Abstract
Stats (for signal transducers and activators of transcription) are a family of transcription factors that regulate cell growth and differentiation. Their activity is latent until phosphorylation by receptor-associated kinases. A sizable body of data from cell lines, mouse models, and human tissues now implicates these transcription factors in the oncogenesis of breast cancer. Because Stat activity is modulated by several posttranslational modifications and protein-protein interactions, these transcription factors are capable of integrating inputs from multiple signaling networks. Given this, the future utilization of Stats as prognostic markers and therapeutic targets in human breast cancer appears likely.
Since their identification 12 years ago,1 the Stat (for signal transducers and activators of transcription) family of transcription factors have been recognized as critical integrators of cytokine and growth factor receptor signaling required for cell growth, survival, differentiation, and motility. A role for two members of this family, namely Stat3 and Stat5, in the pathogenesis of human breast cancer is increasingly appreciated. As presented below, several lines of evidence from breast cancer cell lines, mouse models, and primary human tissues have implicated these transcription factors in mammary oncogenesis. With these findings as a backdrop, this review will explore the multiple mechanisms through which Stat activity is regulated, mechanisms that provide new targets of potential prognostic and therapeutic utility.
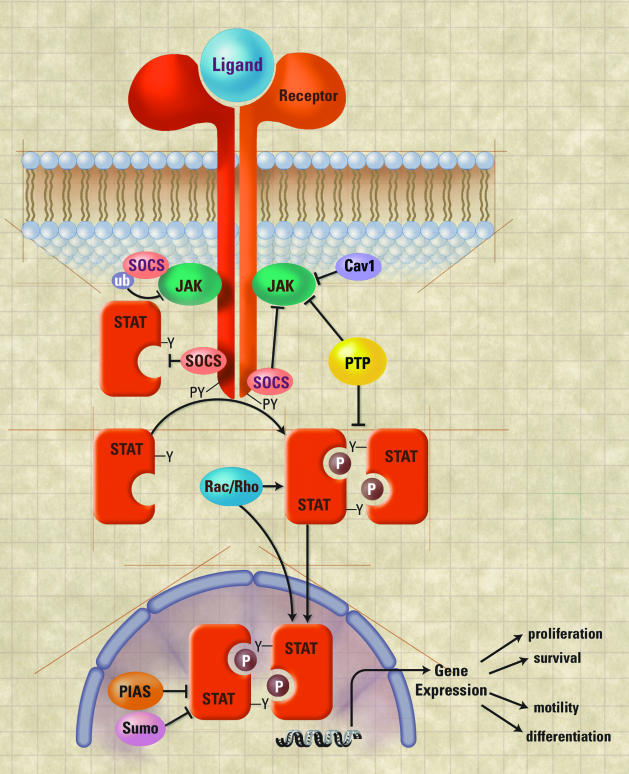
Stats exist within the cytoplasm in a latent or inactive state. After cell-surface receptor activation by ligand, Stats are activated by receptor-associated tyrosine kinases (Figure 1). Significant in this regard are the Jak family (Jaks 1 to 3 and Tyk2) of tyrosine kinases that are rapidly (within 1 minute) activated by autophosphorylation, presumably triggered by ligand-induced receptor dimerization/multimerization. In turn, activated Jak kinases phosphorylate the receptor to which it has bound, and receptor-associated signaling proteins. Receptor phosphorylation enables Stat docking to this complex, via binding of a Stat SH2 domain to a receptor phosphotyrosine residue. This event permits Stat phosphorylation by the juxtaposed Jaks and other receptor-associated kinases. Phosphorylation of a tyrosine residue present in the Stat C-terminus triggers its release from receptor, and homo (and in some cases hetero-)-dimerization of phosphorylated Stat proteins briskly ensues. Dimerized Stats rapidly translocate to the nucleus, where binding to gene promoters bearing cognate DNA-binding sequences occurs. Once bound, Stats engage several elements of the transcriptional apparatus, stimulating gene expression. In the context of breast cancer, Stat activation has been found to occur both in vitro and in vivo after the binding of ligand to several receptors implicated in the pathogenesis of breast cancer; a listing of receptor-Stat affiliations is presented in Table 1.
Figure 1.
Activation and regulation of Stat-mediated transcription. After ligand-induced receptor dimerization, activated Jak sequentially phosphorylates receptor and Stat. This induces the release and dimerization of phosphorylated Stat, enabling its translocation into the nucleus and subsequent DNA binding. As indicated in the figure, Stat activity is regulated at many levels; events that stimulate Stat activity are noted with lines terminating in arrowheads, whereas inhibitory events are marked with lines ending with bars. Abbreviations: ub, ubiquitin; pY, phosphotyrosine; PTP, phosphatase. For the sake of simplicity some Stat regulatory events (ie, serine phosphorylation, protein-protein interactions, and so forth) have been omitted.
Table 1.
Stat Activation Mediated by Cell-Surface Ligand/Receptor Complexes Relevant to Breast Cancer
| Stat3 | Stat5 |
|---|---|
| Epidermal growth factor | Epidermal growth factor |
| Platelet-derived growth factor | Platelet-derived growth factor |
| Hepatocyte growth factor | Prolactin |
| Insulin-like growth factor | Growth hormone |
| Insulin | Erythropoietin |
Identification and Functional Role of Stats
The Stat family of transcription factors was first identified through the careful analysis of the molecular requirements of interferon (IFN)-triggered gene expression.2 Both IFN-α and IFN-γ trigger gene expression within 15 to 30 minutes of IFN receptor stimulation; analysis of the promoter regions of these IFN-activated genes revealed that specific motifs (the GAS motifs) with a consensus DNA sequence of TT(C/A)YNR(G/T)AA were required for this expression. These motifs were used in DNA affinity pull-down of cell lysates; coupled with biochemical purification, these approaches enabled the isolation and sequencing of the first Stats (Stat1α, Stat1β, and Stat2).1 Using DNA-binding sequences obtained from the β-casein promoter, Stat5 was subsequently purified, sequenced, and cloned in a manner similar to Stats 1 and 2.3 In contrast, Stat3 was identified by cDNA library screen with the SH2 domain of Stat1.4 As previous studies had demonstrated that IFN-induced gene expression was inhibited by tyrosine kinase inhibitors, it was quickly recognized that the targets of this phosphorylation were the Stats. As noted above, tyrosine phosphorylation is required for Stat dimerization and nuclear translocation. Additional analysis revealed that Stats’ members were activated by several disparate cell surface receptors (Table 1).5 Contemporaneously, with the recognition of Stats as transcription factors, came the identification and characterization of the Jak family of tyrosine kinases.6–8 The signaling connection between the Jak and Stat families, however, was not known a priori; the breakthrough for this connection was provided by somatic cell genetics.9 Using mutagenesis and complementation approaches, cell clones defective in IFN-induced signaling regained IFN-responsiveness when Jaks or Stats were reintroduced, establishing linkage between these two families. Subsequent analysis with intact, nonmutagenized cells demonstrated the critical, transient association between Jak and Stat family members, required for Stat phosphorylation and activation.2,6
Stat Structure/Function
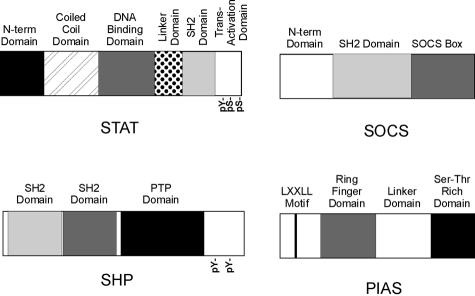
Extensive structural mutagenesis studies have revealed that the Stat proteins consist of numerous distinct functional domains (Figure 2).2,10 The N-terminal domain has been found necessary for the interaction of Stats 3 and 5 with a number of co-activators, such as CBP/p300, c-Jun, and Nmi;11 in addition, this domain is necessary for higher-order associations between Stat5 dimers (ie, tetramerization) that contribute to enhanced Stat5 activity on certain promoters.12 A central DNA-binding domain is required for recognition of cognate binding sequences. This domain is coupled by a flexible linker to a SH2 domain that is necessary for the recognition of phosphotyrosine residues that contribute to Stat dimerization and the recognition of cell-surface receptor binding sites. Between the SH2 motif and the C-terminal transactivation domain resides a conserved tyrosine residue whose phosphorylation is required for the dimerization of all Stat family members. Aside from its critical role in coordination with the transcriptional apparatus, the transactivation domain contains a serine residue(s) that also regulates the activity of this domain.
