Abstract
Genes for peripheral tissue-restricted self-antigens are expressed in thymic and hematopoietic cells. In thymic medullary epithelial cells, self-antigen expression imposes selection on developing autoreactive T cells and regulates susceptibility to autoimmune disease in mouse models. Less is known about the role of self-antigen expression by hematopoietic cells. Here we demonstrate that one of the endocrine self-antigens expressed by human blood myeloid cells, proinsulin, is encoded by an RNA splice variant. The surface expression of immunoreactive proinsulin was significantly decreased after transfection of monocytes with small interfering RNA to proinsulin. Furthermore, analogous to proinsulin transcripts in the thymus, the abundance of the proinsulin RNA splice variant in blood cells corresponded with the length of the variable number of tandem repeats 5′ of the proinsulin gene, known to be associated with type 1 diabetes susceptibility. Self-antigen expression by peripheral myeloid cells extends the umbrella of “immunological self” and, by analogy with the thymus, may be implicated in peripheral immune tolerance.
Keywords: variable number of tandem repeats, insulin, type 1 diabetes, siRNA, self-antigen
T cells with high-avidity receptors for self-antigens are deleted developmentally in the thymus. This central mechanism of immune tolerance appears to depend on the ectopic expression of self-antigens by thymic medullary epithelial cells (1–4). However, in both the mouse (5) and human (6, 7) thymus, self-antigen RNA transcripts or peptide epitopes are also expressed by cells bearing markers of dendritic and monocytic antigen-presenting cells. In human thymus and spleen (6), and more recently in human blood (8), Pugliese and colleagues have described cells with myeloid lineage markers that stain with antibodies to the pancreatic β cell autoantigens in type 1 diabetes [i.e., proinsulin, glutamic acid decarboxylase (GAD), and tyrosine phosphatase-like insulinoma antigen (IA-2)] and transcribe the proinsulin gene. Hematopoietic progenitor cells have been shown to transcribe genes and express proteins for several neural autoantigens, including myelin basic protein (9) and GAD65 (10), and bone marrow-derived cells in extra-pancreatic tissues have been reported to express genes for pancreatic hormones (11). A role for ectopic self-antigen expression in hematopoietic cells was suggested by Zheng et al. (12), who demonstrated activation-induced death of self-reactive T cells in response to bone marrow cells expressing self-antigen.
Proinsulin is a key autoantigen that drives pancreatic β cell destruction in type 1 diabetes (13). In the human thymus, proinsulin mRNA abundance correlates with allelism of the variable number of tandem repeats (VNTR) upstream of the proinsulin coding region on chromosome 11 (14, 15), identified as the IDDM2 susceptibility locus for type 1 diabetes (16, 17). In addition to abundance of expression, the nature of self-antigen in lymphoid tissues may be important for the acquisition of immune tolerance. For example, Klein et al. (18) found that thymic expression of proteolipid protein (PLP), the major myelin sheath protein and an autoantigen in experimental autoimmune encephalomyelitis, was mainly restricted to a shorter RNA splice variant that did not encode the neuronal peptide recognized by T cells in susceptible mice. Similarly, Diez et al. (19) found that human thymus and spleen exclusively expressed a shorter RNA splice variant of IA-2 lacking the exon that in pancreatic β cells encodes the dominant autoantigenic peptide in type 1 diabetes. Thus, the differential expression of self-antigen in lymphoid tissues could educe an autoimmune T cell repertoire selectively directed against extralymphoid tissues.
Central tolerance is not absolute, and self-reactive T cells (e.g., to proinsulin and GAD) can be readily detected in the peripheral blood of healthy individuals (20). The mechanisms that avert activation of peripheral self-reactive T cells and prevent autoimmune disease, namely T cell deletion, T cell anergy, and the induction of regulatory T cells, depend classically on the uptake and presentation of self-antigens by specialized antigen-presenting dendritic cells (21). Extrapolating from the thymic paradigm, however, self-antigens could also be expressed constitutively by peripheral cells with antigen-presenting properties. Here we demonstrate that endocrine self-antigens expressed by human blood myeloid cells include an immunoreactive epitope for proinsulin encoded by an RNA splice variant of the proinsulin gene, the abundance of which corresponds with the length of the proinsulin VNTR.
Results
Blood Myeloid Cells Express Proinsulin and Other Self-Antigens.
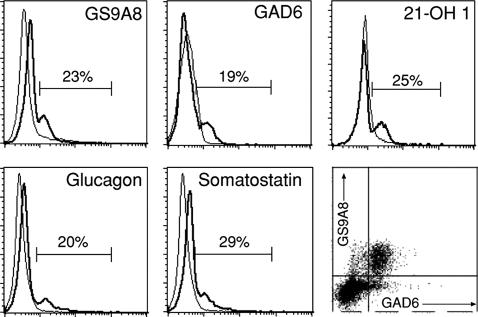
A subpopulation of live-gated peripheral blood mononuclear cells (PBMC) was delineated with monoclonal antibodies (mAbs) specific for proinsulin, GAD, 21-hydroxylase (21-OH), glucagon, or somatostatin, in all samples obtained from 15 healthy individuals (Fig. 1). Dual color staining revealed that proinsulin and GAD were coexpressed by the same cells (Fig. 1 Lower Right). The proinsulin mAb we used, GS9A8, recognizes the threonine/serine/arginine residues at the junction of the insulin B chain and the connecting (C) peptide in proinsulin (22). Cells were also stained with mAb KL1 to the B–C junction in proinsulin but not with mAb 2B7 that recognizes a conformational epitope comprising the N- and C-terminal regions of C-peptide (data not shown).
