Abstract
ADAMs (a disintegrin and metalloproteinases) are multifunctional molecules involved in cell-cell fusion, cell adhesion, membrane protein shedding, and proteolysis. In the present study, we examined the mRNA expression of 13 different ADAM species with putative metalloproteinase activity in human astrocytic tumors, nonneoplastic brain tissues, and other intracranial tumors by reverse transcriptase-polymerase chain reaction, and found that prototype membrane-anchored ADAM12 (ADAM12m) is predominantly expressed in glioblastomas. Real-time quantitative polymerase chain reaction indicated that the expression level of ADAM12m is remarkably at least 5.7-fold higher in glioblastomas (n = 16) than in nonneoplastic brain tissues (n = 6), low grade (n = 7) and anaplastic astrocytic tumors (n = 9) (P < 0.05 for each group), and intracranial neurinomas (n = 5) (P < 0.01). In situ hybridization showed that glioblastoma cells are responsible for the gene expression. By immunohistochemistry, ADAM12m was predominantly immunolocalized on the cell membranes of glioblastoma cells. Immunoblotting analysis demonstrated that ADAM12m is expressed as an activated N-glycosylated form of ∼90 kd in glioblastoma tissues. There was a direct correlation between the mRNA expression levels of ADAM12m and proliferative activity (MIB1-positive cell index) of gliomas (r = 0.791, P < 0.0001; n = 32). Protein bands consistent with the soluble form of heparin-binding epidermal growth factor, a substrate of ADAM12m, were observed by immunoblotting in glioblastoma samples with the ADAM12m expression, and inhibited by treatment with ADAM inhibitor of the glioblastomas. These data demonstrate for the first time that among the 13 different ADAM species, ADAM12m is highly expressed in human glioblastomas, and suggest the possibility that ADAM12m plays a role in the prominent proliferation of the glioblastomas through shedding of heparin-binding epidermal growth factor.
ADAMs (a disintegrin and metalloproteinases) are a gene family of multidomain membrane-anchored proteins comprising of more than 30 members in various animal species (see http://www.people.virginia.edu/∼jw7g/Table_of_the_ADAMs.html) and are implicated in pathophysiological conditions, which include neuronal development,1 cancer development and progression,2,3 and inflammatory responses4 through proteolysis, cell adhesion, cell fusion, and cell-matrix interaction.5,6 They contain several distinct domains with structural homology to the reprolysin/adamalysin family of snake venom metalloproteinases.7 A typical ADAM protein includes an N-terminal signal peptide, and propeptide, metalloproteinase, disintegrin, cysteine-rich, epidermal growth factor-like, transmembrane, and cytoplasmic domains. The metalloproteinase domains of several ADAMs have a catalytic site with the conventional zinc-dependent metalloproteinase sequence (HEXGHXXGXXHD), which is highly homologous to that of the matrix metalloproteinases (MMPs).8 Thus, these ADAMs are supposed to have proteinase activities to various proteins such as precursors of growth factors and cytokines, receptors and membrane-anchored proteins and extracellular matrix macromolecules, all of which play important roles in various biological events. In humans, ADAM8, ADAM9, ADAM10, ADAM12, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28, and ADAM30 have the conventional metalloproteinase sequence, although only a few of them have been demonstrated to exhibit proteolytic activity. Among them, ADAM17 has been most extensively examined and is known to release soluble tumor necrosis factor-α (TNF-α) from its membrane precursor, thus being called TNF-α converting enzyme (TACE).9 ADAM17 also cleaves TNF p75 receptor, L-selectin, transforming growth factor-α (TGF-α),10 amyloid precursor protein, and ErbB4/HER4. ADAM10 is reported to cleave myelin basic protein, amyloid precursor protein, and type IV collagen. ADAM19 processes membrane-anchored neuregulin,11 and ADAM28 digests myelin basic protein. ADAM9 is implicated in the ectodomain shedding of membrane-anchored heparin-binding epidermal growth factor (proHB-EGF).12 ADAM12 also acts as a sheddase for proHB-EGF13 and cleaves insulin-like growth factor-binding protein (IGFBP)-3 and -5.14 Human ADAM12 exists in two forms that arise from alternate splicing; the prototype membrane-anchored protein (ADAM12m) and the shorter secreted type form (ADAM12s).15 Overexpression of ADAM12m has been observed in human carcinomas including breast carcinomas,16 in which ADAM12 may play a role in cell-cell adhesion through the interaction between its cysteine-rich domain and syndecans.6,16 However, little or no information is available for the expression and localization of ADAM species other than ADAM12 in human malignant tumor tissues.
Glioblastoma, which is one of the most intractable tumors in human malignancies, is characterized by the remarkable proliferative activity and the invasive growth to the surrounding normal brain tissue. Signaling of EGF through the EGF receptor (EGFR) is implicated in the proliferative mechanism of glioblastomas.17 This hypothesis is supported by the following data: 1) The EGFR gene is amplified in 30 to 40% of glioblastomas18 and approximately half of the glioblastomas with the EGFR amplification show genomic rearrangements of the EGFR gene, resulting in the expression of a truncated receptor19–21 that is constitutively active without binding to the ligand;22,23 2) glioblastomas express the EGFR ligands, ie, EGF, TGF-α, and HB-EGF,23,24 and EGFR and HB-EGF are co-expressed in the human glioblastomas;24 and 3) glioma cell proliferation is promoted by the addition of HB-EGF and suppressed by the treatment with anti-HB-EGF blocking antibodies.24 Several proteinases including ADAM912 and ADAM1213,25 are known to process proHB-EGF to soluble, active HB-EGF. However, no studies have been performed so far to elucidate the relationship between the ADAM expression and HB-EGF shedding in human glioma tissues.
