Abstract
Two mAbs generated against rhodopsin kinase (RK) were characterized for their epitopes. Both antibodies recognize short peptide sequences, overlapping but distinct, close to the carboxyl terminus. Binding of RK to the antibodies is slow. Attempts were made to use the antibodies immobilized on protein A-Sepharose beads to bind and purify the enzyme. Time-dependent inactivation of the enzyme occurred after its binding to the antibodies. Studies using different conditions to maintain the enzyme in the active form during binding or to reactivate the purified inactivated enzyme were unsuccessful.
Keywords: G-protein-coupled receptor kinases, epitope mapping, surface plasmon resonance, protein folding, immunoaffinity purification
Because of our interest in studying the mechanism of activation of rhodopsin kinase (RK) on binding to the light-activated rhodopsin (1, 2), we have investigated systems for the expression of the RK gene and methods for purification of the expressed enzyme (3). Concurrently with this work, we undertook preparation and characterization of mAbs against RK for their potential use in immunoaffinity purification as well as in structure–function studies.§ In this report, we describe two mAbs specific for RK and our attempts to use them in purification of the enzyme. The antibodies recognize short overlapping amino acid sequences near the carboxyl terminus. They bind RK slowly without its prior denaturation. RK expressed in COS-1 cells bound to the antibodies immobilized by crosslinking to protein A on Sepharose beads. The binding resulted in time-dependent inactivation of the enzyme. Extensive attempts to stabilize the enzyme during binding to the antibodies and to reactivate the resulting pure inactive enzyme have been unsuccessful so far. Thus, the carboxyl-terminal segment is required for stabilization of the folded active enzyme. Its binding to an antibody promotes folding to an alternative inactive form of the enzyme.
Materials and Methods
These were as in the accompanying paper (3) with the following additions.
Materials.
The mAbs against RK were generated at the Hybridoma Core Facility (Mount Sinai Medical Center, New York). The peptides in RK (Table 1) and the carboxyl-terminal rhodopsin undecapeptide were synthesized at the Biopolymers Laboratory (Massachusetts Institute of Technology). Cell growth media were: media 1 and 2 (3) and medium 3, HB-GRO (Irvine Scientific) supplemented with 1% FBS.
Table 1.
Synthetic peptides corresponding to the carboxyl-terminal sequences in RK
| Peptide | Amino terminal | Amino acid sequence in RK | Carboxyl terminal |
|---|---|---|---|
| Trideca | 527 | DLNVWRPDGQMPDD | 540 |
| Dodeca | 528 | LNVWRPDGQMPDD | 540 |
| Undeca | 529 | NVWRPDGQMPDD | 540 |
| Deca | 530 | VWRPDGQMPDD | 540 |
| Nona | 531 | WRPDGQMPDD | 540 |
| Octa | 532 | RPDGQMPDD | 540 |
| Hepta | 533 | PDGQMPDD | 540 |
The buffers were: buffer A, 25 mM Tris, pH 7.5/250 mM NaCl/5 mM EDTA/1% Triton X-100; buffer B, 10 mM 1,3-bis[Tris(hydroxymethyl)methylamino]propane (BTP), pH 7.5/0.05% n-dodecyl β-d-maltoside (DM)/150 mM KCl; buffer C, 10 mM BTP, pH 7.5/0.05% DM/250 mM KCl; buffer D, 10 mM BTP, pH 7.5/150 mM CKl/0.05% DM; buffer E, 20 mM BTP, pH 7.5/5 mM MgCl2.
In RK reactivation experiments, the buffers used were: buffer 1, 50 mM bicine, pH 6/5 mM DTT/4% polyethylene glycol (PEG), 75 mM ammonium acetate/0.4% MEGA-8; buffer 2, 100 mM sodium acetate, pH 5.5/1.1 M ammonium sulfate/10 mM DTT/10% glycerol/0.05% DM; buffer 3, 5 mM Mes-Bis-Tris, pH 6.8/75 mM LiCl/0.1 mM EDTA/1 mM DTT/1.5 mM MEGA-8; buffer 4, 10 mM Hepes, pH 7.4/15 mM NaCl/25 mM ammonium acetate/5% PEG/1 mM DTT/0.05% DM; buffer 5, 40 mM Hepes, pH 7.4/200 mM NaCl/5 mM DTT/17.5% ammonium sulfate/0.05% DM; buffer 6, 100 mM Tris, pH 7.5/18% PEG/100 mM KCl/1 mM DTT/0.05% DM; buffer 7, 32.2 mM Tris, pH 7.5/10% PEG/50 mM ammonium sulfate/5 mM CaCl2/2.5 mM MgCl2/1 mM DTT/0.05% DM; buffer 8, 50 mM Mes, pH 5.9/0.2 M ammonium sulfate/18% PEG/5 mM DTT/0.05% DM; buffer 9, 100 mM Hepes, pH 7.4/0.2 M sodium citrate/1 mM DTT/0.05% DM; control, 10 mM BTP, pH 7.5/150 mM KCl/1 mM DTT/0.05% DM. These buffers were used with or without, (i) 100 μM ATP/2 mM MgCl2, (ii) 100 μM rhodopsin carboxyl-terminal undecapeptide/50 μM mastoparan, and (iii) 0.2 M urea.