Fig. 1.
A subpopulation of blood mononuclear cells expresses epitopes for endocrine self-antigens. PBMC purified from healthy individuals were labeled with FITC-conjugated mAbs to proinsulin (GS9A8), GAD65 (GAD6), 21-OH 1, glucagon, or somatostatin and with isotype control antibodies (thin lines in the histogram plots) and analyzed by flow cytometry. Dead cells were identified by propidium iodide nuclear staining and excluded from the analysis. The Lower Right plot shows labeling of the same cell population by biotin (streptavidin-PE)-conjugated antibody to proinsulin and FITC-conjugated antibody to GAD65. The data shown are representative of 15 individuals studied.
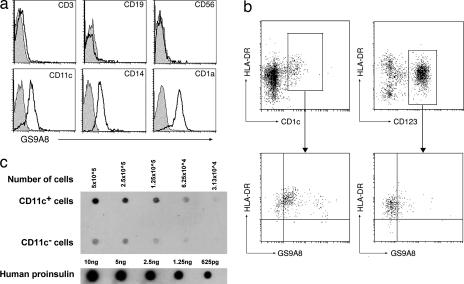
Cells staining for proinsulin displayed markers of dendritic cells (CD11c, CD1a) and monocyte/macrophages (CD11c, CD14) but not of T cells (CD3), B cells (CD19), or NK cells (CD56) (Fig. 2a). Analysis of blood dendritic cell subsets revealed that proinsulin was preferentially expressed by CD11c+ myeloid rather than by CD123+ plasmacytoid dendritic cells (Fig. 2b). No staining was observed with three different isotype control mAbs, and blocking of Fc receptors by the addition of pooled mouse serum and human IgG had no effect on the staining of cells by mAbs to proinsulin or GAD.
Fig. 2.
Myeloid lineage cells express an epitope for proinsulin. (a) PBMC purified from healthy individuals were labeled with PE-conjugated mAbs to T cell (CD3), B cell (CD19), NK cell (CD56), dendritic cell (CD11c, CD1a), and monocyte/macrophage cell (CD11c, CD14) markers and with FITC-conjugated mAb (GS9A8) to proinsulin and analyzed by flow cytometry. Binding of isotype control antibodies is represented by the shaded histograms. (b) Dendritic cells were purified from buffy coat-derived PBMC as described in Materials and Methods, labeled with markers of myeloid (HLA-DR, CD1c) or plasmacytoid (HLA-DR, CD123) dendritic cell subsets and with FITC-conjugated antibody (GS9A8) to proinsulin, and analyzed by flow cytometry. (c) FACS-sorted CD11c+ and CD11c− PBMC (each 5 × 106 per milliliter) were solubilized in 8% SDS and spotted onto nitrocellulose membrane, in parallel with serial dilutions of recombinant human proinsulin, and immunoblotted with the antiproinsulin mAb KL1.
To further define the specificity of proinsulin expression, PBMC were FACS-sorted into cells either positive or negative for the myeloid marker, CD11c. The cells were then lysed in 8% SDS, spotted onto nitrocellulose membrane, and immunoblotted with proinsulin mAb KL1 (Fig. 2c). Proinsulin immunoreactivity was observed predominantly in CD11c+ cells. By densitometric comparison with serial dilutions of proinsulin (Fig. 2), the number of proinsulin-equivalent binding sites per CD11c+ cell was estimated to be on the order of 60,000.
A small amount of proinsulin is cosecreted with insulin from the pancreatic β cell. Proinsulin has a low affinity for the insulin receptor, and binding sites specific for proinsulin itself have been described on lymphocytes (23). To exclude any contribution of exogenous proinsulin to the staining observed with proinsulin mAb, we studied four individuals with long-standing type 1 diabetes and undetectable circulating immunoreactive proinsulin C-peptide. The proportion of these patients' PBMC that stained with proinsulin mAb GS9A8 (22.6%, 22.0%, 16.0%, and 14.9%) was similar to that of age- and sex-matched healthy controls (23.5%, 13.8%, 21.7%, and 22.6%), indicating that staining was not due to the uptake of circulating proinsulin preferentially onto myeloid cells.
Blood Cells Transcribe Self-Antigen Genes, Including a Proinsulin RNA Splice Variant.