In the present study, we examined the expression of 13 different ADAM species with putative metalloproteinase activity, tissue localization of ADAM12m, correlation between the expression level of ADAM12m and proliferative activity, and expression of HB-EGF species in the human malignant astrocytic tumors. The results demonstrate for the first time that ADAM12m is selectively overexpressed in the glioblastomas, and suggest the possibility that the ectodomain shedding of proHB-EGF by ADAM12m is implicated in the proliferation of the glioblastoma cells.
Materials and Methods
Tissue Samples and Histology
Intracranial tissue samples were obtained from 48 patients who underwent surgery at the Keio University Hospital, Tokyo, Japan. There were 7 low-grade astrocytomas, 9 anaplastic astrocytomas, 16 glioblastomas, 5 intracranial neurinomas, 5 metastatic carcinomas (lung adenocarcinomas), and 6 nonneoplastic control brain tissues. The classification of human brain tumors is based on the revised World Health Organization criteria for the central nervous system,17 and low-grade astrocytomas, anaplastic astrocytomas, and glioblastomas correspond to grades II, III, and IV astrocytomas, respectively. All of the tumor tissues were obtained at primary resection, and none of the patients had been subjected to chemotherapy or radiation therapy before resection. Nonneoplastic control brain tissues were obtained from marginal sites of the tumors. The samples were divided into two parts, which were subjected to histopathological examination and experiments for RNA extraction and protein analysis. For the experimental use of the surgical specimens, informed consent was obtained from the patients according to the hospital ethical guidelines.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from the tissue samples by Isogen (Nippon Gene Co. Ltd., Toyama, Japan) and evaluated by using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). The tissue samples, the 28s/18s ribosomal RNA ratios of which were larger than 1.0, were used. The RT reaction was performed at 42°C for 50 minutes, followed by heating at 70°C for 10 minutes for inactivation of the enzyme. The cDNAs were amplified by PCR with primers specific to ADAM8, ADAM9, ADAM10, ADAM12 (two alternate splicing variants, ie, shorter secreted type ADAM12s and prototype membrane-anchored ADAM12m; GenBank numbers NM_021641 and NM_003474, respectively), ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28 (two alternate splicing variants, ADAM28s and ADAM28m; GenBank numbers NM_021777 and NM_014265, respectively), ADAM30, HB-EGF, and housekeeping gene β-actin. Sequences of the specific primers used for PCR are shown in Table 1. PCR amplification by Takara ExTaq (Takara Bio. Inc., Shiga, Japan) was performed on a thermal cycler after an initial denaturation at 94°C for 3 minutes by running 30 cycles under the following conditions: denaturation for 1 minute at 94°C; annealing for 1 minute at 52°C (HB-EGF), 54°C (ADAM8), 56°C (ADAM12m), 60°C (ADAM10, ADAM12s, ADAM19, ADAM20, ADAM21, ADAM28m, ADAM28s, and ADAM30), or 67°C (ADAM9, ADAM15, ADAM17, and β-actin); and extension for 1 minute at 72°C, followed by a final extension for more 2 minutes. The expected sizes of the amplified cDNA fragments are shown in Table 1. The products were electrophoresed on 2% agarose gels and stained with ethidium bromide. The expression levels of the ADAM species in the samples were classified to three groups (negative, low, and high levels) by measuring the relative expression to β-actin, which was determined by analyzing the images captured by FAS II mini (Toyobo Co. Ltd., Osaka, Japan) using public domain NIH Image software (developed at the National Institutes of Health; http://rsb.info.nih.gov/nih-image/) according to the modification of our previous methods.26 As for positive controls, total RNA was extracted from CaR-1 human rectal carcinoma cells (JCRB0207; Health Science Research Resources Bank, Osaka, Japan) for ADAM8; U251 human glioblastoma cells (American Type Culture Collection, Manassas, VA) for ADAM9, ADAM10, ADAM12m, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28s, and ADAM28m; SVGp12 human fetal glial cells (American Type Culture Collection) for ADAM30; or human full-term placenta for ADAM12s, since our preliminary study showed that these human cell lines and placenta expressed the corresponding ADAM species. The specific PCR amplification from the target mRNAs was confirmed by sequencing the PCR products using DYEnamic ET terminator cycle sequencing kit and MegaBACE 1000 sequencer (Amersham Biosciences Corp., Piscataway, NJ).
Table 1.