Methods.
Growth of the hybridoma cell lines and purification of the antibodies.
The hybridoma cell lines were grown to confluence in 15-cm dishes containing 25 ml of medium 3. The total supernate (400 ml) from the culture medium was made 3 M in NaCl, the pH was adjusted to 8.8 with NaOH and, after 1 h at room temperature, the solution was filtered (0.45-μm pore). The filtrate was applied to a protein A- Sepharose column (4 ml, diameter 1 cm) at 0.3 ml/min flow rate. After a wash with 20 bed volumes of PBS (pH 8.8) + 3 M NaCl, the antibodies were eluted with 0.1 M citric acid (pH 4) + 150 mM NaCl in three 3.5-ml fractions. The eluates were neutralized to pH 7 with NaHCO3 and stored at −20°C.
Purification of synthetic peptides.
Crude synthetic peptides (25 mg) in water (400 μl) were applied to a C-18 HPLC column (20 ml). Elution was with a two-step linear acetonitrile gradient; Step 1, 2–40% in 160 ml, and Step 2, 40–70% in 20 ml, both containing 0.1% trifluoroacetic acid. The flow rate was 2–4 ml/min. The nona- and undecapeptides eluted at about 35% acetonitrile concentration. The eluates were lyophilized and the peptides dissolved in water (600 μl). The concentrations of the peptides (one tryptophan each) were calculated from absorption at 280 nm.
Crosslinking of mAb-6D8 to protein A-Sepharose.
Protein A-Sepharose (2 ml, 50% settled beads) preequilibrated in phosphate buffer, pH 8.5, was mixed with purified mAb-6D8 (5 mg in citrate buffer, pH 7) in PBS (15 ml), pH 9. The antibody/protein A complex formation was monitored by depletion of absorption at 280 nm in the supernates of aliquots. Binding of the mAb to protein A was 95% in 1 h. The crosslinking reaction was carried out according to Schneider et al. (4) by using dimethyl suberimidate. The mAb-6D8-protein A-Sepharose matrix contained about 2 mg mAb-6D8/ml beads.
Cloning and expression of RK gene deletion mutants.
The deletion mutants were made by site-directed mutagenesis by using appropriate primers containing the desired mutation and a BamHI or HindIII restriction site. The primers and the pCMV5-RK vector (3) were used in amplification reactions to generate the DNA fragments containing the RK deletion mutants. The fragments after digestion with BamHI and HindIII were subcloned into the Escherichia coli expression vector pQE16 (Qiagen, Chatsworth, CA) under the control of the lac promoter. The resulting constructs were used to transform XL1-Blue E. coli cells (5). The transformants were grown for 3 h in LB containing 50–100 μg/ml ampicillin and 15–30 μg/ml tetracycline. The expression of the RK mutant genes was induced by 0.5 mM isopropyl β-d-thiogalactopyranoside, the cells being grown for 3 h. After centrifugation, the cells were lysed in Laemmli sample buffer. The proteins were separated by SDS/PAGE, and responses of the mutants to mAb-6D8 or mAb-1C3 were analyzed by immunoblotting.
Identification of the epitopes for mAb-6D8 and mAb-1C3 by using synthetic peptides.
Peptides (0.5 nmol each) in water were incubated in buffer A (150 μl) with 0.25 nmol of purified antibody (37.5 μg) and 20 μl of protein A-Sepharose (50% beads). After mutation at 4°C overnight, the samples were pelleted, and the amounts of peptides in the supernates were determined by reverse-phase HPLC. The peptides were eluted by using a two-step linear acetonitrile gradient containing 0.1% trifluoroacetic acid (Step 1, 2–45% in 20 ml; Step 2, 45–75% in 5 ml) (flow rate, 1 ml/min).
Purification of RK from Transfected COS-1 Cells.
Step 1 was on a heparin–Sepharose column (3).
Step 2: Chromatography on mAb-6D8–protein A-Sepharose.
The pooled fractions from Step 1 were applied to a mAb-6D8-protein A-Sepharose column (2 ml, 8 mm diameter) preequilibrated with buffer B, the effluent being recycled 2.8 ml/h for 16 h at 4°C. The column was washed four times with 1 ml of buffer C and 1 ml of buffer B. RK was eluted in 1-ml fractions by using buffer B containing 0.5 mM of the dodecapeptide (Table 1). The flow was reversed during elution resulting in sharpening of the elution peak.