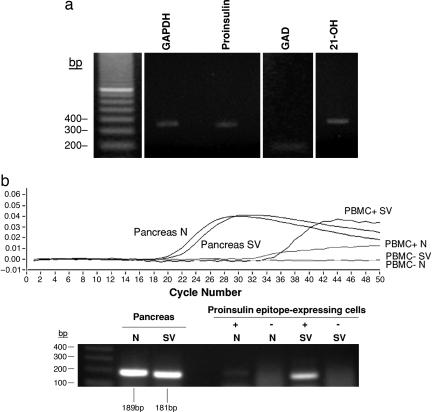
RNA was extracted from PBMC, and RT-PCR was performed with intron-spanning nested primer sets (see Table 1, which is published as supporting information on the PNAS web site). cDNA could be amplified for proinsulin, GAD65, and 21-OH in each of five healthy individuals examined (example shown in Fig. 3a), and product specificity was confirmed by DNA sequencing. Real-time PCR was then used to compare the expression of proinsulin transcripts between human pancreas, thymus, and spleen and PBMC and of 21-OH transcripts between human adrenal gland and PBMC. Amplicon-specific hybridization probes were used to quantify PCR product by fluorescence resonance energy transfer. PCR-amplified proinsulin or 21-OH cDNAs were standardized against the low copy number porphobilinogen deaminase (PBGD) housekeeping gene and expressed as a proportion of titrated pancreatic proinsulin or adrenal 21-OH DNA, respectively. The abundance of proinsulin transcripts in thymus, spleen, and PBMC was 0.01–0.1% of that in pancreas; the abundance of 21-OH transcripts in PBMC was ≈0.05% of that in adrenal gland (data not shown).
Fig. 3.
Blood cells transcribe self-antigen genes, including a proinsulin RNA splice variant. (a) RNA was freshly isolated from PBMC, reverse-transcribed into cDNA with random hexamer oligonucleotides, and amplified by PCR using nested sets of sequence-specific primers that spanned introns to avoid amplifying genomic DNA. GAPDH was used as a housekeeping gene. Electrophoresis in 1% agarose and staining with ethidium bromide demonstrated cDNAs for proinsulin, GAD65, and 21-OH in all five individuals examined (one example shown). Product identities were confirmed by DNA extraction and sequencing. (b) Native (N) and splice variant (SV) proinsulin RNAs from the pancreas and flow-sorted PBMC were quantified by real-time PCR (Upper), and the PCR products were identified by staining with ethidium bromide after electrophoresis in 1% agarose (Lower).
The amount of proinsulin mRNA detected was low relative to immunoreactive proinsulin. Others have reported RNA splice variants for self-antigens in the thymus (18, 19) and for proinsulin in the pancreas (24). Therefore, we looked for proinsulin splice variants in lymphoid tissues and PBMC. The human preproinsulin gene comprises three exons and two introns and, because the start codon is in exon 2, exon 1, intron 1, and part of exon 2 are in the 5′ untranslated region. A variant splice site was reported for pancreatic proinsulin in intron 1 (24). We designed a 5′ forward primer (Table 1) that crosses the variant splice site and does not amplify native proinsulin or genomic DNA. By using this primer and the 3′ reverse primers and hybridization probes used to quantify expression of native proinsulin, we detected the pancreatic proinsulin splice variant in human thymus, spleen, and PBMC. The mean percentage (± SD) of the splice variant in PBMC from 29 fasting, healthy individuals was 1.38 ± 0.94 of that in the pancreas. To compare the relative amounts of native and splice variant RNAs and at the same time demonstrate correspondence between cell surface immunoreactive proinsulin and proinsulin gene transcription, we sorted proinsulin mAb GS9A8+ and GS9A8− cells from PBMC for RNA extraction and real-time PCR. Compared with native proinsulin transcripts, splice variant transcripts were more abundantly expressed in GS9A8+ cells and were not detected in GS9A8− cells (Fig. 3b). Although the splice variant was the minor transcript in the pancreas, it was the major transcript in proinsulin mAb-positive blood cells, being 20-fold more abundant than the native transcript.
Introduction of Proinsulin Small Interfering RNA (siRNA) Reduces Expression of Immunoreactive Proinsulin.
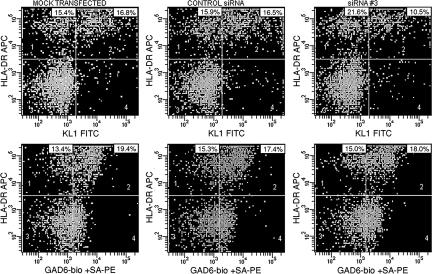
To confirm the specificity of immunoreactive proinsulin on myeloid cells, adherent monocytes were transfected with siRNAs to the second exon of insulin or with control siRNA. Cells were then incubated for 48 h and analyzed by flow cytometry for expression of proinsulin or GAD65. The numbers of proinsulin- or GAD65-positive cells and their mean fluorescence intensities were no different after mock transfection or after transfection with the control siRNA (Fig. 4). However, compared with mock or control transfections, transfection with the targeted siRNA3 reduced the number of immunoreactive proinsulin-expressing cells (Fig. 4 Upper Right). Results were expressed as the percentage of cells positive for immunoreactive proinsulin or GAD relative to the mock transfection. The maximum reduction in each of three separate experiments (63%, 65%, and 70%) was seen at 500 nM siRNA (P= 0.03; one-way ANOVA). Transfection with proinsulin-targeted siRNAs had no effect on the expression of immunoreactive GAD.