Primers for RT-PCR*
| Primer’s name | Oligonucleotide sequence | Product size | |
|---|---|---|---|
| ADAM8 | Forward | 5′-GCCGTCTTCAGGCCTCGGCCCGGGGACTCT-3′ | 651 bp |
| Reverse | 5′-AGGGGCGTTGGCGAGGCACACCGACTGCGG-3′ | ||
| ADAM9 | Forward | 5′-GCTGTCTTGCCACAGACCCGGTATGTGGAG-3′ | 604 bp |
| Reverse | 5′-TGGAATATTAAGAAGGCAGTTTCCTCCTTT-3′ | ||
| ADAM10 | Forward | 5′-ATCCAGTCATGTTAAAGCGATTGATACAATTTAC-3′ | 434 bp |
| Reverse | 5′-TCCAAAGTTATGTCCAACTTCGTGAGCAAAAGTAA-3′ | ||
| ADAM12s | Forward | 5′-GCACCTCCCTTCTGTGACAAGTTT-3′ | 504 bp |
| Reverse | 5′-TGAAAGGCCAGACTTTTGAGTCCT-3′ | ||
| ADAM12m | Forward | 5′-GCACCTCCCTTCTGTGACAAGTTT-3′ | 643 bp |
| Reverse | 5′-CTTGGTGTGGATATTGTGGAGCAG-3′ | ||
| ADAM15 | Forward | 5′-CTGGGACAGCGCCACATTCGCCGGAGGCGG-3′ | 688 bp |
| Reverse | 5′-TCCGCAGAAAGCAGCCATAGGGGGTAGGCT-3′ | ||
| ADAM17 | Forward | 5′-AGAGCTGACCCAGATCCCATGAAGAACACG-3′ | 777 bp |
| Reverse | 5′-GCGTTCTTGAAAACACTCCTGGGCCTTACT-3′ | ||
| ADAM19 | Forward | 5′-TGTGGGAAGATCCAGTGTCA-3′ | 500 bp |
| Reverse | 5′-AGAGCTGAGGGCTTGAGTTG-3′ | ||
| ADAM20 | Forward | 5′-AAAATAGCACACCAGATGGAGTTGCAATTG-3′ | 702 bp |
| Reverse | 5′-ATTCCCACAGTACTTCAGTCTAAATATATT-3′ | ||
| ADAM21 | Forward | 5′-TCTGGCTTGGGGTATTTTTG-3′ | 500 bp |
| Reverse | 5′-TTGGCGTGCTACTTCCTTCT-3′ | ||
| ADAM28s | Forward | 5′-GCTGTGATGCTAAGACATGT-3′ | 644 bp |
| Reverse | 5′-GTTTATGATCTTAGTAGGGTTGCC-3′ | ||
| ADAM28m | Forward | 5′-GCTGTGATGCTAAGACATGT-3′ | 871 bp |
| Reverse | 5′-TGAACAGCCTTTACCATCTG-3′ | ||
| ADAM30 | Forward | 5′-AACCAGGTGCCAACTGTAGC-3′ | 496 bp |
| Reverse | 5′-CCCATGGGTTTCATGGATAG-3′ | ||
| HB-EGF | Forward | 5′-ACAAGCACTGGCCACACCAAACAAG-3′ | 299 bp |
| Reverse | 5′-CCACGATGACCAGCAGACAGACAGA-3′ | ||
| β-actin | Forward | 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′ | 661 bp |
| Reverse | 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′ |
Primers for ADAM species, HB-EGF, and β-actin. The estimated product sizes are shown in this setting.
Real-Time Quantitative PCR
For quantitative analysis of ADAM12m expression, cDNA was used as template in a TaqMan real-time PCR assay by the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. Cycling conditions were 50°C for 2 minutes, 95°C for 10 minutes, and then 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. The primers and TaqMan probe for ADAM12m were chosen with the assistance of the computer program Primer Express (Applied Biosystems) as follows: forward primer 5′-CCTCAGACCTGCTCCACAATATC-3′, reverse primer 5′-TTGAAAAAAGGTGTCGGCTTCT-3′ and TaqMan probe 5′-FAM-CCAAGTGCCCAGATCCACCCACA-TAMRA-3′. Sample data were normalized by 18S ribosomal RNA, which was selected as endogenous control using the TaqMan Human Endogenous Control Plate (Applied Biosystems) according to manufacturer’s protocol. The total gene specificity of the nucleotide sequences chosen for the primers and probe and the absence of DNA polymorphisms were ascertained by BLASTN and Entrez on web sites (http://www.ncbi.nlm.nih.gov/).
In Situ Hybridization for ADAM12
The glioblastoma samples (five cases) that showed high ADAM12m expression by real-time PCR were used for in situ hybridization according to the modification of our previous methods.26 Briefly, single-stranded sense and anti-sense digoxigenin-labeled RNA probes were generated by in vitro transcription of the cDNA with SP6 or T7 RNA polymerase using the DIG RNA labeling kit (Roche Diagnostics GmbH, Mannheim, Germany) following the protocol from the manufacturer. Template DNA was a cDNA fragment encoding the propeptide domain of human ADAM12, nucleotides 708 to 905 (198 bp), which was subcloned into pGEM-11Zf (+) Vector (Promega Corp., Madison, WI). Serial paraffin sections (4 μm thick) were hybridized with the digoxigenin-labeled RNA anti-sense or sense probes,26 and then subjected to immunostaining using mouse anti-digoxigenin antibody (1/500 dilution, Roche Diagnostics GmbH) followed by the peroxidase-labeled avidin:biotin complex method (ABC method) (1/100 dilution; DakoCytomation Norden A/S, Glostrup, Denmark). After the reactions, the sections were counterstained with hematoxylin.