Figure 2.
Structural maps of the Stat, SOCS, SHP, and PIAS protein families. Phosphorylated tyrosine and serine residues implicated in protein function are, respectively, designated pY and pS. The phosphatase domain is noted in the C-terminus of SHP (PTP). The signature leucine-rich motif conserved in the PIAS family is designated in the N-terminus of PIAS3 (LXXLL).
The structure of DNA-engaged STAT dimers has been resolved at the crystallographic level for both Stats 1 and 3.13,14 It should be noted that the data from these crystal structures is limited because they do not include either the functionally critical N-terminal domain or the C-terminal transactivation motif. On a topological level the crystal structures of Stats 1 and 3 resemble a nutcracker, with the interactions between the respective Stat phosphotyrosine/SH2 domains serving as the hinge and the DNA acting as the nut. These structures have revealed a twofold axis of dyadic symmetry surrounding the DNA-binding sites used in these studies. The structures are similar and consist of four domains: a four α-helical bundle within the N-terminus that is presumed to contribute to protein-protein interactions, an eight-stranded β-barrel in which the DNA-binding domain resides, an α-helical connector, and a classic SH2 domain. The Stat homodimer envelopes the bound DNA with distinct amino acid residues contacting both the phosphate moieties and sugar residues of the DNA backbone, as well as specific nucleotides. Interestingly, the engaged DNA is actually bent by 40° as the result of this interaction. The C-terminal phosphotyrosine residues are engaged by their counterparts’ SH2 motif, providing a pivot point for DNA binding.
Role of Stats in Normal Mammary Biology
Mouse Models
During pregnancy, the expression and activity of the Stat family is variably modulated, with the expression and activity of Stat5 closely linked to the onset of lactation, whereas the expression and activity of Stats 1 and 3 are associated with the virgin animal and involution.15 Gene targeting studies have revealed phenotypes for Stats 3 and 5a during mammary differentiation; the Stat5a knockout demonstrates profound loss of alveolar maturation and milk production;16 indeed the mammary phenotype of the Stat5a−/− mouse is similar to that observed in both the prolactin (PRL) and prolactin receptor (PRLr) knockout mice.17,18 In contrast, Stat3 mice with a conditional knockout of Stat3 within the mammary gland demonstrate delayed mammary involution after cessation of pregnancy.19 These data would suggest that Stat5 plays a critical role in lobuloalveolar proliferation, differentiation, and expansion, whereas Stat3 regulates lobuloalveolar apoptosis during pregnancy, lactation, and involution.
Role of Stats in Breast Cancer
Data from Human Tissues—Insights into Biology
Several studies have used different approaches to assess the expression, localization, phosphorylation, and DNA binding ability of Stats 3 and 5 in breast tissues. Immunohistochemical approaches have successfully examined total and phosphorylated Stat 3 and 5 levels in breast tissues, and correlated this expression with intranuclear localization. In this context, both the phosphorylation and nuclear localization of Stats 3 and 5 are presumed to correlate with the level of transcriptional activity of these proteins. The advantage of an immunohistochemical approach is its ease in application to large numbers of patient specimens. The disadvantage to this approach is the presumption that phosphorylation and nuclear localization equate to transcriptionally active Stat; as discussed below there are several mechanisms through which intranuclear Stat function can be repressed. Alternatively, the level of Stat DNA binding can be measured by electrophoretic mobility shift analysis (EMSA); although this is perhaps a better measure of Stat activation, it is time consuming and not amenable to small tissue samples. Ideally, both immunohistochemical analysis and EMSA would be compared on the same cohort of human samples; unfortunately, only one study has made such direct comparison.
Significant elevations in the DNA-binding activity of both Stat3 and Stat515,20 were noted in a small sample of malignantly transformed breast tissues when compared to normal tissues. Although this pioneering study provided some of the first evidence for alterations in Stat DNA-binding activity in breast cancer, it was limited by its lack of normalization to total Stat protein levels (ie, the EMSA analysis was not normalized), small patient cohort, and lack of associated outcomes analysis.
At the immunohistochemical level, two studies have examined Stat3 localization and phosphorylation. The first study21 examined 62 malignant breast cancers and found increased levels of nuclear localized Stat3 in comparison to normal surrounding tissues; no association with clinicopathological or outcomes data were provided. The second study22 assessed a microarray containing tissues from 346 node-negative breast cancer patients with both anti-Stat3 and anti-phospho-Stat3 immunohistochemistry and correlated these results with clinicopathological and survival data. This study revealed that nuclear Stat3 and phospho-Stat3 was noted in 23% and 44% of patients, respectively; however, a rationale for the discrepancy between these percentages was not commented on. In addition, the study found that nuclear phospho-Stat3 expression was correlated with a modest, but statistically significant, improvement in patient survival both at 5 and 20 years. Multivariate analysis of these data revealed that this parameter was statistically more predictive of outcome than tumor size, nuclear grade, age, estrogen/progesterone receptors and Her2 status, and Ki-67 positivity.
A small study examining the expression of phosphorylated Stat5 in human breast tissues by immunohistochemical approaches has revealed significant intranuclear Stat5 in both normal and lactating human breast tissues; parallel studies in mice confirmed that a basal level of Stat5 phosphorylation can be noted in normal virgin, pregnant, and lactating mammary glands.23 This phosphorylation may be because of the local production of PRL by breast tissues.24,25 Whether the phosphorylated Stat5 in the nonpregnant, normal breast is actually bound to DNA and transcriptionally active (or bound to a repressor such as PIAS3, see below) remains to be determined. In another study, immunohistochemical analysis of 83 primary human breast cancers for both Stat5 and tyrosine-phosphorylated Stat5 revealed phosphorylation and nuclear localization in 76% of malignancies examined; no associations with several recognized prognostic markers (tumor size, nodal and estrogen receptor status, percent S phase, and so forth), save for histological differentiation.26 This study also confirmed the presence of a basal level of Stat5 phosphorylation in normal breast tissues. A more recent study examining more than 1300 breast cancers has found a strong association between Stat5 nuclear localization/phosphorylation and improved overall and disease-free survival.27
Only a single study to date has examined the relationship between Stat phosphorylation and DNA binding status28 in vivo. This study, which evaluated Stats 1, 3, and 5, found a strong correlation between Stat 1 and 3 phosphorylation (as measured by immunoblot analysis) and DNA binding (as measured by EMSA) in human tissues, but could not detect Stat5 phosphorylation (as noted above), possibly secondary to antibody difficulties. Although the study found a correlation between Stat1 activation/phosphorylation and survival (but not Stats 3 or 5), the numbers of this study were small (68 samples including both node-negative and node-positive), the data were not normalized to total Stat content, and Stat phosphorylation was not assessed by immunohistochemistry (ie, the contribution of inactive Stat from the surrounding stromal tissues may have been significant). As a final note, no study to date has reported on Stat localization or phosphorylation in the nonepithelial compartment of normal or malignant breast tissues, or in the various histological subtypes of breast cancer.
