, and any differences in results must arise from the inevitable difference between the environments in the research facilities external to the test situation itself. Comparisons of reasonably similar tests given to nearly identical strains in different years in different laboratories, however, evaluate the robustness of behavioral test results to methodological differences, including small genetic differences, that exist between most published reports. Details of the test situation usually differ to some extent between laboratories (13), and the field is principally concerned with this category.
We selected for historical comparisons studies based on previously published reports of tests given to inbred strains from The Jackson Laboratory. First, they had to provide data on at least five of the same substrains examined by at least one of our four laboratories. Second, they had to provide sufficient information in the published article that we could determine the sample size, mean score, and standard deviation for each strain, so that an unweighted-means ANOVA could be performed to compare studies (14). Third, they had to measure essentially the same behavior on a scale that allowed meaningful comparisons.
Results
Brain Weight.
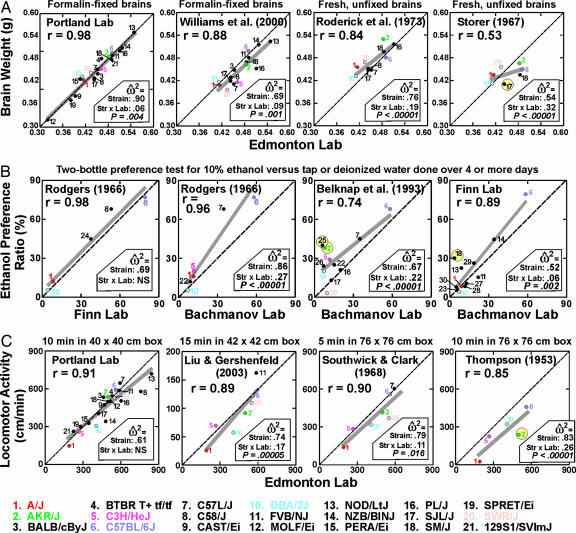
Unlike behavior, weight of a dead and fixed brain does not fluctuate from moment to moment or day to day, and it is measured on the same scale in grams in all studies. Fig. 1A compares data collected in the Edmonton laboratory with strain means in four other data sets. ANOVAs comparing all possible pairs of data sets and details of methods for measuring brains in each study are provided in Tables 1 and 2 and Supporting Text, which are published as supporting information on the PNAS web site. The Edmonton and Portland strain means were very similar indeed (r = 0.98), and average weights across all strains were almost identical at the two sites (0.452 and 0.456 g, respectively) (15). Methods of fixation were very similar in the Williams (16) study, and the correlation of Edmonton data with their strain means was very high (r = 0.88). The two older studies (17, 18) both weighed unfixed brains, and fresh brain weights are expected to be slightly larger than fixed brain weights, but there is no reason to expect major strain-dependent differences in shrinkage (19).The similarity between the Edmonton data and the Roderick et al. means collected 30 years earlier on the same strains (17) was very high (r = 0.84), whereas the Storer (18) and Edmonton data were notably less similar (r = 0.54).
Fig. 1.
Correlations between means of inbred strains observed in different laboratories. The dashed line indicates identical results for two laboratories, and the gray line is the best fit of data from the y axis to data on the x axis, plotted for the actual range of data. When this line is above the dashed line, the means scores in laboratory Y were generally greater than for the corresponding strains tested in laboratory X. Numbers 1–21 correspond to strains shown at the bottom of the figure, with the most common strains shown as colored numerals. Effect sizes from the ANOVAs are shown for the strain main effect and strain-by-laboratory interaction. The significance (P) of the interaction effect is also indicated. NS denotes an interaction not significant at P = 0.05. (A) Brain weight measured in the Edmonton laboratory versus four other laboratories. Data for Edmonton and Portland were collected in 2002 and published in ref. 15. (B) Preference for 10% ethanol solution versus tap water. Nine additional strains not studied in the D.W. and J.C.C. laboratories were included in this set of comparisons (22, BALB/cJ; 23, BUB/BnJ; 24, C57BLKS/J; 25, CBA/J; 26, CE/J; 27, I/LnJ; 28, LP/J; 29, RIIIS/J; 30, SEA/GnJ). (C) Locomotor activity in Edmonton versus four other laboratories, scaled to centimeters per minute.