Immunohistochemistry, Immunoblotting, and Immunoprecipitation
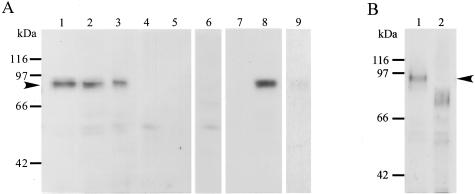
Serial paraffin sections (4 μm thick) were treated with 0.3% hydrogen peroxide/0.1% NaN3 to block endogenous peroxidase activity. For immunostaining of Ki-67, they were also treated in a microwave oven for 5 minutes at 500 W using a citrate buffer (pH 6.0). After blocking nonspecific binding with 10% horse serum for ADAM12m staining or 10% goat serum for Ki-67 staining, they were incubated with mouse monoclonal antibodies against ADAM12m (283-6H3, 5 μg/ml) or Ki-67 (MIB1, 1/50 dilution; DakoCytomation Norden A/S). Subsequently, the specimens were incubated with biotinylated horse antibodies against mouse IgG (1/200 dilution; Vector Laboratories, Inc., Burlingame, CA) followed by the ABC method for ADAM12m or with goat antibodies against mouse IgG conjugated to horseradish peroxidase-labeled dextran polymer (no dilution, EnVision+ Peroxidase Mouse; DakoCytomation, California Inc., Carpinteria, CA) for Ki-67. After immunostaining, the sections were counterstained with hematoxylin. Monoclonal antibody against ADAM12m (283-6H3) was developed by using a synthetic peptide corresponding to the amino acid sequence of the cytoplasmic domain of human ADAM12m (residues 893 to 909, PQYPHQVPRSTHTAYIK-C)15 as an antigen according to the methods described previously.27 After screening 10 candidate clones by enzyme-linked immunosorbent assay with the synthetic peptide, clone 283-6H3 was selected. Monospecificity of the clone was further examined by absorption test of the antibody with the antigen peptide and by immunoblotting of U251 glioblastoma cells and CaR-1 cells (see Figure 5), which showed positive and negative expression of ADAM12m, respectively.
Figure 5.
Immunoblotting of ADAM12m in glioblastoma tissues. A: Homogenates (20 μg/lane) from glioblastoma (lanes 1 to 3 and 6) and nonneoplastic brain tissues (lanes 4 and 5) and cell lysates (20 μg/lane) of CaR-1 (lane 7, a negative control) and U251 (lane 8, a positive control; lane 9, absorption test) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins in the gels were transferred onto polyvinylidene difluoride membranes and the membranes immunostained with anti-ADAM12m antibody (2 μg/ml) (lanes 1 to 5, 7, and 8), the antibody (2 μg/ml) absorbed with antigen peptide (3 μg/ml) (lane 9), or nonimmune mouse IgG (2 μg/ml) (lane 6) as described in Materials and Methods. Note that a positive band of ∼90 kd is found in the glioblastoma samples (lanes 1 to 3) and the U251 cell lysates (lane 8), whereas no such species is recognized in the nonneoplastic brain samples (lanes 4 and 5) or CaR-1 cell lysates (lane 7). Only a background staining is present with nonimmune IgG (lane 6), and a negligible band is observed with the antibody absorbed with the peptide (lane 9). B: Deglycosylation of ADAM12m. Homogenates were treated with N-glycosidase F and subjected to immunoblotting for ADAM12m as described in Materials and Methods.
For immunoblotting of ADAM12m and HB-EGF, tissue samples were homogenized on ice in 1 ml of lysis buffer; 50 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 10 mmol/L CaCl2, and 0.05% Brij35 containing a cocktail of proteinase inhibitors (Roche Diagnostics, GmbH). The supernatants of the homogenates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (8% and 15% total acrylamide for immunoblotting of ADAM12m and HB-EGF, respectively) under reduction after determining protein concentrations by the dye-binding method (Proteostain Protein Quantification Kit Wide Range; Dojindo Laboratories, Kumamoto, Japan). The resolved proteins were transferred onto polyvinylidene difluoride membranes that were reacted with mouse monoclonal antibody specific to ADAM12m (283-6H3, 2 μg/ml), goat polyclonal antibodies specific to HB-EGF (1/500 dilution; R&D Systems Inc., Minneapolis, MN) or nonimmune mouse IgG and goat IgG after blocking nonspecific reaction with 0.5% skim milk. The membranes were incubated with goat antibodies against mouse IgG conjugated to horseradish peroxidase-labeled dextran polymer (1/200 dilution, EnVision+ Peroxidase Mouse; DakoCytomation California Inc.) for ADAM12m or horseradish peroxidase-conjugated rabbit antibodies against goat IgG (1/20,000 dilution; Chemicon International Inc., Temecula, CA) for HB-EGF. Immunoreactive protein bands were detected with enhanced chemiluminescence immunoblotting reagents (Amersham Biosciences Corp). As for controls of ADAM12m, cell lysates of U251 glioblastoma cells (positive control) and CaR-1 cells (negative control) were also subjected to immunoblotting. Recombinant HB-EGF (Calbiochem, San Diego, CA) was used as a positive control for immunoblotting of HB-EGF. To examine the glycosylation of ADAM12m, tissue homogenates of glioblastomas and lysates of U251 cells in phosphate-buffered saline (pH 7.4) were mixed and boiled with sodium dodecyl sulfate-reduction sample buffer and digested with 10 U/ml N-glycosidase F (Roche Diagnostics GmbH) in the buffer containing 0.5% IGEPAL CA-630 (Sigma-Aldrich, Inc., St. Louis, MO) for 16 hours at 37°C. The samples were then subjected to immunoblotting for ADAM12m.