Data from Mouse Models of Breast Cancer
In vivo, the effect of hemizygous loss of Stat5a in a SV40-T antigen transgenic mouse model of mammary cancer has been tested after cross-breeding. The resultant progeny (Stat5a+/−+ Tag) demonstrated a significant reduction in the percentage of tumor-bearing mice, as well as decreased tumor size, delayed first tumor appearance, and increased apoptotic indices.29 The role of Stat3 in murine mammary cancer models remains to be determined, and awaits cross-breeding of the conditional Stat3 mammary knockout mouse with other murine models of mammary cancer.19
Data from Cell Lines—Insights into Function
Stat3 tyrosine phosphorylation and DNA-binding activity of several breast cancer lines has been found to be elevated; conversely, pharmacological or dominant-negative inhibition of Stat3 activity has been found to block the proliferation and survival of breast cancer cells in vitro.30–32 Stat5 has been demonstrated to stimulate the transcriptional activity of the cyclin D locus.33 Transfection of a constitutively activated Stat5 resulted in growth factor-independent growth of the murine Ba/F3 pro-B cell line34 and overexpression of Stat5b has been found to potentiate v-src-mediated transformation of NIH-3T3 cells.35 As noted below, repression of Stat5 DNA-binding activity by PIAS3 inhibits breast cancer cell growth.36 Collectively, these studies have argued for a significant role for Stats in breast cancer proliferation, survival, and differentiation.
The mechanisms through which the Stats promote these protumorigenic events has been an active area of investigation; several lines of evidence indicate that Stats 3 and 5 can activate the transcription of genes associated with cell-cycle progression, cell survival and transformation, and angiogenesis, as elaborated in recent reviews.37,38 Several reports have implicated Stat3 and Stat5 activity in the up-regulation of expression of the cell cycle-regulatory proteins cyclin D1 and D2 in several nonmammary cell lines. However, only a single report to date has suggested that Stats may have similar effects in breast epithelial cells.39 Regulation of anti-apoptotic members of the Bcl-2 family, specifically Bcl-XL, has also been associated with Stat3 and Stat5 activity37,40 in several nonmammary cell lines. Stat 3- and Stat5-induced transcription has also been implicated in c-myc proto-oncogene overexpression;40 like Bcl-XL, little data currently demonstrates whether the activity of the Stats contributes to the regulation of these proteins in mammary epithelial cells. Stat3 activity has also been recently implicated in up-regulated vascular endothelial growth factor expression, an event that may further serve to promote tumor progression.41 Although collectively these data are intriguing from a mechanistic standpoint, further research is required because little data exists regarding Stat-mediated gene up-regulation in malignant breast tissues; furthermore, the specific contributions of the cyclins, Bcl-XL, c-myc, and vascular endothelial growth factor during the pathogenesis of human breast cancer remain to be completely elucidated.
Parallels in Prostate
Many signaling pathways thought to contribute to breast neoplasia are also activated in prostate cancer, and Stats 3 and 5 are no exception. One study of 42 human prostate carcinomas with matched normal controls found significant elevations of Stat3 tyrosine phosphorylation and DNA-binding activity in the malignant specimens;42 no correlations with Gleason grade or prostate-specific antigen levels were noted. Interestingly, surrounding normal tissues associated with prostate cancers were also noted to have elevated levels of Stat3 activity, suggesting that Stat3 activation may precede frank histopathologically detectable prostate cancer. Similarly, 65% of human prostate cancers were noted to have nuclear localized, tyrosine-phosphorylated Stat5.43 Introduction of a dominant-negative Stat construct into the human prostate cancer cell lines CWR22Rv and LnCap was also found to induce cellular apoptosis.43 Interestingly, Stat5a knockout mice demonstrated acinar cyst formation and cell degeneration reminiscent of benign prostatic hypertrophy, however, epithelial hyperplasia or increased prostate size were not noted.44 These data suggest, that, as in the mammary gland, Stat5 contributes to the maintenance of normal tissue architecture and function in the prostate.
Regulation of Stat Function
Tyrosine Phosphorylation
Stat activation requires C-terminal tyrosine phosphorylation by a receptor-associated Jak kinase.1,45 This phosphorylation event occurs adjacent to the transcriptional activation domain within Stats (at position 705 in Stat3 and Y694 and Y699 in Stat5a and Stat5b, respectively) and induces dimerization/multimerization and nuclear translocation of the Stat complex where it engages its cognate DNA-binding sequence, resulting in promoter transactivation under appropriate conditions.2,9,13,46,47 Tyrosine phosphorylation is necessary for Stat activity; replacement of this tyrosine residue at Y694/699/705 results in an inactive Stat incapable of nuclear translocation or transactivation. However, other tyrosine kinase-signaling pathways may also impact Stat activation because both Stats 3 and 5 can be phosphorylated by Src family members.32,35,48 Such Src-mediated phosphorylation by itself, however, does not always results in Stat activation because only Src-induced phosphorylation of Stat5b, but not Stat5a, induced nuclear translocation.48 However, it is clear that Stats are downstream of Src-family-mediated signaling because transcriptionally inactive Stat3 blocks Src-mediated transformation.32 As noted above, enhanced levels of Stat3 and Stat5 tyrosine phosphorylation have been noted in human breast cancers.20,26
Serine Phosphorylation
All Stat family members, with the exception of Stat2, undergo serine phosphorylation after receptor-mediated signaling. Serine phosphorylation at residue 727 of Stats 1 and 3 results in a significant up-regulation of the transcriptional activity of these Stats.49,50 This event appears to be mediated by several converging kinases including MAPK, p38, JNK, and protein kinase Cδ.51–53 Serine phosphorylation of Stat3 appears essential in vivo for postnatal survival and growth because knock-in of a mutant Stat3 cDNA bearing an alanine for serine replacement at position 727 into Stat3 knockout mice failed to compensate the noted phenotype.54 As discussed at greater length below, both of the small GTP binding proteins Rac and Rho are capable of up-regulating Stat3 serine phosphorylation and transactivation; indeed dominant-negative Rac inhibits Stat3 activity, and conversely, dominant-negative Stat3 blocks mutant oncogenic RhoA-induced cell transformation.55,56
In contrast, serine phosphorylation at positions 725 and 779 in Stat5a and position 730 in Stat5b down-regulates the transcriptional activity of Stat5.53,57,58 Serine phosphorylation of both Stat5a and Stat5b appears to be increased during late pregnancy and lactation.57 The phosphorylation of serine 725 and 779 of Stat5a is co-operative and mediated by both MAPK and non-MAPK signaling pathways; however, the suppressive effects of this phosphorylation appears to be mitigated in part by co-stimulation of glucocorticoid receptors in MCF7 breast cancer cells.57 Despite these tantalizing data regarding the serine phosphorylation of Stats 3 and 5, the exact levels of serine phosphorylation of these transcription factors during the pathogenesis of human breast cancer remains to be determined.