There were some discrepancies between older and more recent data. The clearly greater brain size of C57BL/6J than A/J in our study and the Williams study was not seen in the Roderick et al. (17) and Storer (18) studies. On the other hand, the tight clustering of means for strains AKR/J, C57BL/6J, and SM/J was seen in all four studies spanning a range of 35 years. Strain SWR/J brains were considerably heavier (by 46 mg) than strain SJL/J in the Storer data, in contrast with the other three studies. Results from the ANOVAs (Table 1) reveal that the strain-by-laboratory interaction was in all but one case significant, yet it was substantial by our criteria (interaction effect size more than half the strain main effect size; see Methods) only for the comparisons of the Storer data (18) with brain weights from the Wahlsten laboratory (15).
Ethanol Preference.
The remarkable preference of the C57BL/6J mouse strain for a 10% ethanol solution versus tap water and the distaste of DBA/2J for ethanol was originally discovered in 1959 (20), and the technique of measuring ethanol preference was thoroughly explored by Fuller (21). In 1966 Rodgers (22) used the two-bottle preference test to document a wide range of preference scores for 15–19 inbred strains, and another large survey of 15 strains was reported 27 years later by Belknap et al. (23). The older data are compared here with two new inbred strain surveys done by A.B. (28 strains) and D.A.F. (22 strains) (Table 3, which is published as supporting information on the PNAS web site). We present data from each study where a two-bottle preference test of 10% ethanol versus tap water was done with bottle position changed systematically over a period of at least 4 days. Methods of testing preference are described in detail in Table 4, which is published as supporting information on the PNAS web site. The results of D.A.F. could not be compared with Belknap et al. (23) because the two studies included only two identical substrains (C57BL/6J and DBA/2J). The strain correlations among the four studies done 30 or more years apart are impressive indeed (Fig. 1B and Table 1) and for three data sets versus Rodgers (22) are slightly greater than the strain correlations seen for brain weight in independent studies. A considerable number of other studies with fewer inbred strains has been published over the years, and, to our knowledge, the high ethanol preference of C57BL/6J versus the pronounced aversion to ethanol of DBA/2J has been confirmed in every report. Thus, this behavioral difference is highly robust with respect to laboratory environments and the fine details of methods used to assess preference.
Although strain correlations are generally very high, the ANOVAs (Table 1) point to several significant strain-by-laboratory interactions. These two indicators of replicability are not in conflict, because the interaction effects are invariably much smaller that the massive main effects of strain. Nevertheless, closer inspection of strain distributions (Fig. 1B) reveals some noteworthy differences for certain pairs of strains. If an investigator works with only a few inbred strains and happens to include one or two that are especially sensitive to conditions of rearing and/or testing, these kinds of interaction effects could undermine replicability.
Locomotor Activity.
Depending on the test conditions, locomotion is used to index complex traits such as general health, exploratory drive, novelty seeking, anxiety, and the potential future preference for drugs of abuse. The first large survey of 15 inbred strains in 1953 by Thompson (24) involved a simple count by a human of the lines crossed by the mouse in a box divided into 5 × 5-inch squares. Fifteen years later, Southwick and Clark (25) used the same size enclosure but counted entries into 6 × 6-inch squares. Fifty years after Thompson, Liu and Gershenfeld (26) used a computer to monitor beam breaks of photocells spaced 2.5 cm apart, whereas the J.C.C. and D.W. laboratories used video tracking at 25 frames per second. Further details of the test in all laboratories are specified in Table 5, which is published as supporting information on the PNAS web site. Results are compared in Fig. 1C and Table 1. To aid comparisons among studies, we converted all data to a common measurement scale, centimeters traveled per minute.
Fig. 1C and Table 1 indicate that results of studies done 40–50 years apart are remarkably similar. Every laboratory found that A/J was the least active and C57BL/6J was among the most active strains, and the same result has been reported in virtually every other published study involving fewer strains. In only one comparison was the correlation notably lower and the interaction term really large relative to the strain main effect: Liu and Gershenfeld (26) versus Thompson (24). Thompson (24) found large differences between C3H/HeJ and A/J and between BALB/cJ and C57BL/6J, whereas Liu and Gershenfeld (26) observed only minor differences between these strain pairs. This was not a consequence of the great time span between the studies, because the Thompson data (24) were highly correlated with results of Southwick and Clark (25) as well as our own very recent data.