To study the effects of ADAM inhibitor on the processing of cell surface proHB-EGF, glioblastoma tissues that expressed ADAM12m were cut into pieces (∼3 × 3 × 2 mm) and cultured for 12 to 24 hours in the presence and absence of 1 μmol/L KB-R7785, an inhibitor to ADAM13 (a kind gift by Dr. Koichiro Yoshino, Carnabioscience, Kobe, Japan). Supernatants of the homogenates (50 μg/lane) were prepared and subjected to immunoblotting as described above. After stripping bound antibodies, the membranes were reprobed with anti-β-actin antibody (1 μg/ml, A5441; Sigma-Aldrich, Inc.) to confirm that similar amounts of samples were applied in each lane. In addition, ADAM12m-expressing U251 glioblastoma cells were cultured in the presence and absence of 1 μmol/L KB-R7785 for 6 hours, and cell surface proteins were biotinylated with 0.1 mg/ml sulfo-NHS-biotin in 20 mmol/L HEPES, pH 7.5, 0.4 mol/L NaCl on ice. Then, cell lysates (1.5 × 106 cells/lane) were prepared and HB-EGF was immunoprecipitated by reacting with goat anti-HB-EGF polyclonal antibodies (5 μg) or nonimmune goat IgG (5 μg) according to the methods described by Goishi and colleagues.28 The immunoprecipitated proteins were electrophoresed, transferred to polyvinylidene difluoride membranes, and reacted with avidin-conjugated horseradish peroxidase (Vector Laboratories Inc.). To evaluate the amounts of proteins loaded to lanes, the same amount of each cell lysate was subjected to immunoblotting for β-actin. The protein bands on the membranes were detected with enhanced chemiluminescence immunoblotting reagents as described above.
MIB1-Positive Cell Index
To assess proliferative activity of astrocytic tumors, MIB1-positive cell index was determined by counting nuclei immunoreactive with MIB1 antibody in five different fields at a magnification of ×200 without knowledge of clinical data. Any nuclear staining, regardless of intensity, was considered positive for MIB1. The MIB1-positive cell index was expressed as a percentage of immunoreactive tumor cells to the total counted tumor cells.
Statistics
Statistical analyses were performed using Kruskal-Wallis test and subsequent Bonferroni/Dunn procedure as a post hoc test to assess the significance among astrocytic tumors and nonneoplastic control brain tissues. The correlations between the mRNA expression levels of ADAM12m and the MIB1-positive cell index were studied using the Pearson correlation coefficient. P values less than 0.05 were considered significant.
Results
mRNA Expression of ADAM Species in Astrocytic Tumors
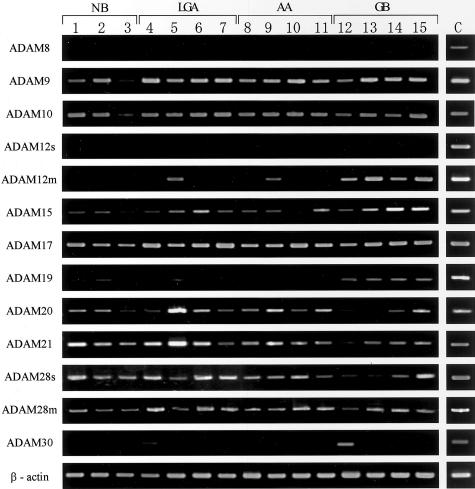
The mRNA expression of ADAM8, ADAM9, ADAM10, ADAM12s, ADAM12m, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28s, ADAM28m, and ADAM30 was first screened by RT-PCR in astrocytic tumors (n = 21), intracranial neurinomas (n = 5), metastatic carcinomas (n = 5), and nonneoplastic brain tissues (n = 5). As shown in Figure 1, PCR products for all of the ADAM species except for ADAM8 and ADAM12s were detected at least in one of the tissue samples examined. Although ADAM8 and ADAM12s were undetectable in these samples, availability of RT-PCR for these ADAM species was demonstrated with RNA samples extracted from CaR-1 cells and human placental tissue (Figure 1). The data on the expression of each ADAM gene are summarized in Table 2. ADAM9, ADAM10, ADAM17, ADAM28s, and ADAM28m were constitutively expressed in all of the tumors and nonneoplastic brain tissues (Figure 1 and Table 2), and the expression of ADAM15, ADAM20, and ADAM21 was observed in more than 92% of the samples (Table 2). In contrast, ADAM8 and ADAM12s were not expressed in the tumors or nonneoplastic control brain tissues, and ADAM30 was randomly expressed in a few tumors and control brain tissues (Table 2). On the other hand, the expression pattern of ADAM12m and ADAM19 in the astrocytic tumors appeared to be selective in the glioblastomas (Table 2). However, because the nonneoplastic brain tissues expressed ADAM19 but not ADAM12m (Figure 1 and Table 2), it seemed likely that only the expression of ADAM12m is glioblastoma-selective. Thus, we focused on ADAM12m and further analyzed its expression levels by real-time quantitative PCR.