Immunoprecipitation of RK.
Protein extract from COS-1 cells (50 μl, 0.03–0.05 nmol RK) was mixed with mAb-6D8 (40–200 μg) and 100 μl of protein A-Sepharose beads in buffer D (total volume, 250 μl). After incubation at room temperature for 30 min, 1 h or 2 h, the beads were pelleted, and the supernatant was analyzed by SDS/PAGE and immunoblotting with mAb-6D8. The formation of mAb-6D8-protein A complex was complete within 30 min.
Antibody-mediated inactivation of RK.
RK (about 0.7 pmol) purified on heparin–Sepharose (3) was incubated in sets of three tubes with mAb-6D8 (50-fold molar excess) in buffer D. The tridecapeptide (Table 1) (0.6 mM) was added to the third tube. The samples (6 μl) were incubated at 4°C for 30 min or 5 h. The tridecapeptide (0.6 mM) was then added to tube no. 4, and the samples were treated in the dark with 4 μl of 100 μM [γ-32P]ATP (1,000–2,000 cpm/pmol) plus 50 μM rod outer segments in buffer E (3).
Experiments on stabilization of RK during interaction with the antibodies.
RK from heparin–Sepharose (3) (28 μl, about 0.6 μg) was mixed with 10 μl of mAb-6D8-protein A-Sepharose beads (2 mg/ml) in each well of a 96-well microplate. Solutions representing 12 separate groups of reagents were added (10 μl) to the 96 wells, and the samples were incubated for 5 h at 4°C. The beads were then collected by centrifugation and washed twice with 250 μl of the corresponding solution. Five microliters of each supernate was applied to a nitrocellulose filter by using a microdot system, and the amount of unbound RK was quantitated after immunoblotting by phosphoimaging. Known amounts of purified RK served to calibrate the signal. RK was eluted in 30 μl of the corresponding solutions supplemented with 250 μM of the dodecapeptide (Table 1). Aliquots from each well were used in assay of RK activity.
Attempted reactivation of RK after immunopurification.
In 40 wells of a 96-well microplate, heparin-purified RK samples (40 μl) were mixed with 40 μl of mAb-6D8-protein A-Sepharose beads (2 mg/ml). After 2 h at 4°C, 38 μl of the solution was removed from each well, and the beads were washed with 200 μl of buffer E. The beads were then washed twice with 200 μl of one of the 40 “reactivation” buffers (see Materials); the beads were collected by centrifugation between each wash. Elution of RK from the beads was performed over 30 min in 40 μl of each one of the “reactivation” buffers containing 250 μM of dodecapeptide (Table 1). Aliquots (20 μl) from each well were transferred to new wells in two fresh 96-well microplates, and the plates were put in a humid chamber at 4°C and 16°C, respectively. After 5, 24, and 45 h of incubation, 4 μl from each well was used in RK activity assay. After 24 h, 5 μl of the samples containing urea were transferred to microdialysers, and dialysis was performed for 24 h, at 4°C or 16°C, against the corresponding buffers containing no urea.
Surface plasmon resonance.
The instrument used was BIACORE 3000 (BIACORE). Rabbit anti-mouse IgG Fc (RamFc) antibody (BIACORE) was immobilized onto a CM5 sensor chip surface via amine coupling. RK antibody was then captured onto the RamFc surface. RK(His)6 (3) was incubated at 25°C for 45 min before use. Experiments were at 25°C in buffer D. A 10-mM HCl solution was used for surface regeneration at 2 × 0.5 min. The kinetic data were analyzed by using biaevaluation 3.0 software (BIACORE).
Results
MAbs 6D8 and 1C3.
Two hydribomas producing the anti-RK antibodies, 6D8 and 1C3, were isolated from mice immunized with RK (6). When grown in 15-cm dishes, the hybridomas produced 35–50 μg of the antibodies per milliliter of culture supernatant. The antibodies were purified as in Methods.
mAb-6D8 and mA-1C3 are Specific to RK in Immunoblotting.
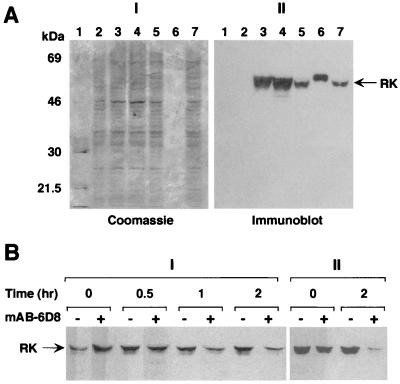
COS-1 cells producing RK (Methods) were solubilized in Laemmli sample buffer, and the proteins were separated on SDS/PAGE and visualized by Coomassie blue staining and by immunoblotting with mAb-6D8 (Fig. 1A). Of the total proteins in the extract (Fig. 1AI), only one band corresponding to RK (64 kDa) was observed on the immunoblot (Fig. 1AII). The same result was obtained on immunoblotting with mAb-1C3 (not shown).