Elevated Plus Maze.
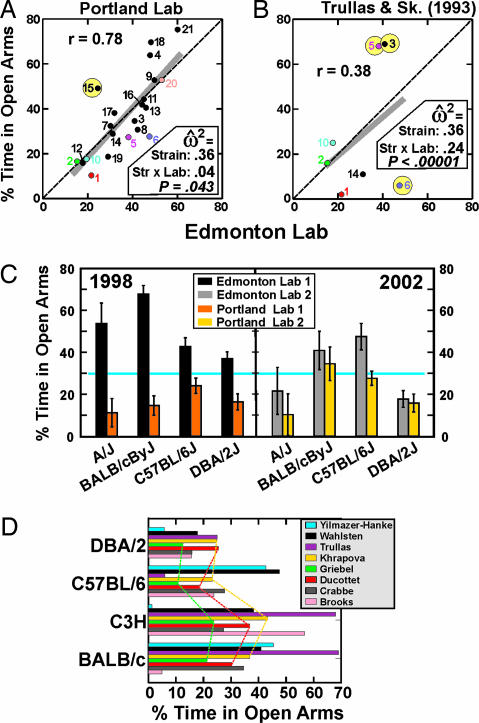
Anxiety or anxiety-like behavior, as assessed by the elevated plus maze (27, 28), is very sensitive to environmental variation. In our previous study of eight mouse strains on the elevated plus maze in three laboratories, average anxiety-like behavior levels were much lower in Edmonton than Portland for almost all strains (1), even though we used physically identical apparatus and test protocols. Five years later we ran the same test in Edmonton and Portland using the same mazes and protocols from the previous study, expanding the sample to 21 inbred strains. This time, our two laboratories obtained very similar results (Fig. 2A). ANOVA indicated very large differences among strains, no significant laboratory difference, and a very small strain-by-laboratory interaction (Table 1).
Fig. 2.
Percent time spent exploring the two open arms of the elevated plus maze. Lines, effect sizes, and strain numbers in A and B have the same meanings as in Fig. 1. (A) For data in the Edmonton and Portland laboratories, the value for the wild-derived inbred strain PERA/Ei (number 15 in yellow circle) in Portland was considerably greater than in Edmonton, but other means were generally very similar for the two laboratories. (B) There were remarkable discrepancies for the strains BALB/cByJ (no. 3), C3H/HeJ (no. 5), and C57BL/6J (no. 6) in the Edmonton laboratory versus the study of Trullas and Skolnick (30). (C) Four of the same strains were tested in 1998 (1) and again in 2002 as part of the Mouse Phenome Project using the same apparatus and protocols as in 1998, but the actual laboratories had moved to new locations in each university, resulting in substantially different data, especially in Edmonton, where every strain was above 30% (blue line) in 1998. (D) Four inbred strains were tested recently in eight laboratories, indicated in the figure by the surname of the first author; complete references (30–35) are provided in the text. The substrains were not identical across all laboratories shown in D. Dashed lines show that the profile of strain means was very similar for three studies [Ducottet and Belzung (32), Griebel et al. (33), and Khrapova et al. (34)], but remarkable differences between other laboratories are evident.
Four of the strains in the current study were also tested in our earlier study (1). As shown in Fig. 2C, open arm exploration increased markedly for strain BALB/cByJ in Portland, whereas it decreased considerably for strains A/J, BALB/cByJ, and DBA/2J in Edmonton. The apparatus and protocols were the same, but laboratory environments changed considerably at both sites. The colony and testing rooms in Edmonton were moved to another wing of the same building, into an area where several other mammalian species were housed in a bustling, centralized animal facility, whereas the testing laboratory in Portland was moved from a large facility where many mouse studies were being done at the same time to a more isolated location in another building where the mice for this study were housed in the same room with the test apparatus.
The elevated plus maze test was originally devised for use with rats and later was adapted for use with mice (29), then applied to several inbred strains by Trullas and Skolnick (30). Our physical maze and procedures and those used by Trullas and Skolnick (30) were almost the same (Table 6, which is published as supporting information on the PNAS web site), because we found their version to be serviceable and therefore copied it. The results of our current study and the Trullas and Skolnick (30) study were drastically different for strains C3H/HeJ, BALB/cByJ, and C57BL/6J (Fig. 2B), and the strain-by-laboratory interaction was large relative to the main effect of strain.