Figure 1.
mRNA expression of ADAM8, ADAM9, ADAM10, ADAM12s, ADAM12m, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28s, ADAM28m, and ADAM30 in astrocytic tumors and nonneoplastic brain tissues by RT-PCR. Total RNA was extracted from the nonneoplastic control brain tissues (lanes 1 to 3, NB), low-grade astrocytomas (lanes 4 to 7, LGA), anaplastic astrocytomas (lanes 8 to 11, AA), and glioblastomas (lanes 12 to 15, GB), and reverse-transcribed into cDNA followed by a PCR reaction as described in Materials and Methods. Fifteen representative samples are shown. Each amplification of ADAM species and β-actin was performed at least in triplicate. As for positive controls (C), total RNAs from CaR-1 cells (for ADAM8 and β-actin), U251 cells (for ADAM9, ADAM10, ADAM12m, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28s, and ADAM28m), SVGp12 cells (for ADAM 30), and human full-term placental tissue (for ADAM12s) were used for RT-PCR.
Table 2.
Expression of ADAM Species in Human Astrocytic Tumors, Intracranial Tumors and Nonneoplastic Brain Tissues by RT-PCR*
| Sample no. | Tissue histology | Density of PCR Products
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADAM8 | ADAM9 | ADAM10 | ADAM12s | ADAM12m | ADAM15 | ADAM17 | ADAM19 | ADAM20 | ADAM21 | ADAM28s | ADAM28m | ADAM30 | ||
| 1 | NB† | −‡ | + | + | − | − | + | ++ | + | + | ++ | ++ | ++ | + |
| 2 | NB | − | ++ | ++ | − | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − |
| 3 | NB | − | ++ | ++ | − | − | + | ++ | + | + | + | ++ | ++ | − |
| 4 | NB | − | ++ | ++ | − | − | + | ++ | + | + | + | ++ | ++ | − |
| 5 | NB | − | + | + | − | − | + | ++ | − | + | − | ++ | + | − |
| 6 | LGA | − | ++ | ++ | − | − | + | ++ | − | + | ++ | ++ | ++ | + |
| 7 | LGA | − | ++ | ++ | − | + | + | ++ | − | ++ | ++ | ++ | ++ | − |
| 8 | LGA | − | ++ | ++ | − | − | + | ++ | − | ++ | ++ | ++ | ++ | − |
| 9 | LGA | − | ++ | ++ | − | − | + | ++ | + | ++ | ++ | ++ | ++ | + |
| 10 | LGA | − | ++ | ++ | − | + | ++ | ++ | − | ++ | ++ | ++ | ++ | − |
| 11 | LGA | − | ++ | ++ | − | − | ++ | ++ | − | + | + | ++ | ++ | − |
| 12 | LGA | − | ++ | ++ | − | − | + | ++ | + | + | + | ++ | ++ | − |
| 13 | AA | − | ++ | ++ | − | − | − | ++ | − | + | + | + | ++ | − |
| 14 | AA | − | ++ | ++ | − | − | + | ++ | + | ++ | ++ | ++ | ++ | + |
| 15 | AA | − | ++ | ++ | − | + | + | ++ | − | + | + | ++ | ++ | + |
| 16 | AA | − | ++ | ++ | − | − | + | ++ | − | + | ++ | ++ | ++ | − |
| 17 | AA | − | ++ | ++ | − | − | + | ++ | + | ++ | + | ++ | ++ | + |
| 18 | AA | − | ++ | ++ | − | − | + | ++ | − | + | + | ++ | ++ | − |
| 19 | AA | − | ++ | ++ | − | − | + | ++ | + | ++ | + | ++ | ++ | − |
| 20 | GB | − | ++ | ++ | − | ++ | − | ++ | + | + | + | + | + | ++ |
| 21 | GB | − | ++ | ++ | − | + | ++ | ++ | + | ++ | + | + | + | − |
| 22 | GB | − | ++ | ++ | − | + | + | ++ | + | − | + | + | − | − |
| 23 | GB | − | ++ | ++ | − | ++ | + | ++ | ++ | + | ++ | ++ | ++ | − |
| 24 | GB | − | ++ | ++ | − | ++ | ++ | ++ | + | ++ | ++ | ++ | ++ | + |
| 25 | GB | − | ++ | ++ | − | ++ | ++ | ++ | + | ++ | ++ | ++ | ++ | − |
| 26 | GB | − | ++ | + | − | ++ | ++ | ++ | + | ++ | + | ++ | ++ | − |
| 27 | Neu | − | ++ | ++ | − | − | ++ | ++ | − | + | + | + | ++ | + |
| 28 | Neu | − | ++ | ++ | − | − | ++ | ++ | − | + | + | ++ | ++ | + |
| 29 | Neu | − | ++ | ++ | − | − | ++ | ++ | − | + | + | ++ | ++ | + |
| 30 | Neu | − | ++ | ++ | − | + | ++ | ++ | − | + | + | + | ++ | − |
| 31 | Neu | − | ++ | ++ | − | − | + | ++ | − | + | + | ++ | − | − |
| 32 | Met | − | ++ | ++ | − | + | + | ++ | − | + | + | + | + | − |
| 33 | Met | − | ++ | ++ | − | + | ++ | ++ | − | ++ | ++ | + | + | + |
| 34 | Met | − | ++ | ++ | − | − | + | ++ | − | ++ | ++ | + | + | + |
| 35 | Met | − | ++ | ++ | − | + | − | ++ | − | ++ | + | + | + | − |
| 36 | Met | − | ++ | ++ | − | − | + | ++ | − | − | − | + | + | − |
Density of PCR products was semiquantitatively estimated according to the method described in Materials and Methods.