Dephosphorylation
Several phosphatases, notably SHP1, SHP2, CD45, PTP1B, and TCPTP59 have been demonstrated to regulate Jak kinase activity, and many of these have also been found in association with the Stat family. A structural example of a SHP phosphatase is presented in Figure 2; typically, receptor-associating phosphatases contain tyrosine-binding SH2 motifs, a phosphatase domain, and regulatory tyrosine residues that undergo variable phosphorylation. TCPTP has been found to associate with Stat5a and Stat5b within the nucleus and induce their dephosphorylation and inactivation; interestingly the phosphatase activity of TCPTP is not required for the inactivation of Stat5-mediated gene expression (ie, the association of TCPTP in this regard may be sufficient).60 The overexpression of PTP1B also has similar effects on Stat5a and Stat5b phosphorylation and activity; however, this phosphatase is found within the cytoplasm.61 SHP2 has been found in association with Stat5a,62,63 and migrates with Stat5 as a complex into the nucleus. Interestingly, the intact phosphatase activity of SHP2 is required for Stat5 phosphorylation, suggesting that this phosphatase is not directly involved in the dephosphorylation/deactivation of Stat5, but instead indirectly with the activation of this Stat. SHP1 has also been reported to associate with Stat5b in the liver.64 Although the effects of TCPTP, PTP1B, and SHP2 on Stat activity have been reported in mammary epithelial cell lines, these studies have singularly used overexpression methodologies. Furthermore, to date no study has examined either the expression or activity of these phosphatases in the context of primary human neoplasms. Thus, additional studies examining the role of the phosphatases during the pathogenesis of human breast cancer may prove highly illuminating.
SOCS Proteins
The SOCS (suppressors of cytokine signaling) family consists of eight members comprised of SOCS1 to SOCS7 and CIS (cytokine-inducible SH2 domain proteins). Each family member demonstrates three domains: a poorly conserved amino terminus, a central phosphotyrosine-binding SH2 domain, and a conserved, carboxy terminal SOCS-box motif, that may mediate posttranslational ubiquitination (Figure 2).65–67 In resting cells, SOCS proteins are expressed at low levels; after receptor-mediated Stat family signaling, SOCS levels dramatically increase within 20 to 40 minutes. This increase is principally mediated at the transcriptional level,68,69 however posttranslational mechanisms may also contribute (ie, phosphorylation) to SOCS protein stability.70 SOCS proteins serve as classic negative counterregulatory inhibitors of Stat activation. This occurs by several mechanisms. SOCS1 blocks Stat phosphorylation and activation by directly binding to phosphorylated JAK, whereas SOCS3 inhibits JAK activity by first binding receptor. In contrast, CIS blocks Stat activation by blocking Stat-binding sites on receptors.71–73 In addition, the ability of the SOCS box to engage elements of the ubiquitination pathway may significantly contribute to the down-regulation of receptor activated Jak kinase.74 Analysis of mice bearing knockouts of specific SOCS family members has revealed dysregulation of cytokine and growth hormone signaling in vivo.59 Modulation of both CIS and SOCS3 levels in mammary epithelial cells has been found to differentially regulate Stat5 signaling.75 In parallel, knockout of a single SOCS1 allele resulted in correction of the lactational deficiency presented in PRLr+/− heterozygous mice; in contrast knockout of SOCS1 in an IFN-γ−/− mouse resulted in accelerated lobuloalveolar development and precocious lactation.76 Together these knockout data suggest that SOCS1 has a considerable role in dampening PRL-activated Stat signaling in mammary tissues. A single report examining small numbers of primary human breast cancer has found elevations in SOCS family transcripts and protein,77 an event that may contribute to elevated Stat family signaling in breast cancer.
Caveolin
Caveolins are necessary and sufficient for the formation of caveola that serve to concentrate and internalize lipids and cell surface receptor-signaling complexes. Recently, the observation that caveolin-1 shares homology with the SOCS family, led to the hypothesis that this protein may also function to down-regulate Jak-Stat signaling.78 Indeed, when examined, caveolin-1 was found to associate specifically with Jak2 and down-regulate Jak2-Stat5a signaling. When caveolin-1 knockout mice were generated, these animals during pregnancy were found to have accelerated development of the lobulo-alveolar compartment of the mammary gland, premature milk production, and enhanced phosphorylation of Stat5a.78 These finding indicate that caveolin-1 may function in a manner analogous to the SOCS family. Additional matings of the caveolin-1−/− mice with the mammary tumor-prone polyoma middle T transgenic mice, revealed that the offspring had significant acceleration in the development of dysplastic mammary lesions.79 As loss and/or mutation of the caveolin-1 genomic locus has been reported in breast cancer, these findings suggest that caveolin-1, acting through the Jak-Stat pathway may contribute to the pathogenesis of human breast cancer.
Rac/Rho
Recent evidence has indicated that the Rho family of small GTP-binding proteins may influence Stat activation at multiple levels. Overexpression studies have indicated that the Rho family member Rac1 can directly bind to Stat3;55 use of both constitutively active and dominant-negative forms of Rac suggested that Rac activity may influence both the tyrosine and serine phosphorylation of Stat3, as well as its activity. This regulation may occur at multiple levels because data has also suggested that Rac can influence Jak2 activity,55 stress-activated kinase 4,80 and the autocrine elaboration of cytokines, such as interleukin-6,81 that also may indirectly influence Stat phosphorylation and activity. Similarly, RhoA can modulate the tyrosine and serine phosphorylation status and activity of Stat3 through the tyrosine kinases Jak2 and Src, and the serine kinase JNK.56 It is uncertain whether RhoA directly interacts with Stat3, and it is unclear whether the actions of either RhoA or Rac are exerted purely at the cytoplasmic level or whether these two GTP-binding proteins accompany Stat3 into the nucleus. Recent data has also indicated that like Stat3, Stat5 may also be activated by RhoA.82 These data suggest that RhoA may increase the tyrosine and decrease the serine phosphorylation levels of Stat5 and contribute to the process of epithelial to mesenchymal transition. In addition, recent data from our laboratory has indicated that the guanine nucleotide exchange factor, Vav2, can bind Stat5 directly and up-regulate the activity of both Rac1 and Stat5 (Miller SL, DeMaria JE, Freier D, Riegal AM, Clevenger CV, submitted for publication). Several lines of evidence now indicate that both Rac1 and RhoA protein expression is elevated in breast malignancies, particularly those of high-grade or advanced stage;83–85 preliminary evidence suggests these proteins are not mutated and that the mechanisms of their overexpression is posttranscriptional.85 Thus, the activation of small GTP-binding proteins may provide an additional mechanism for additional signal integration and activation of the Stat5 complex.
Ligand/CypB
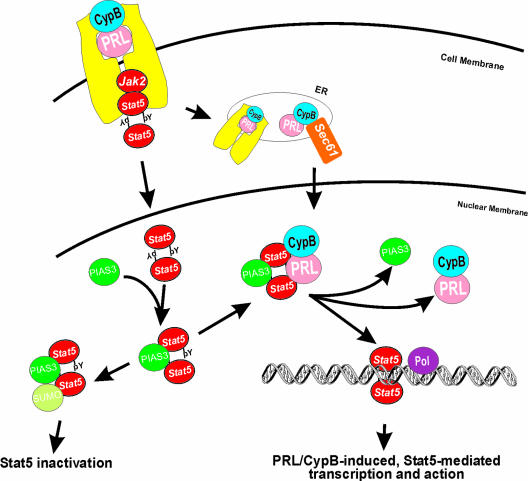
Classic theory dictates that peptide hormones regulate Stat family activity at a distance via cell-surface receptors, ie, by receptor-mediated activation of Jak2 and MAPK, which in turn phosphorylate and regulate Stat family activity. A growing body of evidence, however, has indicated an intranuclear function for polypeptide hormones with relevance to breast cancer, including PRL, growth hormone, epidermal growth factor, nerve growth factor, platelet-derived growth factor, fibroblast growth factor, interleukin 5, and insulin.86 In parallel with the cell-surface activation of Stat, these hormones are endocytosed and retrotranslocated to the nucleus after receptor binding.87–89 The relationship between the nuclear actions of polypeptide hormones and Stats had been unclear until the recent discovery by my laboratory of the role of the peptidyl prolyl isomerase cyclophilin B (CypB) in the nuclear transport and function of PRL.90 Our studies revealed that complex between PRL and CypB exists in human serum. After receptor-mediated transport of the PRL/CypB, this complex is retrotransported to the nucleus via a N-terminal nuclear localization sequence present in CypB (Figure 3). Within the nucleus the PRL/CypB complex directly interacts with Stat5. PRL/CypB acts as a transcriptional inducer of Stat5 by facilitating the interaction of this transcription factor with DNA by inducing the release of a repressor of Stat5, namely PIAS3.36 The actions of the PIAS family of proteins with respect to Stat activity are discussed below. By inducing the release of this Stat repressor, intranuclear PRL can directly potentiate the activity of Stat5. These observations would suggest that considerable parallels exist between steroid and peptide hormones in their respective genomic and nongenomic actions, and like steroid hormones, may suggest that polypeptide ligands contribute to their signaling specificity through their intranuclear functions. Although preliminary data from our laboratory90 also indicate that CypB can potentiate the activity of growth hormone, its role with respect to other polypeptide hormones implicated in the pathogenesis of breast cancer remain unclear. Whether such mechanisms contribute to the regulation of Stat3 activity in breast tissues also remains to be determined.