Studies with 6–11 inbred strains on the elevated plus maze have been reported from several laboratories (31–35), but unfortunately the number of specific substrains in common with our experiment or the study of Trullas and Skolnick (30) was four or fewer in every instance. By treating substrains from different suppliers as effectively the same strain, it was possible to compute correlations involving three to six strains in common. Mean scores for four strains that were examined in all eight laboratories are shown in Fig. 2D. Three studies yielded strain means that were positively correlated for four to six strains in common (32–34), whereas the results of Yilmazer-Hanke et al. (35) were negatively correlated with each of these three studies but positively correlated with the data from Edmonton and Portland.
Many factors can influence mouse anxiety (36). Consequently, the experimenter needs to choose test parameters very carefully. At the same time, the great similarity of apparatus and procedures in the synchronized Edmonton and Portland experiments and the independent test by Trullas and Skolnick (30) did not yield similar data (Fig. 2B). Furthermore, we could detect no major differences or similarities in methods, as described in Table 6, that could account for the patterns of strain correlations among the eight studies (Fig. 2D). This suggests that variations in the local laboratory environments had important influences on the manifestation of mouse anxiety.
Other Behaviors.
Agonistic behavior of male mice appears to be strongly influenced by rearing and testing conditions (37). Two early studies found opposite rank orderings of two strains (38, 39), an interaction that Ginsburg (40) later attributed to forceps handling of young mice before weaning in one of the laboratories. Maxson et al. (41) demonstrated Y chromosome effects on agonistic behavior and their interaction with genetic background (41), but the consomic line differences vanished when the strains were moved into a specific pathogen-free laboratory, a gene–environment interaction that was eventually traced to the acidified water (37). Selective breeding is very effective in creating large line differences if the males are reared in isolation, but the line difference largely disappears when they are instead reared in groups (42–44). Recent inbred strain surveys (45–48) used very different test methods and/or different substrains, and they differed substantially in many details from the 1968 multiple-strain comparison by Southwick and Clark (25). The available data did not warrant a formal statistical comparison between studies, and we could not evaluate the stability of scores over decades.
A number of multiple strain surveys of behavior have been done recently and are reported in the Mouse Phenome Database (www.jax.org/phenome). At the present time, however, the Mouse Phenome Database includes few instances where the same phenotype was examined by different laboratories. We have compared data on the accelerating rotarod and water escape learning in two laboratories, but our refined versions were considerably different from previous tasks. In the future, it will be informative to assess replicability by carefully copying the apparatus and protocols of earlier studies.
Discussion
It is evident from these comparisons among studies that inbred strain differences for ethanol preference and locomotor activity are highly replicable across laboratories and even decades, whereas strain differences in elevated plus maze exploration are strongly dependent on local conditions. Data on agonistic behavior suggest that it is also very sensitive to pretesting environment, but stability over decades cannot yet be fairly judged. Thus, there is no basis for viewing mouse behavior as inherently unstable or unsuitable for genetic analysis in any general sense. Some of the classical strain differences in behavior that were discovered 40–50 years ago are highly robust and still evident today. Other kinds of tests are more labile. These findings do not imply that tests of anxiety and agonistic interactions are bad tests. On the contrary, the tests are very sensitive to genuine individual and strain differences in the constructs they are designed to assess, but the processes underlying those constructs are also very sensitive to environmental conditions.
The data do not indicate that any one strain is especially stable or labile across a wide range of behavioral phenotypes. Certain extreme strain differences on certain tasks, such as C57BL/6J versus DBA/2J on ethanol preference and C57BL/6J versus A/J on motor activity, are highly replicable over laboratories and decades, but those strains show considerable variation for other phenotypes such as anxiety-related and agonistic behaviors. C57BL/6J quickly develops obesity and diabetes on a high-fat diet (49) and as a neonate is easily primed by loud noise so that it later suffers audigenic seizures (50). C57BL/6J serves as a good choice when backcrossing a new mutation onto a standard strain background, not because it is markedly stable phenotypically, but because it breeds reasonably well and is so widely used as a de facto standard in different laboratories.