NB, nonneoplastic brain tissue; LGA, low grade astrocytoma; AA, anaplastic astrocytoma; GB, glioblastoma; Neu, intracranial neurinoma; Met, metastatic carcinoma.
Symbols: −, negative; +, low density band; ++, high density band.
Real-Time Quantitative PCR of ADAM12m
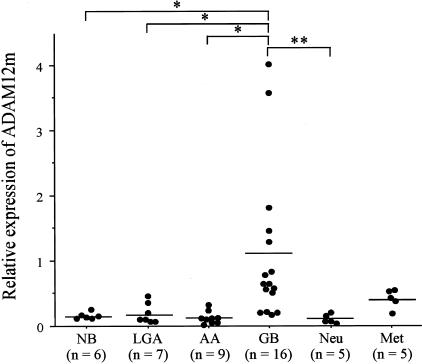
To evaluate the expression levels of ADAM12m in the astrocytic tumors (n = 32), other intracranial tumors (n = 10), and nonneoplastic brain tissues (n = 6), real-time quantitative PCR was performed. The values in each sample were standardized for sample-to-sample variations using 18S ribosomal RNA as normalization. As shown in Figure 2, the relative mRNA expression level of ADAM12m in glioblastomas (1.09 ± 1.16, mean ± SD; n = 16) was significantly at least 5.7-fold higher than that in nonneoplastic brain tissues (0.16 ± 0.08, n = 6), low-grade astrocytomas (0.19 ± 0.16, n = 7), or anaplastic astrocytomas (0.14 ± 0.10, n = 9) (P < 0.05 for each group). In addition, the level was significantly ∼10-fold higher in glioblastomas than in intracranial neurinomas (0.11 ± 0.07, n = 5) (P < 0.01). The expression level in glioblastomas tended to be higher than that in metastatic carcinoma (0.41 ± 0.15, n = 5), although the difference was not significant.
Figure 2.
The mRNA expression levels of ADAM12m in nonneoplastic brain tissues (NB), low-grade astrocytomas (LGA), anaplastic astrocytomas (AA), glioblastomas (GB), intracranial neurinomas (Neu), and metastatic carcinomas (Met). Relative mRNA expression levels of ADAM12m to 18S ribosomal RNA in each sample were measured by the real-time quantitative PCR as described in Materials and Methods. Bars indicate mean value. *, P < 0.05; **, P < 0.01.
In Situ Hybridization
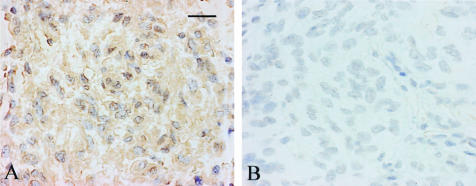
Cells expressing ADAM12 mRNA in glioblastoma tissues were identified by in situ hybridization. With the anti-sense probes, the signal for ADAM12 was observed in the glioblastoma cells (Figure 3A). The sense probes gave only a background signal in the glioblastoma tissues (Figure 3B).
Figure 3.
In situ hybridization of ADAM12 in glioblastomas. In situ hybridization was performed as described in Materials and Methods. A: Note positive signals for ADAM12 in the cytoplasm of glioblastoma cells with the anti-sense probes. B: The sense probes give only a background signal in the glioblastomas. Scale bar, 25 μm.