Fig. 4.
Introduction of proinsulin siRNA reduces expression of immunoreactive proinsulin. As described in Materials and Methods, PBMC (5 × 106 per milliliter) in RPMI medium 1640 containing 10% FCS were incubated in uncoated plastic flasks in 5% CO2/air at 37°C for 3 h. Adherent cells (5 × 106) were resuspended in transfection medium for human monocytes with or without (“mock transfection”) 500 nM siRNA and transfected. Cells, including some not transfected, were then incubated in transfection medium for human monocytes in 5% CO2/air at 37°C for 48 h. After washing and the addition of FcR blocking reagent, cells were stained with KL1-FITC (proinsulin) (Upper) or biotinylated-GAD6-streptavidin-PE (GAD65) (Lower) and HLA-DR-APC mAbs or isotype control mAbs and analyzed by flow cytometry. siRNA1–siRNA3 are to the second exon of insulin; the −ve siRNA, used as a negative control, is a scrambled version of siRNA1.
Proinsulin mRNA Splice Variant Expression Corresponds with the Length of the VNTR at the INS Susceptibility Locus for Type 1 Diabetes.
The INS or IDDM2 susceptibility locus for type 1 diabetes maps to a VNTR 5′ of the proinsulin gene coding region (16, 17). The short (class I) VNTR allele is associated with less thymic proinsulin mRNA (14, 15) and a higher risk for type 1 diabetes (17), whereas the long (class III) allele is associated with more thymic proinsulin mRNA and a lower risk for type 1 diabetes. Genotyping for the −23 Hph I polymorphism, the alleles of which are in linkage disequilibrium with the class I and III alleles, was performed on 28 healthy subjects in whom the expression of the proinsulin mRNA splice variant in PBMC was measured by real-time PCR. Consistent with the reported allele frequencies (25), only 2 of the 28 subjects were class III/III. The class I/I genotype was associated with lower expression of proinsulin mRNA splice variant in PBMC (median = 1.33% of pancreas, n = 20) compared with the class I/III genotype (median = 2.11% of pancreas, n = 6) (P = 0.03, unpaired t test with Welch correction).
Discussion
We show that the proinsulin gene is transcribed preferentially as an RNA splice variant by human blood myeloid cells, which express immunoreactive proinsulin and epitopes for other self-antigens. Proinsulin detected on myeloid cells was not due to the uptake of circulating proinsulin. All nucleated cells have receptors for insulin that bind proinsulin with low affinity, and lymphocytes have been reported to have specific receptors for proinsulin (23), but we did not observe staining of T or B lymphocytes with mAbs to proinsulin. Furthermore, proinsulin staining was similar in healthy controls and individuals with type 1 diabetes who had undetectable circulating proinsulin C-peptide. Importantly, we found a direct correspondence in sorted cells between proinsulin gene transcription and surface immunoreactive proinsulin, and the latter was specifically reduced by the introduction of insulin-specific siRNAs.
The relatively high expression of proinsulin immunoreactivity compared with native proinsulin mRNA prompted us to search for proinsulin mRNA splice variants. The splice variant we identified was reported by Shalev et al. (24) as a minor transcript in human pancreas, but in peripheral blood cells it appears to be the major transcript. It is more efficiently translated than the native gene (24), which may allow for even greater expression of proinsulin protein. Because splicing is initiated in the 5′ untranslated region, the variant RNA encodes full-length proinsulin protein and could therefore generate epitopes recognized by antibodies raised to the native protein. In contrast to splice variants of the autoantigens PLP (18) and IA-2 (19), which encode truncated proteins in the thymus but full-length proteins elsewhere, the proinsulin splice variant would not obviously lead to different forms of proinsulin protein being expressed in myeloid cells and pancreatic β cells. However, alternate splicing may serve as a mechanism for ectopic expression of proinsulin, which otherwise would only be expressed in β cells with their specialized transcriptional machinery.
Self-antigen expression by thymic medullary epithelial cells is considered to be a requirement for the negative selection and deletion of high-affinity, self-reactive T cells (3, 4). Several investigators have also reported self-antigen transcription and protein expression by cells with dendritic and monocytic phenotypes in the mouse and human thymus (5–7) and in the periphery (8–12). The mAbs we used to detect proinsulin are to the B–C chain junction, which appears to be an important immunogenic region in proinsulin. Most of the available mouse mAbs to proinsulin recognize this region, which comprises a T cell epitope in autoimmune diabetes-prone nonobese diabetic (NOD) mice (26, 27) and humans at risk for type 1 diabetes (13, 28). In the NOD mouse, an epitope spanning the B–C junction binds to I-Ag7, the MHC class II molecule, and induces regulatory, antidiabetogenic CD4 T cells when administered to the nasorespiratory mucosa (27). On the other hand, we cannot exclude the possibility that other regions of proinsulin are also accessible on the surface of myeloid cells, including other epitopes implicated in the pathogenesis of autoimmune diabetes in humans (29, 30) and NOD mice (31). How proinsulin or proinsulin peptide is “presented” on myeloid cells, and the role of peripheral self-antigen expression in immune homeostasis, are key questions to be addressed.