Figure 1.
(A) Specificity of mAb-6D8 to RK. Transfected COS-1 cells were solubilized in Laemmli sample buffer and subjected to SDS/PAGE (5 × 104 cells per lane). (I). The proteins were transferred to a poly(vinylidene difluoride) membrane that was immunoprobed by using mAb-6D8 (II) and later stained with Coomassie blue (I). Lane 1, molecular weight markers; lane 2, mock transfection; lanes 3–5, cells transfected with pCMV5-RK (three independent transfections); lane 6, 125 ng of purified RK from baculovirus-infected insect cells; lane 7, 125 ng of purified RK mixed with the mock transfected cell extract. (B) Binding of mAb-6D8 to RK. RK was incubated with protein A-Sepharose beads and with (+) or without (−) a 10-fold (I) or 50-fold (II) excess of mAb-6D8 (Methods). At different time intervals, the RK-mAb-6D8-protein A-Sepharose complex was collected by centrifugation, and the presence of RK in the supernatant was analyzed by immunoblotting.
mAb-6D8 and mAb-1C3 Bind to Native RK.
Immunoprecipitation of RK by using the total protein extract from COS-1 cells producing RK was carried out with mAb-6D8 (Methods). Two concentrations of the antibody, 10-fold (Fig. 1AI) and 50-fold excess (Fig. 1AII) relative to RK were used. As seen in Fig. 1BI, RK remaining in solution decreased as a function of time. However, the binding to the antibody was not complete in 2 h. By using 50-fold excess of the antibody (Fig. 1BII), the binding was essentially complete at 2 h. Thus, although native RK bound mAb-6D8, binding was slow.
Kinetics and Stability of RK Binding to mAb-1C3 and mAb-6D8 by Using Surface Plasmon Resonance.
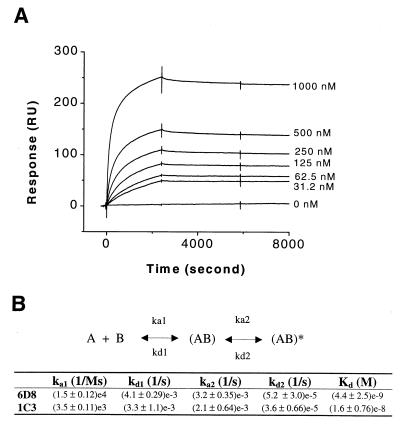
mAb-1C3 or mAb-6D8 was captured, in an oriented manner, onto a RamFc-coated surface (Methods); the complexes formed were stable under the conditions used below for the antibody–kinase interaction studies. As shown in Fig. 2A for mAb-6D8, binding kinetics were slow and, therefore, data were collected at 25°C over prolonged periods for both the association and dissociation phases. The kinetics of RK binding to both antibodies exhibited a double exponential pattern and were analyzed by using a two-state model. This model suggests that a conformational change occurs after the formation of the initial binding complex (Fig. 2B). The selection of this model as a minimal mechanism to describe the present data is supported by accompanying results of the binding studies.
Figure 2.
Kinetic characterization of the binding of RK to mAb-6D8 and mAb-1C3 by using the surface plasmon resonance technique. (A) Sensorgram showing the binding of RK and mAb-6D8. After stabilization of the baseline for 5 min after capturing the antibody onto the RamFc surface, RK was injected, and the binding of RK to the antibody was monitored. Different concentrations of RK used are indicated in the sensorgram. A control surface that contained only RamFc was used. (B) The kinetic data were analyzed by using a two-state model. In this model, (AB) and (AB)* are two different conformations of the binding complexes. Duplicate experiments were run for each RK–antibody binding interaction. The averaged value for each rate constant, together with the standard error, is shown.
The overall affinity constants (7) obtained for mAb-6D8 and mAb-1C3 were 4.4 × 10−9 M and 1.6 × 10−8 M, respectively (Fig. 2B). This 4-fold difference in the affinity values is largely because of the difference in the association rate constant for the formation of the initial complex.
mAb-6D8 and mAb-1C3 Epitopes in the RK Sequence.