Comparisons of current and classical data suggest that minor genetic changes over several decades have been of little consequence for mouse behavioral testing and, ipso facto, demonstrate the robustness of carefully conducted behavioral assays. It appears that substrain differentiation is generally not a major threat to replicability of behavioral data over several decades, provided the substrains are formed after the root strain has already been inbred for many generations. A study of 1,638 single-nucleotide polymorphisms observed that differences among several C57BL/6 substrains represented residual heterozygosity at the time substrains were separated as well as new mutations (51). Allelic differences at 12 of 342 microsatellite loci were noted between the C57BL/6J and C57BL/6NTac substrains that were separated in 1951, >150 generations ago (52). A C57BL/6J mouse from The Jackson Laboratory today will not be identical nucleotide-for-nucleotide to the same strain 50 years ago. It will nevertheless be far more similar to its own ancestor in 1950 than to substrains separated early in the history of a major strain, for example C57BL/10, C57L, and C58 (51), or to another substrain separated around 1950.
All of the phenotypes examined here rank as highly complex traits. Quantitative trait locus analyses have indicated influences of multiple loci for brain weight (16) and for the behavioral traits studied (53). A minor change in one or two of many pertinent genes should not shift the mean score of a specific strain appreciably when it is tested several decades later. There does not seem to be any tendency in the data reviewed here for strains to differ more substantially when the studies were conducted many years apart. For example, the ethanol preference data of A.B. were more similar to those of Rodgers (22) from 40 years earlier than those of Belknap et al. (23) just 10 years earlier. Although the Thompson (24) activity data were not highly correlated with the data from Liu and Gershenfeld (26), they were very similar to more recent data from the Edmonton laboratory.
The importance of many fine details of husbandry and testing is recognized on the basis of carefully controlled experiments conducted within a single laboratory. Phytoestrogens in the diet (54), cage enrichment (4), cage position in the colony room (55), size of the drinking spout orifice (56), shipping before testing (57), and even the specific experimenter administering the test (4, 58) have been found to affect laboratory mouse physiology and behavior. Nevertheless, just as a single gene usually accounts for little variance on its own, it is highly unlikely that a multidimensional laboratory difference in behavioral data can be traced to a single environmental variant. A small environmental effect might be evident within a carefully controlled experiment, whereas the same effect might not be audible amidst the cacophony of multiple laboratory-specific parameters in rearing and testing.
The sample of phenotypes in this systematic comparison of recent and classical data sets is not sufficient to warrant strong conclusions about what kinds of behavior should be most and least robust across laboratories. Very tentatively, we suggest that things more closely associated with sensory input and motor output will tend to be less affected by minor variants in the laboratory environment, whereas behaviors related to emotional and social processes will be more labile. A similar classification regarding degree of genetic influence has been proposed on theoretical grounds by Lipp (59).
Methods
Animals and Laboratories.
Data on brain weight, open field activity, and elevated plus maze were collected simultaneously in the J.C.C. laboratory in Portland and the D.W. laboratory in Edmonton, whereas data on ethanol preference were collected in the A.B. and D.A.F. laboratories as separate studies done within a few months of each other. We all obtained the animals from The Jackson Laboratory at 4–6 weeks of age. The D.W. and J.C.C. laboratories evaluated the same 21 inbred strains, consisting of priority lists A and B of the Mouse Phenome Project (129S1/SvImJ, A/J, AKR/J, BALB/cByJ, C3H/HeJ, C57BL/6J, C57L/J, C58/J, CAST/EiJ, DBA/2J, FVB/NJ, MOLF/EiJ, NOD/LtJ, NZB/BlNJ, PERA/EiJ, PL/J, SJL/J, SM/J, SPRET/EiJ, and SWR/J) plus the strain BTBR T+ tf/J from list D (www.jax.org/phenome), which has an interesting but viable loss of forebrain commissures (15). The A.B. laboratory studied 28 strains, and the D.A.F. laboratory studied 22 strains (Table 3), 10 of which were common to at least two studies but were in addition to those studied by the J.C.C. and D.W. laboratories (129P3/J, BALB/cJ, BUB/BnJ, C57BL/KsJ, CBA/J, CE/J, I/LnJ, LP/J, RIIIS/J, and SEA/GnJ). No attempt was made to equate the housing conditions in the four laboratories, but conditions were nevertheless quite similar, as described in Table 7, which is published as supporting information on the PNAS web site.