Figure 3.
Mechanism for PRL/CypB induction of Stat5-induced gene expression. After receptor-mediated endocytosis, the transport of PRL into the nucleus is facilitated by its interaction with CypB. On entry into the nucleus, this complex interacts with Stat5 inducing the release of a repressor of its DNA-binding activity, namely PIAS3. The PRL/CypB-mediated release of PIAS3 from Stat5 results in significantly enhanced gene expression. Abbreviations: Pol, RNA polymerase II transcriptional apparatus; Sec61, ER transporter apparatus.
Precedent studies have demonstrated ubiquitous expression of CypB. Although the expression of PRL has been classically associated with the pituitary, the local autocrine/paracrine elaboration of this hormone has been documented at multiple sites, including breast tissues.24,25 Recent unpublished studies in our laboratory have also found appreciable PRL within the nucleus of breast malignancies. If the intranuclear actions of PRL contribute to the growth of PRL-responsive tissues, then interference with this pathway may prove useful in the treatment of breast cancer. To test this hypothesis, an enzymatically inactive mutant of CypB was synthesized by recombinant technique and introduced into the culture medium of human breast cancer cells. This treatment resulted in a significant inhibition of the growth of such cells,36 at concentrations 100- to 1000-fold less than any previously reported PRL antagonist.91 Collectively, these data would suggest that pharmacological manipulation of intranuclear actions of polypeptide hormones may inhibit Stat activity and be of therapeutic utility in the treatment of breast cancer.
PIAS/Sumoylation
The PIAS (for peptide inhibitors of activated Stats) family of proteins (PIAS1, PIAS3, PIASx, and PIASy) have been found to bind Stat family members and block their binding to DNA and/or transcriptional activity.92–94 PIAS proteins demonstrate three regions of protein homology: a signature N-terminal LXXLL motif, thought to contribute to nuclear receptor interactions, a central ring finger domain, whose function was unknown, until recently (see below), and a serine-rich C-terminus (Figure 2). Unlike the cytokine-induced expression of the SOCS/CIS family, PIAS proteins are constitutively expressed within the nucleus and appear to act as constitutive repressors of Stat activity.94 Each PIAS member associates and modulates the function of distinct subset of transcription factors, ie, PIAS1 binds to Stat1 and p53;93,95 PIAS3 associates with Stats 3, 5a, 5b, and Gfi;36,92,96 PIASx with the androgen receptor;97 and PIASy with LEF1.98 While the nature of the interaction between PIAS proteins and transcription factors is undergoing elucidation, recent evidence examining the PIAS1-Stat1 association indicates that the N-terminal domain of Stat1 binds to the region between the ring finger and serine/threonine-rich domains of PIAS1, a region termed the linker domain.99 How the PIAS family modulates transcription factor function has been uncertain, however, recent data have indicated that PIAS proteins serve as small ubiquitin-like modifier (SUMO) E3 ligases.95,98,100,101
Sumoylation is the process whereby one of three SUMO (for small ubiquitin-like modifiers) peptides (termed SUMO1 to SUMO3), consisting of ∼100 amino acids (ie, comparable in length to ubiquitin) is added to a consensus sumoylation site (ΨKXE) within a peptide. Like ubiquitin conjugation, sumoylation requires an E1-activating enzyme and an E2 conjugase.101 Although this process can occur without an E3 ligase, it is not efficient. The existence of mammalian E3 ligases was not recognized until two separate studies using yeast two-hybrid approaches suggested that PIAS family members were such SUMO ligases. These studies revealed that PIAS1 and PIASy in vivo and in vitro promoted the sumoylation of p53 and LEF1, respectively.95,98,102 Unlike ubiquitin conjugation, sumoylation appears to modify protein function not through degradation, but instead by altering function, localization, or extent of ubiquitylation.100 PIAS-induced sumoylation requires the ring finger motif of PIAS because deletion or modification of this motif eliminates sumoylation of PIAS-binding partners.98,102 Interestingly, the functional effects of PIAS association and PIAS-induced sumoylation appear to be distinct. For instance, removal of sumoylation sites in LEF1 did not alter its targeting to nuclear bodies, whereas deletion of the ring motif in PIASy did block such targeting.98 Loss of the ring finger domain from PIAS1 did not block p53-mediated transcription in one study,103 although other studies have suggested that this effect may be cell-type specific.102 These data suggest that functional modulation by PIAS proteins may result as a consequence of PIAS interaction, subsequent sumoylation, or both in a given cellular context.
Collectively, it appears that either the direct association of PIAS family members, or the SUMO conjugation they induce, can significantly modify the function of transcription factors that include members of the Stat family. Recent data from our laboratory has indicated that PIAS3 overexpression in breast cancer cell lines can significantly modulate Stat5-mediated gene expression and induce cellular apoptosis.36 The extent of PIAS and SUMO family expression in breast tissues remains unclear, although preliminary evidence from our laboratory suggests that dysregulation of PIAS expression does occur in human breast cancers.
Transcriptional Co-Regulators
The activity of Stats 3 and 5 has been shown to be up-regulated by its interaction with transcription factors/co-activators including CBP, Nmi, CPAP, c-jun, SMRT, and the glucocorticoid receptor53,94,104–109 and histone deactylases.110 Although the association of some of these proteins, such as CBP, with Stat family proteins is thought to be rate limiting to transactivation, the precise function and expression of most of these proteins with respect to Stat5 activity in breast cancer remains to be determined.
Future Opportunities—From the Lab to the Bedside
The central role that Stats may play in breast cancer relates to their intrinsic ability to integrate the incoming signals from activated receptors and their associated signal transduction networks. As such they are ripe for additional studies, both as prognostic markers and therapeutic targets.
As noted above, preliminary data indicate that the levels of Stat3 and Stat5 expression and/or phosphorylation may relate to the differentiation and/or favorable outcome. As in many initial studies, limitations as to sample size, outcome follow-up, and experimental design found in the immunohistochemical and EMSA studies with Stats 3 and 5 to date will require additional follow-up investigation. Because Stat transcriptional activity may be the central measure of the biological function of Stats 3 and 5, additional assays other than measurement of Stat phosphorylation and DNA binding may need to be applied to clinical specimens. Furthermore, the analysis of Stat activity will need to be targeted to the pluripotent tumor stem cells that may require Stat activity for survival and/or growth.111
With the development of high-throughput screening assays for Stat-protein interaction and Stat transcriptional activity,112 the ability to screen sizable libraries for small pharmacological inhibitors of Stats has become a distinct possibility. It is conceivable that an effective inhibitor of Stat function may interrupt its ability to interact with DNA, alter its posttranslational modifications (phosphorylation/sumoylation), or block/enhance its interaction with the transcriptional co-activators or co-repressors.113 Given the existent data from cell lines and animal models overviewed above, it is probable that a future pharmacological agent targeting Stat function will be of use in the treatment of human breast cancer.