(i) Localization by using deletion mutants To determine the location of the epitopes for mAb-6D8 and mAb-1C3, the reactivity of the antibodies to a series of RK deletion mutants was examined. RK gene deletion mutants (Fig. 3A) were constructed and expressed in E. coli (Methods). The observed immunoreactivity profiles of the mutants to the antibodies are shown in Fig. 3B. The large amino-terminal deletion did not affect the response to either of the two antibodies, whereas the carboxyl-terminal deletions, ΔC84, ΔC61, ΔC44, ΔC36, and ΔC29, all abolished response to both antibodies. However, the deletion mutant, ΔC22, gave a positive response to both the antibodies. The results suggested that the epitopes of both antibodies are either located between the deletion sites ΔC29 and ΔC22 or are centered around the ΔC29 deletion site.
Figure 3.
Deletion mutants of RK and their responses to mAb-6D8 and mAb-1C3. (A) The three domains in RK sequence are shown (Top). The seven deletion mutants (I–VII) are shown with the deleted sequences in black. (B) Immunoresponses of the mutants to the antibodies. The mutant proteins were produced in E. coli and tested for their reactivity toward mAb-6D8 and mAb-1C3, as described in Methods; responses are shown by a plus or minus sign.
Mapping of the Epitopes by Using Synthetic Peptides.
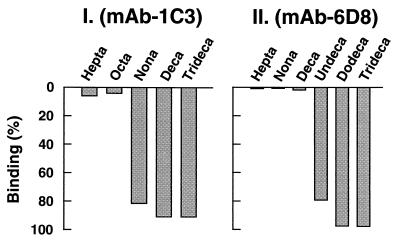
The individual peptides (Table 1) were incubated with complexes of mAb-6D8 or mAb-1C3 with protein A-Sepharose beads (Methods). After centrifugation, the supernates were analyzed for peptides by HPLC. Decreases in the peptide peaks, compared with the control, would indicate binding of the peptides to the antibodies. As seen in Fig. 4I for mAb-1C3, the nonapeptide was the shortest length that bound RK. For mAb-6D8 (Fig. 4II), the undecapeptide bound RK, whereas the decapeptide did not. Thus, the epitopes for the two antibodies are distinct but overlapping. More precise determination of the epitope boundaries at the carboxyl terminus was not carried out.
Figure 4.
Mapping of the mAb-6D8 and mAb-1C3 epitopes by using synthetic peptides. Individual peptides (Table 1) were incubated in the presence of antibody and protein-A Sepharose beads as in Methods. After centrifugation, the presence of peptides in the supernates was analyzed by HPLC. The binding of each peptide to either antibody was measured as the decrease in the peptide peak intensity relative to parallel controls in the absence of antibody. The extents of binding are shown in bars as percent of the control.
Immunoaffinity Chromatography Yields Pure but Inactive RK by Using the mAbs.
The two-step procedure described below gave pure RK (>90% homogeneous) in inactive state. Of the two antibodies, mAb-6D8 gave better results, presumably because of its higher affinity for RK (Fig. 2B).
Step 1: Chromatography on Heparin–Sepharose.
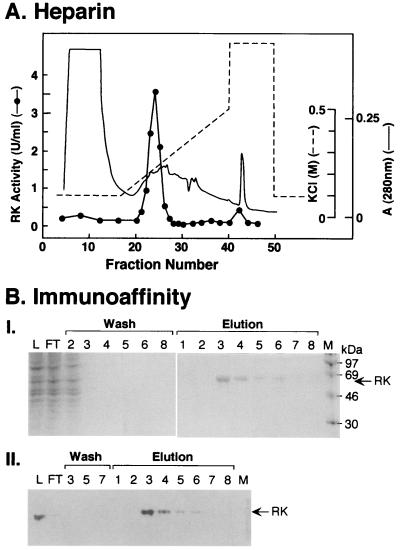
The protein extract from transfected COS-1 cells (1.2 × 108) was applied to a heparin–Sepharose column (Methods). The separation obtained is shown in Fig. 5A. RK activity eluted as a single peak centered at 0.3 M salt. Purification was about 7-fold with 70% recovery.
Figure 5.
Purification of RK from transfected COS-1 cells. (A) Chromatography on heparin–Sepharose. RK purification procedure from transiently transfected COS-1 cells was as in Methods. Elution profiles show absorption at 280 nm KCl, concentration and the RK activity. (B) Chromatography on mAb-6D8-protein A-Sepharose. Fractions 22–25 from A were pooled and applied to an immunoaffinity column as in Methods. Proteins from the loading sample (L), the flow-through (FL), the high-salt wash (wash 2–4), the low-salt wash (wash 5, 6, 8), and the elution fractions (elution 1–8) were analyzed by SDS/PAGE (Coomassie staining) (I). Selected fractions were analyzed by immunoblotting by using mAb-6D8 (II; M, molecular weight markers).
Step 2: Chromatography on mAb-6D8/Protein A-Sepharose.