Data Analysis.
There currently is no standard in the field for what size a strain correlation denotes a bona fide replication of results. Instead, we rely strongly on close inspection of the data by those having extensive experience with a particular kind of test. Strictly speaking, replication of results in two laboratories requires a large strain difference but no strain-by-laboratory interaction effect. The magnitude of a strain difference can be expressed as partial ω2, the proportion of variance attributable to the differences among strain means when laboratory differences are removed from the equation. The partial ω2 can also be estimated for an interaction effect (1). If ω2 for an interaction effect is considerably less than ω2 for a strain main effect, then we consider the interaction to be relatively small, even if its statistical significance (P value) is beyond reproach. Likewise, to be considered large or substantial in a strain-by-laboratories study, the interaction effect size should be at least half the strain main effect size when data are compared across two laboratories.
Supplementary Material
Acknowledgments
We thank Naomi Yoneyama, Andrea Wetzel, Maria Theodorides, Sue Burkhart-Kasch, Janet D. Dorow, Jason R. Sibert, Jason P. Schlumbohm, Charlotte D. Wenger, Chia-Hua Yu, Pamela Metten, Brandie Moisan, Sean F. Cooper, Tera Mosher, Tim Frigon, and Elizabeth Munn for assistance in collecting the data. This work was supported in part by National Institutes of Health grants to D.W. (Grant AA12714), A.B. (Grant AA11028), J.C.C. (Grant AA10270 and Integrative Neuroscience Initiative on Alcoholism Consortium Grant AA13519), and D.A.F. (Integrative Neuroscience Initiative on Alcoholism Consortium Grants AA134785, AA10760, AA12439, and AA13478); Department of Veterans Affairs grants (to J.C.C. and D.A.F.); and Natural Sciences and Engineering Research Council Grant 45825 (to D.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The data reported in this paper have been deposited in the Mouse Phenome Database, www.jax.org/phenome (accession no. MPD108).
References
- 1.Crabbe JC, Wahlsten D, Dudek BC. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- 2.Crestani F, Martin JR, Möhler H, Rudolph U. Nat Neurosci. 2000;3:1059. doi: 10.1038/80553. [DOI] [PubMed] [Google Scholar]
- 3.Kafkafi N, Benjamini Y, Sakov A, Elmer GI, Golani I. Proc Natl Acad Sci USA. 2005;102:4619–4624. doi: 10.1073/pnas.0409554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewejohann L, Reinhard C, Schrewe A, Brandewiede J, Haemisch A, Gortz N, Schachner M, Sachser N. Genes Brain Behav. 2006;5:64–72. doi: 10.1111/j.1601-183X.2005.00140.x. [DOI] [PubMed] [Google Scholar]
- 5.Pfaff D. Proc Natl Acad Sci USA. 2001;98:5957–5960. doi: 10.1073/pnas.101128598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grubb SC, Churchill GA, Bogue MA. Bioinformatics. 2004;20:2857–2859. doi: 10.1093/bioinformatics/bth299. [DOI] [PubMed] [Google Scholar]
- 7.Chesler EJ, Lu L, Shou SM, Qu YH, Gu J, Wang JT, Hsu HC, Mountz JD, Baldwin NE, Langston MA, et al. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 8.Green EL. Genetics and Probability in Animal Breeding Experiments. New York: Oxford Univ Press; 1981. [Google Scholar]