Proinsulin is a dominant autoantigen driving β cell destruction by autoreactive T cells in type 1 diabetes (13). Previously, we reported that syngeneic transplantation of hematopoietic stem cells encoding proinsulin completely prevents autoimmune diabetes in NOD mice (32), an effect most likely mediated by the expression of proinsulin in resting dendritic cells in peripheral lymphoid tissues (33). In the present study, we found a relationship between the abundance of proinsulin mRNA splice variant expression in circulating cells and the length of the INS VNTR (IDDM2 susceptibility locus). The same relationship has been reported for native proinsulin RNA in the human thymus (14, 15). The shorter (class I) allele, which is associated with a greater risk of type 1 diabetes than the longer (class III) allele (17), corresponds with a lower abundance of proinsulin RNA splice variant in the thymus. In the mouse thymus, a lower abundance of proinsulin RNA was associated with higher peripheral T cell reactivity to proinsulin (34). These observations lend weight to the hypothesis that type 1 diabetes susceptibility reflects the degree to which proinsulin-reactive T cells are edited in the thymus, as determined by the VNTR allele-regulated level of proinsulin gene transcription. Therefore, the association we describe between the VNTR alleles and peripheral expression of the proinsulin RNA splice variant could also contribute to the maintenance of antigen-specific tolerance. In humans, this potential mechanism of tolerance can now be directly investigated in peripheral blood. “Immunological self” may extend from a static population of thymic medullary epithelial cells to circulating, bone marrow-derived myeloid cells, linking central thymic and peripheral tolerance.
Materials and Methods
Human Tissues.
Heparinized venous blood was obtained, after an overnight fast, from 28 healthy, young adult individuals and 4 young adults who had had type 1 diabetes for a median duration of 9.2 years. Fresh white cell buffy coats were provided by the Australian Red Cross with Human Ethics Committee approval and informed consent. PBMC were isolated by density centrifugation over Ficoll–Hypaque Plus (Amersham Biosciences, Castle Hill, Australia). Human pancreas tissue was obtained from organ donor subjects with Human Ethics Committee approval. Total RNA from human thymus, spleen, and adrenal gland was purchased from Becton Dickinson (North Ryde, Australia).
C-Peptide Assay.
Plasma immunoreactive C-peptide was measured with the Immulite 2000 system (Diagnostics Products, Los Angeles, CA).
Antibodies.
The mouse mAb, clone GS9A8, recognizes the junction between insulin B chain and the connecting (C) peptide in human proinsulin (22). A mouse mAb specific for a conformational epitope comprising the N- and C-terminal regions of human C-peptide separated by a β turn at position 47–50 of proinsulin (clone 2B7) was purchased from Advanced Immunochemical (White City, CA). Mouse mAbs to human proinsulin were also generated in the Monoclonal Antibody Laboratory of The Walter and Eliza Hall Institute. An IgG1 clone, termed KL1, was used in the current study. Epitope mapping with overlapping synthetic peptides spanning the full length of human proinsulin revealed that precipitation of 125I-proinsulin by KL1 was blocked by proinsulin B24-C36 (FFYTPKTRREAED). Thus, the KL1 epitope, similarly to clone GS9A8, spans the B–C chain junction of proinsulin.
GAD6, a mouse IgG2a mAb that reacts with human GAD65, an autoantigen in type 1 diabetes, was a gift of David Gottlieb (Washington University School of Medicine, St. Louis, MO). 21-OH 1, a mouse IgG mAb to human 21-OH, an autoantigen in Addison's disease, was a gift of RSR Ltd (Cardiff, U.K.). IgG mAbs to somatostatin and glucagon were purchased from Novo Biolabs (Bagsvaerd, Denmark). Antibodies to proinsulin and other self-antigens were either conjugated to fluorescein isothiocyanate (FITC) or biotinylated for use with streptavidin-fluorochromes. Phycoerythrin (PE)-conjugated antibodies to CD11c, CD14, CD1a, CD3, CD19, and CD56, and PerCP, or APC-conjugated antibodies to HLA-DR and CD45, were purchased from BD PharMingen (Palo Alto, CA). PE-conjugated antibodies to CD1c (BDCA-1) and CD123 were purchased from Miltenyi Biotec (North Ryde, Australia). FITC- or PE-conjugated IgG1 clone MOPC (Becton Dickinson, Mountain View, CA), FITC-conjugated IgG1 clone OX6 and IgG2a clone 10.2.16, and nonconjugated IgG1 clone 2D6, produced in our mAb facility, were used as isotype controls.
Proinsulin.
Recombinant human proinsulin was expressed in Escherichia coli, extracted and refolded, and purified to homogeneity by HPLC-mass spectrometry in collaboration with Kaku Nakagawa (Kaneka Corporation, Takasago, Japan). Recombinant GAD65 was a gift of Peter Van Endert (Hôpital Necker, Paris, France).
Flow Cytometry.