Footnotes
Address reprint requests to Charles V. Clevenger, M.D., Ph.D., Department of Pathology and Laboratory Medicine, University of Pennsylvania, 513 SC Labs, 422 Curie Blvd., Philadelphia, PA 19104. E-mail: clevengc@mail.med.upenn.edu.
Supported by the National Cancer Institute, National Institutes of Health (grants R01 CA69294, CA92265, and CA102682).
References
- Schindler C, Shuai K, Prezioso VR, Darnell JE. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- Darnell JE. The JAK-STAT pathway: summary of initial studies and recent advances. Rec Prog Horm Res. 1996;51:391–404. [PubMed] [Google Scholar]
- Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Wen Z, Darnell JE. Stat3: a Stat family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- Sadowski HB, Shuai K, Darnell JE, Gilman MZ. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- Ihle JN, Kerr IM. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- Silvennoinen O, Witthuhn BA, Quelle FW, Cleveland JL, Yi T, Ihle JN. Structure of the murine Jak2 protein tyrosine-kinase and its role in interleukin 3 signal transduction. Proc Natl Acad Sci USA. 1993;90:8429–8433. doi: 10.1073/pnas.90.18.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witthuhn BA, Quelle FW, Silvennoinen O, Yi T, Tang B, Miura O, Ihle JN. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Kerr IM, Stark GR. JAK-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- Bromberg JF. Activation of Stat proteins and growth control. Bioessays. 2001;23:161–169. doi: 10.1002/1521-1878(200102)23:2<161::AID-BIES1023>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- John S, Vinkemeier U, Soldaini E, Darnell JE, Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol. 1999;19:1910–1918. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Vinkemeier U, Zhao Y, Jerzalmi D, Darnell JE, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- Watson CJ. Stat transcription factors in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:115–127. doi: 10.1023/a:1009524817155. [DOI] [PubMed] [Google Scholar]
- Liu X, Robinson GW, Wagner K-U, Garrett L, Wyhshaw-Boris A, Henninghausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- Horseman ND, Zhao W, Montecino-Rodriguiez E, Tanaka M, Nakashima K, Engle SJ, Smith F, Markoff E, Dorshkind K. Defective mammopoiesis, but normal hematopoiesis, in mice with a targeted disruption of the prolactin gene. EMBO J. 1997;16:6926–6935. doi: 10.1093/emboj/16.23.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormandy CJ, Camus A, Barra J, Damotte D, Lucas B, Buteau H, Edery M, Brousse N, Babinet C, Binart N, Kelly PA. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 1997;11:167–178. doi: 10.1101/gad.11.2.167. [DOI] [PubMed] [Google Scholar]
- Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, Akira S, Clarke R, Watson CJ. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ, Miller WR. Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br J Cancer. 1995;71:840–844. doi: 10.1038/bjc.1995.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berclaz G, Alternatt HJ, Rohrbach V, Siragusa A, Dreher E, Smith PD. EGFr dependent expression of Stat3 (but not Stat1) in breast cancer. Int J Oncol. 2001;19:1155–1160. doi: 10.3892/ijo.19.6.1155. [DOI] [PubMed] [Google Scholar]
- Dolled-Filhart M, Camp RL, Kowalski DP, Smith BL, Rimm DL. Tissue microarray analysis of signal transducers and activators of transcription 3 (Stat3) and phospho-Stat3 in node-negative breast cancer shows nuclear localization is associated with a better prognosis. Clin Cancer Res. 2003;9:594–600. [PubMed] [Google Scholar]
- Nevalainen MT, Xie J, Bubendorf L, Wagner K-U, Rui H. Basal activation of transcription factor Stat5 in nonpregnant mouse and human breast epithelium. Mol Endocrinol. 2002;16:1108–1124. doi: 10.1210/mend.16.5.0839. [DOI] [PubMed] [Google Scholar]
- Clevenger CV, Chang W-P, Ngo W, Pasha TLM, Montone KT, Tomaszewski JE. Expression of prolactin and prolactin receptor in human breast carcinoma: evidence for an autocrine/paracrine loop. Am J Pathol. 1995;146:695–705. [PMC free article] [PubMed] [Google Scholar]
- Reynolds C, Montone KT, Powell CM, Tomaszewski JE, Clevenger CV. Distribution of prolactin and its receptor in human breast carcinoma. Endocrinology. 1997;138:5555–5560. doi: 10.1210/endo.138.12.5605. [DOI] [PubMed] [Google Scholar]
- Cotarla I, Ren S, Zhang Y, Gehan E, Singh B, Furth PA. Stat5a is activated and nuclear localized in a high proportion of human breast cancers. Int J Cancer. 2003;108:665–671. doi: 10.1002/ijc.11619. [DOI] [PubMed] [Google Scholar]
- Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, Sauter G, Rui H. Signal transducer and activator of transcription-5 (Stat5) activation and breast cancer prognosis. J Clin Oncol. 2004;22:2053–2060. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Widschwendter A, Tonko-Geymayer S, Welte T, Daxenbichler G, Marth C, Doppler W. Prognostic significance of signal transducer and activator of transcription 1 activation in breast cancer. Clin Cancer Res. 2002;8:3065–3074. [PubMed] [Google Scholar]
- Ren S, Cai HR, Li M, Furth PA. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene. 2002;21:4355–4339. doi: 10.1038/sj.onc.1205484. [DOI] [PubMed] [Google Scholar]
- Li L, Shaw PE. Autocrine-mediated activation of STAT3 correlates with cell proliferation in breast carcinoma lines. J Biol Chem. 2002;277:17397–17405. doi: 10.1074/jbc.M109962200. [DOI] [PubMed] [Google Scholar]
- Burke WM, Jin X, Lin HJ, Huang M, Liu R, Reynolds RK, Lin J. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene. 2001;20:7925–7934. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muo-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, Laudano A, Gazit A, Levitzki A, Kraker A, Jove R. Constitutive activation of Stat3 by the Src and Jak tryosine kinases participates in growth regulation of human breast cancer cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- Brockman JL, Schroeder MD, Schuler LA. PRL activates the cyclin d1 promoter via the jak2/stat pathway. Mol Endocrinol. 2002;16:774–784. doi: 10.1210/mend.16.4.0817. [DOI] [PubMed] [Google Scholar]
- Onishi M, Nosaka T, Misawa K, Mui ALF, Gorman DM, McMahon M, Miyajima A, Kitamura T. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazansky AV, Rosen JM. Signal transducers and activators of transcription 5B potentiates v-Src mediated transformation of NIH-3T3 cells. Cell Growth Differ. 2001;12:1–7. [PubMed] [Google Scholar]
- Rycyzyn MA, Clevenger CV. The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. Proc Natl Acad Sci USA. 2002;99:6790–6795. doi: 10.1073/pnas.092160699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Jove R. The Stats of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. Stat proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- Brockman JL, Schroeder MD, Schuler LA. Prolactin activates the cyclin D1 promoter via the JAK2-STAT pathway. Mol Endocrinol. 2002;16:774–784. doi: 10.1210/mend.16.4.0817. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wreszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Pardoll D, Jove R, Yu H. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- Dhir R, Ni Z, Lou W, DeMiguel F, Grandis JR, Gao AC. Stat3 activation in prostatic carcinomas. Prostate. 2002;51:241–246. doi: 10.1002/pros.10079. [DOI] [PubMed] [Google Scholar]