- 9.Beck JA, Lloyd S, Hafexparast M, Lennon-Pierce M, Eppig JT, Festing MFW, Fisher EMC. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 10.Kahan B, Auerbach R, Alter BJ, Bach FH. Science. 1982;217:379–381. doi: 10.1126/science.6953593. [DOI] [PubMed] [Google Scholar]
- 11.Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 12.Threadgill DW, Yee D, Matin A, Nadeau JH, Magnuson T. Mamm Genome. 1997;8:441–442. doi: 10.1007/s003359900464. [DOI] [PubMed] [Google Scholar]
- 13.Wahlsten D, Metten P, Phillips TJ, Boehm SL, II, Burkhart-Kasch S, Dorow J, Doerksen S, Downing C, Fogarty J, Hen R, et al. J Neurobiol. 2003;54:283–311. doi: 10.1002/neu.10173. [DOI] [PubMed] [Google Scholar]
- 14.Winer BJ, Brown DR, Michels KM. Statistical Principles in Experimental Design. 3rd Ed. New York: McGraw-Hill; 1991. [Google Scholar]
- 15.Wahlsten D, Metten P, Crabbe JC. Brain Res. 2003;971:47–54. doi: 10.1016/s0006-8993(03)02354-0. [DOI] [PubMed] [Google Scholar]
- 16.Williams RW. In: Mouse Brain Development. Goffinet AM, Rakic P, editors. New York: Springer; 2000. pp. 21–49. [Google Scholar]
- 17.Roderick TH, Wimer RE, Wimer C, Schwartzkroin PA. Brain Res. 1973;64:345–353. doi: 10.1016/0006-8993(73)90188-1. [DOI] [PubMed] [Google Scholar]
- 18.Storer JB. Exp Gerontol. 1967;2:173–182. [Google Scholar]
- 19.Wahlsten D, Hudspeth WJ, Bernhardt K. J Comp Neurol. 1975;162:519–532. doi: 10.1002/cne.901620408. [DOI] [PubMed] [Google Scholar]
- 20.McClearn GE, Rodgers DA. Q J Stud Alcohol. 1959;20:691–695. [Google Scholar]
- 21.Fuller JL. J Comp Physiol Psychol. 1964;57:85–88. doi: 10.1037/h0043100. [DOI] [PubMed] [Google Scholar]
- 22.Rodgers DA. Psychosom Med. 1966;28:498–513. doi: 10.1097/00006842-196601000-00005. [DOI] [PubMed] [Google Scholar]
- 23.Belknap JK, Crabbe JC, Young ER. Psychopharmacology. 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- 24.Thompson WR. Can J Psychol. 1953;7:145–155. doi: 10.1037/h0083586. [DOI] [PubMed] [Google Scholar]
- 25.Southwick CH, Clark LH. Commun Behav Biol. 1968;1:49–59. [Google Scholar]
- 26.Liu X, Gershenfeld HK. Brain Res Bull. 2003;60:223–231. doi: 10.1016/s0361-9230(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 27.Belzung C, Griebel G. Behav Brain Res. 2001;125:141–149. doi: 10.1016/s0166-4328(01)00291-1. [DOI] [PubMed] [Google Scholar]
- 28.Rodgers RJ. Behav Pharmacol. 1997;8:477–496. doi: 10.1097/00008877-199711000-00003. [DOI] [PubMed] [Google Scholar]
- 29.Lister RG. Psychopharmacology. 1987;92:180–185. doi: 10.1007/BF00177912. [DOI] [PubMed] [Google Scholar]
- 30.Trullas R, Skolnick P. Psychopharmacology. 1993;111:323–331. doi: 10.1007/BF02244948. [DOI] [PubMed] [Google Scholar]
- 31.Brooks SP, Pask T, Jones L, Dunnett SB. Genes Brain Behav. 2005;4:307–317. doi: 10.1111/j.1601-183X.2004.00109.x. [DOI] [PubMed] [Google Scholar]
- 32.Ducottet C, Belzung C. Behav Brain Res. 2005;156:153–162. doi: 10.1016/j.bbr.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Griebel G, Belzung C, Perrault G, Sanger DJ. Psychopharmacology. 2000;148:164–170. doi: 10.1007/s002130050038. [DOI] [PubMed] [Google Scholar]
- 34.Khrapova MV, Popova NK, Avgustinovich DF. Zh Vyssh Nerv Deiat Im I P Pavlova. 2001;51:324–328. [PubMed] [Google Scholar]
- 35.Yilmazer-Hanke DM, Roskoden T, Zilles K, Schwegler H. Behav Brain Res. 2003;145:145–159. doi: 10.1016/s0166-4328(03)00107-4. [DOI] [PubMed] [Google Scholar]
- 36.Hogg S. Pharmacol Biochem Behav. 1996;54:21–30. doi: 10.1016/0091-3057(95)02126-4. [DOI] [PubMed] [Google Scholar]