Flow cytometry was performed on freshly isolated PBMC or on monocytes that had been transfected with siRNAs (as described below). PBMC were washed and suspended at a concentration of 106 per milliliter in 100 μl of FACS buffer (1% FCS in PBS) containing 10 μl of pooled BALB/c mouse serum and 10 μl of pooled human IgG to block binding of mouse antibody to human Fc receptors and were stained with a 2 μg/ml concentration of primary antibody for 30 min on ice. Nonfluorochromated mAbs to 21-OH, somatostatin, and glucagon were detected by using FITC-conjugated F(ab)2 affinity-purified sheep anti-mouse antibodies (Chemicon, Victoria, Australia); in these experiments, mouse serum was not used for blocking the primary antibody, and 10 μl of sheep serum was used to block the secondary antibody. The cells were washed with cold FACS buffer and then analyzed. Analyses were performed on a FACScan (Becton Dickinson) after exclusion of propidium iodide-labeled dead cells, and data were analyzed with CellQuest software (Becton Dickinson).
Cell Sorting and Dendritic Cell Purification.
Healthy donor PBMC labeled with FITC-labeled GS9A8 or KL1 mAb to proinsulin and with PE-labeled mAb to CD11c were sorted in a MoFlo high-speed FACS sorter (DakoCytomation, Fort Collins, CO) to a purity >85%. Lineage-negative CD4+ dendritic cells were purified from buffy coat-derived PBMC with a blood dendritic cell isolation kit (Miltenyi Biotec) and further depleted on the MoFlo sorter of lineage-positive cells labeled with PE-conjugated antibodies to CD3, CD19, CD14, and CD56. Dendritic cells were then identified by staining for HLA-DR and either CD11c (myeloid) or CD123 (plasmacytoid) markers.
Cell Lysis and Immunoblotting.
PBMC were FACS-sorted into CD11c+ and CD11c− cells, and equal numbers (5 × 105) were lysed directly in 100 μl of 125 mM Tris containing 8% SDS, 0.1 M DTT, and 10% glycerol for 10 min at 22°. After centrifugation at 10,000 × g for 10 min, supernatants were spotted in parallel onto nitrocellulose membrane and immunoblotted with biotinylated KL1 mAb detected with streptavidin-HRP. Dilutions of known amounts of human proinsulin were immunoblotted in parallel for densitometric comparison to estimate the proinsulin equivalents expressed per cell.
RNA Isolation and cDNA Synthesis.
RNA was isolated using an RNeasy Mini Kit (Qiagen, Clifton Hill, Australia). RNA integrity was confirmed by spectrophotometry at 260/280 nm and inspection of 28S and 18S band intensities after agarose gel electrophoresis. Total RNA (1 μg) was reverse-transcribed into cDNA with random hexamer primers (Promega, Annandale, Australia) and Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega) in the presence of RNAsin (Promega). cDNA samples were aliquoted and stored at −20°C.
PCR.
Nested PCR was performed in 50 μl of PCR buffer containing final concentrations of 200 nM primers, 2 mM MgCl2, 400 μM dNTPs, 5 units of TaqDNA polymerase, and 125 ng of cDNA template. After 35 cycles of amplification in a FTS-960 thermal cycler (Corbett Research, Mortlake, Australia), 2 μl of the PCR was used as DNA template for a further 35-cycle PCR performed with nested primers. Primers (Table 1) were purchased from Sigma Genosys (Woodlands, TX) and usually spanned introns to avoid amplification of contaminating genomic DNA. A 5′ forward primer was designed to detect the proinsulin splice variant identified by Shalev et al. (24) in human pancreatic islets. Splicing is initiated in intron 1 upstream of the normal start codon in exon 2, retaining 26 bp of intron 1 without altering the coding region. The identity of amplified PCR product was confirmed by electrophoresis in 1% agarose gel, band extraction with the QIAquick gel extraction method (Qiagen), and sequencing by Big Dye Terminator Reaction (Applied Biosystems, Scoresby, Australia).
Real-time PCR was performed in a Lightcycler (Roche Diagnostics, Castle Hill, Australia) with sequence-specific hybridization probes (Table 1) and the QuantiTect probe kit (Qiagen). Master mixtures were prepared in accordance with the manufacturer's instructions to include a 0.5 μM concentration of each primer and a 0.1 μM concentration of each probe. Water-only samples were used as negative controls. Amplification for all primer/probe sets was carried out with initial activation for 15 min at 95°C ramped at 20°C/sec, followed by 45 cycles each of denaturation at 95°C for 10 sec, annealing at 50°C for 30 sec, and extension at 72°C for 30 sec, ramped at 20°C/sec. PCR products were verified by 1% agarose gel electrophoresis. PBGD was chosen as a housekeeping gene because it has no known pseudogenes and is stably expressed at a low copy number. Sequence-specific standard curves were generated for PBGD, proinsulin, proinsulin splice variant, and 21-OH by using serial dilutions of PBMC, thymus, spleen, and pancreas or adrenal cDNA. The expression of product with respect to pancreas or adrenal DNA, each normalized to 100%, was calculated with the Lightcycler RelQuant software (Roche Diagnostics).
Proinsulin Expression After Introduction of siRNA.