- Ahonen T, Xie J, LeBaron MJ, Zhu J, Nurmi M, Alanen K, Rui H, Nevalainen MT. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J Biol Chem. 2003;278:27287–27292. doi: 10.1074/jbc.M304307200. [DOI] [PubMed] [Google Scholar]
- Nevalainen MT, Ahonen TJ, Yamashita H, Chadrashekar V, Bartke A, Grimley PM, Robinson GW, Hennighausen L, Rui H. Epithelial defects in prostates of Stat5a-null mice. Lab Invest. 2000;80:993–1006. doi: 10.1038/labinvest.3780105. [DOI] [PubMed] [Google Scholar]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine J-C, Teglund S, Vanin EF, Bodner S, Calamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Shuai K, Stark GR, Kerr IM, Darnell JE. A single phosphotyrosine residue of Stat1 is required for gene activation by interferon-γ. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- Qureshi SA, Leung S, Kerr IM, Stark GR, Darnell JE. Function of Stat2 protein in transcriptional activation alpha interferon. Mol Cell Biol. 1996;16:288–293. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazansky AB, Kabotyanski EB, Wyszomierski SL, Mancini MA, Rosen JM. Differential effects of prolactin and src/abl kinases on the nuclear translocation of STAT5B and STAT5A. J Biol Chem. 1999;274:22484–22492. doi: 10.1074/jbc.274.32.22484. [DOI] [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- Nagy ZS, Wang Y, Erwin-Cohen RA, Aradi J, Monia B, Wang LH, Stepkowski SM, Rui H, Kirken RA. Interleukin-2 family cytokines stimulate phosphorylation of the Pro-Ser-Pro motif of Stat5 transcription factors in T cells: resistance to suppression of multiple serine kinase pathways. J Leuk Biol. 2002;72:819–828. [PubMed] [Google Scholar]
- Wierenga AT, Vogelzang I, Eggen BJ, Vellenga E. Erythropoietin-induced serine 727 phosphorylation of Stat3 in erythroid cells is mediated by a MEK-, ERK-, and MSK1-dependent pathway. Exp Hematol. 2003;31:398–405. doi: 10.1016/s0301-472x(03)00045-6. [DOI] [PubMed] [Google Scholar]
- Pircher TJ, Peterson H, Gustafsson J-A, Haldosen LA. Extracellular signal-regulate kinase (ERK) interacts with signal transducer and activator of transcription (STAT) 5a. Mol Endocrinol. 1999;13:555–565. doi: 10.1210/mend.13.4.0263. [DOI] [PubMed] [Google Scholar]
- Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DF, Darnell JE. Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol. 2004;24:407–419. doi: 10.1128/MCB.24.1.407-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AR, Vikis HG, Stewart S, Fanburg BL, Cochran BH, Guam KL. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290:144–147. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- Aznar S, Valeron PF, del Rincon SV, Perez LF, Perona R, Lacal JC. Simultaneous tyrosine and serine phosphorylation of STAT3 transcription factor is involved in RhoA GTPase oncogenic transformation. Mol Biol Cell. 2001;12:3282–3294. doi: 10.1091/mbc.12.10.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Nevalainen MT, Xu J, LeBaron MJ, Wagner K-U, Erwin RA, Harmon JM, Hennighausen L, Kirken RA, Rui H. Role of serine phosphorylation of Stat5a in prolactin-stimulated beta-casein gene expression. Mol Cell Endocrinol. 2001;183:151–163. doi: 10.1016/s0303-7207(01)00546-9. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Xu J, Erwin RA, Farrar WL, Kirken RA, Rui H. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J Biol Chem. 1998;273:30218–30224. doi: 10.1074/jbc.273.46.30218. [DOI] [PubMed] [Google Scholar]
- Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- Aoki N, Matsuda T. A nuclear protein tyrosine phosphatase TC-PTP is a potential negative regulator of the PRL-mediated signaling pathway: dephosphorylation and deactivation of STAT5a and 5b by TC-PTP in the nucleus. Mol Endocrinol. 2002;16:58–69. doi: 10.1210/mend.16.1.0761. [DOI] [PubMed] [Google Scholar]
- Aoki N, Matsuda T. A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J Biol Chem. 2000;275:39718–39726. doi: 10.1074/jbc.M005615200. [DOI] [PubMed] [Google Scholar]
- Chughtai N, Schimchowitsch S, Lebrun J-J, Ali S. Prolactin induces SHP-2 association with Stat5, nuclear translocation, and binding to the β-casein gene promoter in mammary cells. J Biol Chem. 2002;277:31107–31114. doi: 10.1074/jbc.M200156200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wen R, Yang S, Schuman J, Zhang EE, Yi T, Feng G-S, Wang D. Identification of SHP2 as a Stat5a phosphatase. J Biol Chem. 2003;278:16520–16527. doi: 10.1074/jbc.M210572200. [DOI] [PubMed] [Google Scholar]
- Ram PA, Waxman DJ. Interaction of growth hormone-activated STATs with SH2-containing phosphotyrosine phosphatase SHP-1 and nuclear JAK2 tyrosine kinase. J Biol Chem. 1997;272:17694–17702. doi: 10.1074/jbc.272.28.17694. [DOI] [PubMed] [Google Scholar]
- Hilton DJ. Negative regulators of cytokine signal transduction. Cell Mol Life Sci. 1999;55:1568–1577. doi: 10.1007/s000180050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr R, Hilton DJ. Negative regulation of the JAK/STAT pathway. Bioessays. 1999;21:47–52. doi: 10.1002/(SICI)1521-1878(199901)21:1<47::AID-BIES6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Yasukawa H, Sasaki A, Yoshimura A. Negative regulation of cytokine signaling pathways. Annu Rev Immunol. 2000;18:143–164. doi: 10.1146/annurev.immunol.18.1.143. [DOI] [PubMed] [Google Scholar]
- Hilton DJ, Richardson RT, Alexander WS, Viney EM, Wilson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci USA. 1997;387:924–929. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, Akira S, Kishimoto T. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- Bram RJ, Hung DT, Martin PK, Schreiber SL, Crabtree GR. Identification of the immunophilins capable of mediating inhibition of signal transduction by cyclosporin A and FK506: roles of calcineurin binding and cellular location. Mol Cell Biol. 1993;13:4760–4769. doi: 10.1128/mcb.13.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Yasukawa H, Shouda T, Kitamura T, Dikic I, Yoshimura A. CIS3/SOCS3 suppresses erythropoietin signaling by binding the EPO receptor and JAK2. J Biol Chem. 2000;275:29338–29347. doi: 10.1074/jbc.M003456200. [DOI] [PubMed] [Google Scholar]
- Tomic S, Chughtai N, Ali S. SOCS-1, -2, -3: selective targets and functions downstream of the prolactin receptor. Mol Cell Endocrinol. 1999;158:45–54. doi: 10.1016/s0303-7207(99)00180-x. [DOI] [PubMed] [Google Scholar]
- Yasukawa H, Misawa H, Sakamoto H, Masahuara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-G, Farley A, Nicholson SE, Willson TA, Zugara LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausman G, Kile BJ, Kent SBH, Alexander WS, Metcalf D, Hilton DJ, Nicola NA, Baca M. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci USA. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonko-Gronmeyer S, Goupille O, Tonko M, Soatroi C, Yoshimura A, Strueli C, Ziemiecki A, Kofler R, Doppler W. Regulation and function of the cytokine-inducible SH2 domain proteins, CIS and SOCS3, in mammary epithelial cells. Mol Endocrinol. 2002;16:1680–1695. doi: 10.1210/mend.16.7.0872. [DOI] [PubMed] [Google Scholar]