Pooled fractions from Step 1 were applied to the immunoaffinity column (Methods). About 85% of RK activity bound to the column. No activity was detected in washes with 4 ml of high salt and 4 ml of low salt. RK eluted as a sharp peak at two bed volumes of the eluant (Fig. 5B). The amount of RK recovered was about 70 μg. No activity was detected in the eluted RK.
Time-Dependent Inactivation of RK on Interaction with the mAbs.
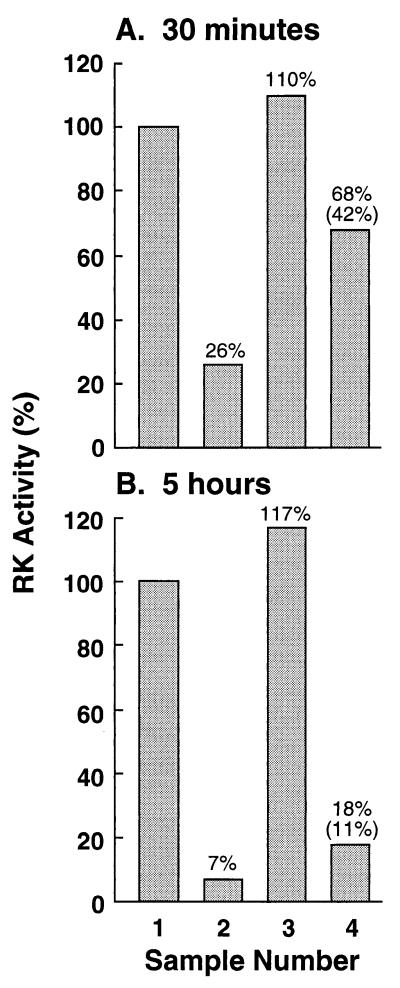
RK was incubated with mAb-6D8 as described in Methods. After 30 min, 26% of the activity remained (Fig. 6A, lane 2; lane 1 is the control). If the tridecapeptide was added at the beginning of the incubation, no loss of activity was observed (Fig. 6A, lane 3). When the tridecapeptide was added at the end of the 30-min incubation, a substantial amount of RK activity (42%; Fig. 6A, lane 4) was observed. On incubation of RK with mAb-6D8 for 5 h (Fig. 6B), 7% of RK activity survived (lane 2). Addition of the tridecapeptide at the beginning again gave full protection of the activity (lane 3). Addition of the tridecapeptide at the end of the incubation resulted in only a small (11%) recovery of the activity (lane 4). Under the conditions used, binding of RK to mAb-6D8 was 93% complete after 5 h. Thus, inactivation of RK when complexed with mAb-6D8 was time dependent.
Figure 6.
Time-dependent inactivation of RK as complexed with mAb-6D8. RK was incubated in the presence of mAb-6D8 (lanes 2–4) as in Methods. The tridecapeptide (Table 1) was absent (lane 2), present throughout (lane 3), or added at the end (lane 4) of the incubation time. After 30 min or 5 h of incubation, RK activity was measured. The results are normalized to the control experiments in the absence of mAb (lane 1).
Attempts to Stabilize the Active Form of RK and to Reactivate Purified Inactive RK.
Destabilization of protein structure during purification, leading to partial or complete loss of activity, is observed often. The presence or addition of certain chemical components to the solubilization medium may help stabilization of the active form. Purified inactive or denatured proteins may refold to an active state under certain conditions (8). Several kinases have been purified or crystallized (9), and conditions have been described for their stabilization. These include formation of secondary or tertiary complexes, addition of polyhydroxyl compounds, detergents, or lipids, and the use of different pH or salt conditions.
(i) Conditions Influencing the Binding of RK to mAb-6D8 and the Stability of RK During Purification.
Stabilization of RK during its interaction with mAb-6D8 was explored first. Following the literature, more than 90 different conditions were tried, which represented 12 different groups of agents (Table 2). Both the kinetics of binding of RK to mAb-6D8 and the kinase activity after purification were measured. Binding kinetics were slowed in the presence of some detergents, lipids, sugars, proteins, polar solvents (such as ethanol), or salts, but were not significantly affected by other agents. However, none of the agents tried prevented the inactivation of RK during its interaction with the antibodies (Table 2). The small protection provided by some agents resulted from slowing down the binding of RK to mAb-6D8 (Fig. 6A).
Table 2.