Three annealed siRNAs to the second exon of proinsulin (siRNA1–siRNA3) and an siRNA consisting of the scrambled sequence of siRNA1 lacking homology to any known gene (negative control) (see Table 2, which is published as supporting information on the PNAS web site) were purchased from Ambion (Austin, TX) at a stock concentration of 100 μM. PBMC (5 × 106 per milliliter) in complete medium (RPMI medium 1640/1 mM sodium pyruvate/0.05 mM 2-mercaptoethanol/0.1 mM nonessential amino acids/10% FCS/2 mM glutamine) were incubated in uncoated plastic flasks in a humidified atmosphere of 5% CO2/air at 37°C for 3 h. After gentle decanting of nonadherent cells, the adherent monocytes were collected by scraping, washed in medium, and counted. Then, 5 × 106 cells were resuspended in 100 μl Amaxa nucleofection solution for monocytes (Human Monocyte Nucleofector Kit; Amaxa Biosystems, Cologne, Germany) containing up to 500 nM siRNA and transfected in an Amaxa nucleofector electroporator on program T-08, in accordance with the manufacturer's instructions. The cells were immediately mixed with 500 μl of prewarmed human monocyte nucleofector medium, transferred into 24-well plates containing 1 ml of this medium per well, and incubated in 5% CO2/air at 37°C for 48 h. After collection and washing in complete medium and the addition of FcR blocking reagent (Miltenyi Biotec) for 10 min at 4°C, cells were stained with FITC-KL1 (proinsulin) or FITC-GAD (GAD65) and APC HLA-DR mAbs or isotype control mAbs in parallel samples and analyzed by flow cytometry. Compensations were performed before addition of propidium iodide to the samples.
INS VNTR Genotyping.
The INS gene −23 Hph I A/T polymorphism shows association with type 1 diabetes susceptibility and is in strong linkage disequilibrium with other susceptibility alleles, including the VNTR polymorphism upstream of the INS gene (16, 17). The −23 Hph I site was genotyped after PCR with 10 pmol of the primers (5′–3′) AGCAGGTCTGTTCCAAGG and CTTGGGTGTGTAGAAGAAGC in a 10-μl reaction containing 1.0 μl of 1 mM dNTPs, 0.6 μl of 25 mM MgCl2, 0.5 units of Taq polymerase, 10% glycerol, and 100 ng of genomic DNA. The PCR was performed in a CR-9600 cycler (Corbett Research) at 94°C for 3 min, followed by 30 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec and a final cycle of 72°C for 5 min. The PCR product was digested for 2 h at 37°C in a 20-μl reaction containing 12 μl pf dH2O, 5 μl of PCR product, 2 μl of Hph I 10× buffer, and 1 unit of Hph I. Under these conditions, the A allele was indicated by the presence of a 120-bp band and the T allele by a 171-bp band.
Supplementary Material
Acknowledgments
We thank Dexing Huang and Cameron Wells for technical assistance and Catherine McLean for valuable secretarial assistance. P.N., J.A.D., and K.P.J. were postdoctoral fellows of the Juvenile Diabetes Research Foundation (JDRF). This work was supported by a JDRF Program Center grant (R.J.S.; to L.C.H.) and by the National Health and Medical Research Council of Australia.
Abbreviations
- GAD
glutamic acid decarboxylase
- PBMC
peripheral blood mononuclear cells
- PE
phycoerythrin
- 21-OH
21-hydroxylase
- VNTR
variable number of tandem repeats.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jolicoeur C, Hanahan D, Smith KM. Proc Natl Acad Sci USA. 1994;91:6707–6711. doi: 10.1073/pnas.91.14.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith KM, Olson DC, Hirose R, Hanahan D. Int Immunol. 1997;9:1355–1365. doi: 10.1093/intimm/9.9.1355. [DOI] [PubMed] [Google Scholar]
- 3.Derbinski J, Schulte A, Kyewski B, Klein L. Nat Immunol. 2001;11:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 4.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 5.Throsby M, Homo-Delarche F, Chevenne D, Goya R, Dardenne M, Pleau JM. Endocrinology. 1998;139:2399–2406. doi: 10.1210/endo.139.5.5989. [DOI] [PubMed] [Google Scholar]
- 6.Pugliese A, Brown D, Garza D, Murchison D, Zeller M, Redondo M, Diez J, Eisenbarth GS, Patel DD, Ricordi C. J Clin Invest. 2001;107:555–564. doi: 10.1172/JCI10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sospedra M, Ferrer-Francesch X, Dominguez O, Juan M, Foz-Sala M, Pujol-Borrell R. J Immunol. 1998;161:5918–5929. [PubMed] [Google Scholar]
- 8.Garcia CA, Prabakar KR, Diez J, Cao ZA, Allende G, Zeller M, Dogra R, Mendez A, Rosenkranz E, Dahl U, et al. J Immunol. 2005;175:2111–2122. doi: 10.4049/jimmunol.175.4.2111. [DOI] [PubMed] [Google Scholar]