- Lindeman GJ, Wittlin S, Lada H, Naylor MJ, Santamaria M, Zhang JG, Starr R, Hilton DJ, Alexander WS, Ormandy CJ, Visvader J. SOCS1 deficiency results in accelerated mammary gland development and rescues lactation in prolactin receptor-deficient mice. Genes Dev. 2001;15:1631–1636. doi: 10.1101/gad.880801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raccurt M, Tam SP, Lau P, Lambert A, Garcia-Cabellero T, Li H, Brown RJ, McGuckin MA, Morel G, Waters MJ. Suppressor of cytokine signaling gene expression is elevated in breast carcinoma. Br J Cancer. 2003;89:524–532. doi: 10.1038/sj.bjc.6601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Lee H, Frank PG, Razani B, Nguyen AV, Parlo AF, Russel RG, Hulit J, Pestell RG, Lisanti MP. Caveolin-1-deficient mice show accelerated mammary gland development during pregnancy, premature lactation, and hyperactivation of the Jak2-Stat5a signaling cascade. Mol Biol Cell. 2002;13:3416–3430. doi: 10.1091/mbc.02-05-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Cheung MW, Park DS, Razani B, Cohen AW, Muller WJ, Di Vizio D, Chopra NG, Pestell RG, Lisanti MP. Loss of caveolin-1 gene expression accelerates the development of dysplastic mammary lesions in tumor-prone transgenic mice. Mol Biol Cell. 2003;14:1027–1042. doi: 10.1091/mbc.E02-08-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuringa JJ, Jonk LJ, Dokter WH, Vellenga E, Kruijer W. Interleukin-6-induced Stat3 transactivation and Ser727 phosphorylation involved Vav, Rac1 and the kinase SEK1/MKK4 as signal transduction components. Biochem J. 2000;347:89–96. [PMC free article] [PubMed] [Google Scholar]
- Faruqi TR, Gomez D, Bustelo XR, Bar-Sagi D, Reich NC. Rac1 mediates STAT3 activation by autocrine IL-6. Proc Natl Acad Sci USA. 2001;98:9014–9019. doi: 10.1073/pnas.161281298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar Benitah S, Valeron PF, Rui H, Lacal JC. Stat5a activation mediates the epithelial to mesenchymal transition induced by oncogenic RhoA. Mol Biol Cell. 2003;14:40–53. doi: 10.1091/mbc.E02-08-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, Harbeck N, Schmitt M, Lengyel E. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform Rac1b. Oncogene. 2000;19:3013–3020. doi: 10.1038/sj.onc.1203621. [DOI] [PubMed] [Google Scholar]
- Fritz G, Just I, Kaina B. Rho GTPases are over-expressed in human tumors. Int J Cancer. 1999;81:682–687. doi: 10.1002/(sici)1097-0215(19990531)81:5<682::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumors: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87:635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger CV. Nuclear localization and function of polypeptide ligands and their receptors: a new paradigm for hormone specificity within the mammary gland? Breast Cancer Res. 2003;5:181–187. doi: 10.1186/bcr601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger CV, Sillman AL, Prystowsky MB. Interleukin-2 driven nuclear translocation of prolactin in cloned T-lymphocytes. Endocrinology. 1990;127:3151–3159. doi: 10.1210/endo-127-6-3151. [DOI] [PubMed] [Google Scholar]
- Lobie PE, Mertani H, Morel G, Marales-Bustos O, Norstedt G, Waters MJ. Receptor-mediated nuclear translocation of growth hormone. J Biol Chem. 1994;269:21330–21339. [PubMed] [Google Scholar]
- Rao Y-P, Buckley DJ, Olson MD, Buckley AR. Nuclear translocation of prolactin: collaboration of tyrosine kinase and protein kinase C activation in rat Nb2 node lymphoma cells. J Cell Physiol. 1995;163:266–276. doi: 10.1002/jcp.1041630207. [DOI] [PubMed] [Google Scholar]
- Rycyzyn MA, Reilly SC, O’Malley K, Clevenger CV. Role of cyclophilin B in PRL signal transduction and nuclear retrotranslocation. Mol Endocrinol. 2000;14:1175–1186. doi: 10.1210/mend.14.8.0508. [DOI] [PubMed] [Google Scholar]
- Fuh G, Wells JA. Prolactin receptor antagonists that inhibit the growth of breast cancer cell lines. J Biol Chem. 1995;270:13133–13137. doi: 10.1074/jbc.270.22.13133. [DOI] [PubMed] [Google Scholar]
- Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- Rodel B, Tavassoli K, Karsunky H, Schmidt T, Bachmann M, Schaper F, Henirich P, Shuai K, Elsasser HP, Moroy T. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 2000;19:5845–5855. doi: 10.1093/emboj/19.21.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Aittomaki S, Silvennoinen O, Palvima JJ, Janne IA. ARIP3 (androgen receptor-interacting protein 3) and other PIAS proteins differ in their ability to modulate steroid receptor-dependent transcription activation. Mol Endocrinol. 2000;14:1986–2000. doi: 10.1210/mend.14.12.0569. [DOI] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration in to nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Fu Y, Shuai K. Distinct roles of the NH2- and COOH-terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT) 1 (PIAS1) in cytokine-induced PIAS1-Stat1 interaction. Proc Natl Acad Sci USA. 2000;97:5267–5272. doi: 10.1073/pnas.97.10.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK. A new ring for SUMO: wrestling transcriptional responses in nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- Muller S, Hoege C, Pryowolakis G, Jentsch S. SUMO, ubiquitin’s mysterious cousin. Nat Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megidish T, Xu JH, Xu CW. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J Biol Chem. 2002;277:8255–8259. doi: 10.1074/jbc.C200001200. [DOI] [PubMed] [Google Scholar]
- Doppler W, Groner B, Ball RK. Prolactin and glucocorticoid hormones synergistically induce expression of transfected rat beta-casein gene promoter constructs in a mammary epithelial cell line. Proc Natl Acad Sci USA. 1989;86:104–108. doi: 10.1073/pnas.86.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvai AE, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose DW, Rosenfeld MG, Glass CK. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner B. Transcription factor regulation in mammary epithelial cells. Domest Anim Endocrinol. 2002;23:25–32. doi: 10.1016/s0739-7240(02)00142-x. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Brindle PK, Handa M, Ihle JN. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. EMBO J. 2001;20:6836–6844. doi: 10.1093/emboj/20.23.6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wreszczynska MH, Horvath CM, Darnell JE. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol Cell Biol. 1999;19:7138–7146. doi: 10.1128/mcb.19.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Lee L-Y, Luo G, Book ML, Morris SM. Lactogenic hormone signal transduction. Biol Reprod. 1998;58:302–311. doi: 10.1095/biolreprod58.2.295. [DOI] [PubMed] [Google Scholar]
- Xu M, Nie L, Kim S-H, Sun X-H. Stat4-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPβ. EMBO J. 2003;22:893–904. doi: 10.1093/emboj/cdg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingo T, Gregg C, Enwere E, Fujikawa H, Hassam R, Geary C, Cross JC, Weiss S. Pregnancy-stimulated neurogenesis in the adult female forebrain mediated by prolactin. Science. 2003;299:117–120. doi: 10.1126/science.1076647. [DOI] [PubMed] [Google Scholar]
- Seidel HM, Lamb P, Rosen J. Pharmaceutical intervention in the JAK/STAT pathway. Oncogene. 2000;19:2645–2656. doi: 10.1038/sj.onc.1203550. [DOI] [PubMed] [Google Scholar]
- Darnell JE. Transcription factors as targets for chemotherapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]