Conditions influencing the kinetics of binding of RK to mAb-6D8 and stability of RK during purification
| Agents | Extent of binding to mAb-6D8, % | Active RK recovered, % |
|---|---|---|
| mAb–minus | 0 | 100 |
| mAb–plus | 100 | 2 |
| Detergents (DM, Tween 80, cholate, OG, CHAPS*) | 60–100 | 2–21 |
| Phospholipids (asolectin phosphatidyl choline) | 70–75 | 10–19 |
| Sugars (polyethylene glycol, hexanediol, glycerol, adonitol) | 40–80 | 2–19 |
| Peptides/proteins (rhodopsin terminal undecapeptide, BSA, cytochrome C) | 50–70 | 5–13 |
| ATP | 100 | 2 |
| Reducing agents (DTT) | 80 | 6–15 |
| Denaturing agents (urea, GuHCl) | 100 | 2–4 |
| Metal-ions (Zn2+, Cd2+, Ni2+) | 100 | 2–5 |
| Amino acids (phosphoserine, phosphothreonine) | 100 | 2–3 |
| Alcohols (ethanol, propanol, butanol) | 70–90 | 3–28 |
| Di-tricarboxylic acids (acetate, citrate, tartrate, cacodylate) | 100 | 2–10 |
| Salts (CaCl2, MgCl2, NaCl, NH4, Li2SO4,Na-phosphate, K-phosphate) | 50–90 | 3–25 |
The experiments were carried out as in Fig. 6. The basic buffer (10 mM BTP, pH 7.5/150 mM KCl, 0.05% DM) contained additions belonging to one of the groups listed above. The binding of RK to mAb-6D8 was measured as in Fig. 6 (lane 2), and the effects of the groups of agents studied on the extent of RK binding to mAb-6D8 are shown as a percent of the “mAb control” sample. The amounts of active RK recovered after disruption of the antigen/mAb complex by the antipeptide were measured as in Fig. 6 (lane 4) and are shown as percentages of control (100%) in the absence of mAb-6D8.
*3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate.
(ii) Attempts to Reactivate Purified Inactive RK.
Freshly purified inactive RK was incubated under various conditions (Methods) with additional components for stabilization (sugars, detergents) or destabilization (urea). RK was stabilized by complexing it with ATP, the carboxyl-terminal tail peptide of rhodopsin, and with mastoparan, a peptide known to activate RK. The “reactivation” experiments were carried out at two temperatures (4°C and 16°C), and RK activity was measured after 5, 24, and 45 h of incubation. No regain of activity was observed under any of the conditions tested (Table 3). The presence of urea in the buffers improved recovery of kinase activity relative to the buffers alone but did not cause significant reactivation of RK.
Table 3.
Attempted reactivation of inactive RK
| Buffers | RK
activity
|
|||
|---|---|---|---|---|
| No addition | Addition
|
|||
| ATP + MgCl2 | Peptides | Urea | ||
| Control | 7.8 | 6.7 | 9.0 | 9.1 |
| 1 | 0 | 0.2 | 1.2 | 5.6 |
| 2 | 0 | 0 | 3.7 | 11.7 |
| 3 | 0.6 | 2.0 | 2.1 | 4.5 |
| 4 | 5.5 | 6.5 | 6.5 | 9.0 |
| 5 | 2.5 | 2.5 | 5.2 | 5.6 |
| 6 | 2.0 | 4.6 | 5.8 | 11.6 |
| 7 | 4.6 | 4.6 | 5.2 | 7.8 |
| 8 | 1.2 | 2.7 | 3.3 | 3.9 |
| 9 | 6.8 | 8.0 | 11.7 | 15.6 |
Freshly purified inactive RK was incubated in different buffers alone or after supplementing with ATP + MgCl2 or peptides (rhodopsin–carboxyl-terminal undecapeptide + mastroparan) or urea (see Materials and Methods). The activity of RK was measured after 5 h of incubation at 4°C. Similar results were obtained at 16°C and after 24 and 45 h of incubation (not shown).
Discussion
Two mAbs specific to RK have been characterized. The antibodies recognize short overlapping but distinct amino acid sequences near the carboxyl terminus. Although the antibodies are expected to be useful for structure/conformation studies of RK, they could also be potentially useful for studies of RKs, such as human and rat, that have high homology to the bovine RK. The binding of the antibodies to RK is slow, as shown by immunoprecipitation experiments (Fig. 1B), but more quantitatively by surface plasmon resonance (Fig. 2). By this technique, RK–antibody complexes showed high affinity (nanomolar range), but the rates of complex formation were slow (Fig. 2A). The slow interaction is likely because of the inaccessibility of the epitope in native RK. The binding kinetics follow a double exponential best modeled by a two-state binding reaction (Fig. 2B). Because of the observed inactivation of RK on antibody binding, it is possible that the fast and slow kinetic components of the double exponential correspond to the binding and the subsequent inactivating reactions, respectively.