- 37.Maxson SC. In: Techniques for the Genetic Analysis of Brain and Behavior: Focus on the Mouse. Goldowitz D, Wahlsten D, Wimer RE, editors. Amsterdam: Elsevier; 1992. pp. 349–373. [Google Scholar]
- 38.Ginsburg B, Allee WC. Physiol Zool. 1942;15:485–506. [Google Scholar]
- 39.Scott JP. J Hered. 1942;33:11–15. [Google Scholar]
- 40.Ginsburg BE. In: Stimulation in Early Infancy. Ambrose JA, editor. New York: Academic; 1969. pp. 73–96. [Google Scholar]
- 41.Maxson SC, Ginsburg BE, Trattner A. Behav Genet. 1979;9:219–226. doi: 10.1007/BF01071302. [DOI] [PubMed] [Google Scholar]
- 42.Lagerspetz KM, Lagerspetz KY. Scand J Psychol. 1971;12:241–248. doi: 10.1111/j.1467-9450.1971.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 43.Hood KE, Cairns RB. Aggress Behav. 1989;15:361–380. [Google Scholar]
- 44.Nyberg J, Sandnabba K, Schalkwyk L, Sluyter F. Genes Brain Behav. 2004;3:101–109. doi: 10.1111/j.1601-183x.2003.0056.x. [DOI] [PubMed] [Google Scholar]
- 45.Roubertoux PL, Le Roy I, Mortaud S, Perez-Diaz F, Tordjman S. In: Handbook of Molecular-Genetic Techniques for Brain and Behavior Research. Crusio WE, Gerlai RT, editors. Amsterdam: Elsevier Science; 1999. pp. 696–709. [Google Scholar]
- 46.Kulikov AV, Osipova DV, Naumenko VS, Popova NK. Genes Brain Behav. 2005;4:482–485. doi: 10.1111/j.1601-183X.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- 47.Guillot PV, Chapouthier G. Behav Brain Res. 1996;77:211–213. doi: 10.1016/0166-4328(95)00163-8. [DOI] [PubMed] [Google Scholar]
- 48.Tordjman S, Carlier M, Cohen D, Cesselin F, Bourgoin S, Colas-Linhart N, Petiet A, Perez-Diaz F, Hamon M, Roubertoux PL. Behav Genet. 2003;33:529–536. doi: 10.1023/a:1025774716976. [DOI] [PubMed] [Google Scholar]
- 49.Surwit RS, Kuhn CM, Cochrane C, Mccubbin JA, Feinglos MN. Diabetes. 1988;37:1163–1167. doi: 10.2337/diab.37.9.1163. [DOI] [PubMed] [Google Scholar]
- 50.Henry KR, Bowman RE. J Comp Physiol Psychol. 1970;70:235–241. doi: 10.1037/h0028715. [DOI] [PubMed] [Google Scholar]
- 51.Petkov PM, Ding YM, Cassell MA, Zhang WD, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, et al. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bothe GWM, Bolivar VJ, Vedder MJ, Geistfeld JG. Genes Brain Behav. 2004;3:149–157. doi: 10.1111/j.1601-183x.2004.00064.x. [DOI] [PubMed] [Google Scholar]
- 53.Flint J. J Neurobiol. 2003;54:46–77. doi: 10.1002/neu.10161. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Tranguch S, Xie H, Hanley G, Das SK, Dey SK. Proc Natl Acad Sci USA. 2005;102:9960–9965. doi: 10.1073/pnas.0501632102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Izidio GS, Lopes DM, Spricigo L, Jr, Ramos A. Genes Brain Behav. 2005;4:412–419. doi: 10.1111/j.1601-183X.2005.00121.x. [DOI] [PubMed] [Google Scholar]
- 56.Dotson CD, Spector AC. Physiol Behav. 2005;85:655–661. doi: 10.1016/j.physbeh.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 57.Tordoff MG, Alarcon LK, Byerly EA, Doman SA. Physiol Behav. 2005;86:480–486. doi: 10.1016/j.physbeh.2005.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chesler EJ, Wilson SG, Lariviere WR, Rodriguez-Zas SL, Mogil JS. Nat Neurosci. 2002;5:1101–1102. doi: 10.1038/nn1102-1101. [DOI] [PubMed] [Google Scholar]
- 59.Lipp H-P. Behav Processes. 1995;35:19–33. doi: 10.1016/0376-6357(95)00037-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.