- 9.Marty MC, Alliot F, Rutin J, Fritz R, Trisler D, Pessac B. Proc Natl Acad Sci USA. 2002;99:8856–8861. doi: 10.1073/pnas.122079599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goolsby J, Marty MC, Heletz D, Chiappelli J, Tashko G, Yarnell D, Fishman PS, Dhib-Jalbut S, Bever CTJ, Pessac B, Trisler D. Proc Natl Acad Sci USA. 2003;100:14926–14931. doi: 10.1073/pnas.2434383100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima H, Fujimiya M, Matsumura K, Nakahara T, Hara M, Chan L. Proc Natl Acad Sci USA. 2004;101:2458–2463. doi: 10.1073/pnas.0308690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X, Yin L, Liu Y, Zheng P. Eur J Immunol. 2004;34:3126–3134. doi: 10.1002/eji.200425177. [DOI] [PubMed] [Google Scholar]
- 13.Narendran P, Mannering SI, Harrison LC. Autoimmun Rev. 2003;2:204–210. doi: 10.1016/s1568-9972(03)00009-0. [DOI] [PubMed] [Google Scholar]
- 14.Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, Colle E, Polychronakos C. Nat Genet. 1997;15:289–292. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 15.Pugliese A, Zeller M, Fernandez AJ, Zalcberg LJ, Bartlett RJ, Ricordi C, Pietropaolo M, Eisenbarth GS, Bennett ST, Patel DD. Nat Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 16.Lucassen AM, Julier C, Beressi JP, Boitard C, Froguel P, Lathrop M, Bell JI. Nat Genet. 1993;4:305–310. doi: 10.1038/ng0793-305. [DOI] [PubMed] [Google Scholar]
- 17.Bennett ST, Lucassen AM, Gough SC, Powell EE, Undlien DE, Pritchard LE, Merriman ME, Kawaguchi Y, Dronsfield MJ, Pociot F, et al. Nat Genet. 1995;9:284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 18.Klein L, Klugmann M, Nave KA, Tuohy VK, Kyewski B. Nat Med. 2000;6:56–61. doi: 10.1038/71540. [DOI] [PubMed] [Google Scholar]
- 19.Diez J, Park Y, Zeller M, Brown D, Garza D, Ricordi C, Hutton J, Eisenbarth GS, Pugliese A. Diabetes. 2001;50:895–900. doi: 10.2337/diabetes.50.4.895. [DOI] [PubMed] [Google Scholar]
- 20.Mannering SI, Morris JS, Jensen KP, Purcell AW, Honeyman MC, van Endt P, McCluskey J, Harrison LC. J Immunol Methods. 2003;283:173–183. doi: 10.1016/j.jim.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Walker LS, Abbas AK. Nat Rev Immunol. 2002;2:11–19. doi: 10.1038/nri701. [DOI] [PubMed] [Google Scholar]
- 22.Madsen OD. Diabetes. 1987;36:1203–1211. doi: 10.2337/diab.36.10.1203. [DOI] [PubMed] [Google Scholar]
- 23.Jehle PM, Lutz MP, Fussgaenger R. Diabetologia. 1996;394:421–432. doi: 10.1007/BF00400673. [DOI] [PubMed] [Google Scholar]
- 24.Shalev A, Blair PJ, Hoffmann SC, Hirshberg B, Peculis BA, Harlan DM. Endocrinology. 2002;143:2541–2547. doi: 10.1210/endo.143.7.8920. [DOI] [PubMed] [Google Scholar]
- 25.Walter M, Albert E, Conrad M, Keller E, Hummel M, Ferber K, Barratt BJ, Todd JA, Ziegler AG, Bonifacio E. Diabetologia. 2003;46:712–720. doi: 10.1007/s00125-003-1082-z. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Bergerot I, Elliott JF, Harrison LC, Abiru N, Eisenbarth GS, Delovitch TL. J Immunol. 2001;167:4926–4935. doi: 10.4049/jimmunol.167.9.4926. [DOI] [PubMed] [Google Scholar]
- 27.Martinez NR, Augstein P, Moustakas AK, Papadopoulos GK, Gregori S, Adorini L, Jackson DC, Harrison LC. J Clin Invest. 2003;111:1365–1371. doi: 10.1172/JCI17166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudy G, Stone N, Harrison LC, Colman PG, McNair P, Brusic V, French MB, Honeyman MC, Tait B, Lew AM. Mol Med. 1995;6:625–633. [PMC free article] [PubMed] [Google Scholar]
- 29.Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Nature. 2005;435:224–228. doi: 10.1038/nature03625. [DOI] [PubMed] [Google Scholar]
- 30.Mannering SI, Harrison LC, Williamson NA, Morris JS, Thearle DJ, Jensen KP, Kay TW, Rossjohn J, Falk BA, Nepom GT, Purcell AW. J Exp Med. 2005;202:1191–1197. doi: 10.1084/jem.20051251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF, Eisenbarth GS. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steptoe RJ, Ritchie JM, Harrison LC. J Clin Invest. 2003;111:1357–1363. doi: 10.1172/JCI15995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steptoe RJ, Ritchie JM, Jones LK, Harrison LC. Diabetes. 2005;54:434–442. doi: 10.2337/diabetes.54.2.434. [DOI] [PubMed] [Google Scholar]
- 34.Chentoufi AA, Polychronakos C. Diabetes. 2002;51:1383–1390. doi: 10.2337/diabetes.51.5.1383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.