Inactivation after the binding of the antibodies to the carboxyl-terminal segment shows that the latter is involved in stabilization of the folded enzyme in the active form. Thus, interactions between the carboxyl domain and either the catalytic or amino terminal domain, or both, are critical for maintaining the active state of the enzyme. Preliminary data (not shown) suggest that inactivation is not caused by protein aggregation. Rather, it appears that inactivation results from refolding (misfolding) of the enzyme in a time-dependent manner to an inactive state, presumably a consequence of a conformational change.
The inactive conformation of RK persists after the removal of the antibody, and all attempts so far to reverse it have failed. According to criteria defined by Carr et al. (10), this would indicate that the native original conformation of RK is metastable under our conditions. It is noteworthy that pure active RK is unstable and loses its ability to phosphorylate rhodopsin with a half-life of about 3 min at 30°C. Autophosphorylation and addition of adonitol, glycerol, or Tween 80 provide a stabilizing effect, whereas BSA does not. These results could also be explained by the transition of RK from a native active metastable state to an inactive stable state. By analogy with some viral fusion proteins (10), this transition could represent a storage of energy important for the function of RK, for example, during its relocalization from the cytoplasm to the membrane.
Because of their involvement in multiple aspects of regulatory mechanisms, most often the kinases are kept in an inactive state until their activation is signaled. Available information suggests that the maintenance of the kinases in the inactive form is achieved by modulating the position, conformation, or phosphorylation of a few key elements (9, 11), the modulation being brought about via interactions between the catalytic domain and flanking amino- and/or carboxyl-terminal domains or between the kinase and regulatory proteins such as cyclins or cAMP-binding proteins (9, 11, 12).
Numerous reports have described the inhibition of enzyme activities after binding to specific antibodies. However, irreversible inactivation as observed here seems not to have been recorded. Some of the reported inhibitions could nevertheless reflect a similar phenomenon that may not have been investigated. Conversely, it is possible that the antibody-mediated irreversible inactivation process, characteristic of a new subset of catalytic antibodies, constitutes a rare hitherto unrecognized event.
Acknowledgments
We are grateful to Prof. U. L. RajBhandary for reading of the manuscript and for helpful suggestions. We thank Drs. Sandra Smith-Gill and Claudia Lipschultz at the National Cancer Institute of the National Institutes of Health (NIH) for helpful discussions and Ms. Judy Carlin for patient assistance in the preparation of the manuscript. This work was supported by NIH grant GM28289 and National Eye Institute Grant EY11710 (H.G.K.), Human Frontier Science Program, Award no. LT449/96 (C.B.), and National Cancer Institute Training Grant CA091112 (L.N.). We acknowledge the award of a grant (RR13657) from the National Center for Research Resources/NIH Shared Instrumentation Program for the purchase of the BIACORE.
Abbreviations
- RK
rhodopsin kinase
- BTP
1,3-bis[Tris(hydroxymethyl)methylamino]propane
- DM
n-dodecyl β-d-maltoside
- HRP
horse radish peroxidase
- RamFc
rabbit anti-mouse IgG Fc
Footnotes
This is paper 40 in the series “Structure and Function in Rhodopsin.” Paper 39 is ref. 3.
References
- 1.Palczewski K, Buczylko J, Kaplan M W, Polans A S, Crabb J W. J Biol Chem. 1991;266:12949–12966. [PubMed] [Google Scholar]
- 2.Dean K R, Akhtar M. Eur J Biochem. 1993;213:881–890. doi: 10.1111/j.1432-1033.1993.tb17832.x. [DOI] [PubMed] [Google Scholar]
- 3.Bruel C, Cha K, Reeves P J, Getmanova E, Khorana H G. Proc Natl Acad Sci USA. 2000;97:3004–3009. doi: 10.1073/pnas.97.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider C, Newman R A, Sutherland D R, Asser U, Greeves M F. J Biol Chem. 1982;257:10766–10769. [PubMed] [Google Scholar]
- 5.Mandel M, Higa A. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 6.Cha K, Bruel C, Inglese J, Khorana H G. Proc Natl Acad Sci USA. 1997;94:10577–10582. doi: 10.1073/pnas.94.20.10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szedlacsek S E, Duggleby R G. Methods Enzymol. 1995;249:144–180. doi: 10.1016/0076-6879(95)49034-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen G-Q, Gouaux E. Proc Natl Acad Sci USA. 1997;94:13431–13436. doi: 10.1073/pnas.94.25.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor S S, Radzio-Andzelm E. Structure (London) 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 10.Carr C M, Chaudhry C, Kim P S. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng J, Knighton D R, Xuong H-H, Taylor S S, Somadski J M, Ten Eyck L F. Protein Sci. 1993;2:1559–1573. doi: 10.1002/pro.5560021003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaltiel S, Cox S, Taylor S S. Proc Natl Acad Sci USA. 1998;95:484–491. doi: 10.1073/pnas.95.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]