Immunohistochemistry and Immunoblotting for ADAM12m
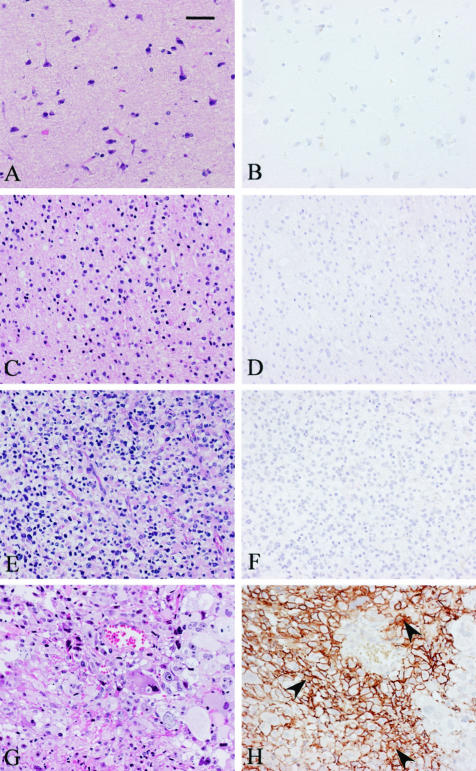
ADAM12m was immunolocalized to the glioblastoma cells in 88% of the glioblastoma cases (14 of 16 cases; Figure 4, G and H), although no staining was seen in nonneoplastic brain tissues (0 of 6 cases; Figure 4, A and B) or low-grade astrocytomas (0 of 7 cases; Figure 4, C and D). It was immunostained in one of the nine anaplastic astrocytomas (data not shown), but another eight cases showed negative staining (one of nine cases; Figure 4, E and F). The immunostaining was predominantly observed on the cell membranes of glioblastoma cells (Figure 4H), but the cytoplasm of some glioblastoma cells showed occasionally weak immunostaining (Figure 4H, arrowheads). Blood vessels in anaplastic astrocytomas and glioblastomas were negatively stained (Figure 4, F and H). No staining was observed in any tissue samples with nonimmune mouse IgG (data not shown).
Figure 4.
Immunolocalization of ADAM12m in glioblastomas. Paraffin sections of nonneoplastic brain tissues (A and B), low-grade astrocytomas (C and D), anaplastic astrocytomas (E and F), and glioblastomas (G and H) were stained with H&E (A, C, E, and G) or immunostained with monoclonal antibody against ADAM12m (B, D, F, and H) as described in Materials and Methods. Note the selective immunostaining on the cell membranes of most glioblastoma cells and weak staining within the cytoplasm of a few glioblastoma cells (arrowheads, H). Blood vessel cells are completely negative (F and H). Scale bar, 50 μm.
By immunoblotting analysis, ADAM12m existed as a major protein band of ∼90 kd in the homogenates of glioblastoma tissues (Figure 5A, lanes 1 to 3), whereas the nonneoplastic brain tissues showed negative bands (Figure 5A, lanes 4 and 5). Specificity of the antibody was confirmed by the following findings: 1) only negligible bands were detected in the glioblastoma tissue samples by nonimmune mouse IgG (Figure 5A, lane 6); 2) the ∼90-kd band was recognized in the cell lysates of U251 cells with strong expression of ADAM12m (Figure 5A, lane 8), but not in those of CaR-1 cells without ADAM12m expression (Figure 5A, lane 7); and 3) immunoreactivity was remarkably reduced to a negligible level by incubating the antibody with the antigen peptide before the immunoblotting (Figure 5A, lane 9 for U251 cell lysates and data not shown for homogenates of glioblastoma tissues). When the tissue homogenates of glioblastomas and U251 cell lysates were treated with N-glycosidase F, which removes all types of N-linked oligosaccharides from glycoproteins, and then immunoblotted with the antibody, the major ∼90-kd protein band was shifted to the band of ∼75 kd (Figure 5B and data not shown for U251 cell lysates). This indicates that the ∼90-kd band is an N-glycosylated protein.
MIB1-Positive Cell Index and Its Correlation with ADAM12m Expression Levels
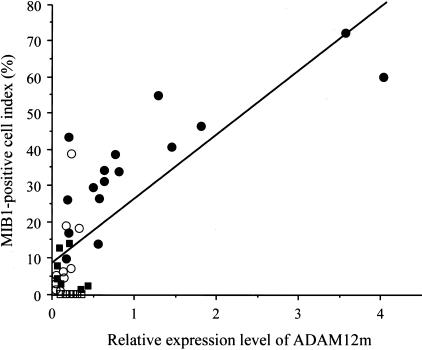
To study whether mRNA expression levels of ADAM12m correlate with proliferation of astrocytic tumor cells, all of the astrocytic tumors (n = 32) were subjected to immunostaining for Ki-67 using MIB1 antibody to obtain MIB1-positive cell index. Immunoreactive cells increased with histological grade (data not shown), confirming the data of previous studies.29 MIB1-positive cell index for astrocytic tumors was as follows: 6.29 ± 5.20% for low-grade astrocytomas (n = 7), 9.82 ± 12.1% for anaplastic astrocytomas (n = 9), and 36.0 ± 16.8% for glioblastomas (n = 16). No staining was observed in nonneoplastic brain tissues (n = 6) or in glioblastoma tissues stained with nonimmune mouse IgG (data not shown). When the MIB1-positive cell index of the astrocytic tumors was plotted against mRNA expression levels of ADAM12m in each case, there was a direct correlation between the index and the ADAM12m expression levels (r = 0.791, P < 0.0001; n = 32) (Figure 6).
Figure 6.
Correlation between MIB1-positive cell index and ADAM12m mRNA expression levels. The relative mRNA expression levels of ADAM12m (ADAM12m/18s ribosomal RNA ratios) in astrocytic tumors were plotted against the MIB1-positive cell index. Note a direct correlation between the mRNA expression levels and the index (r = 0.791, P < 0.0001). ▪, low-grade astrocytomas (n = 7); ○, anaplastic astrocytomas (n = 9); •, glioblastomas (n = 16); □, nonneoplastic brain tissue (n = 6).
mRNA Expression and Immunoblotting of HB-EGF
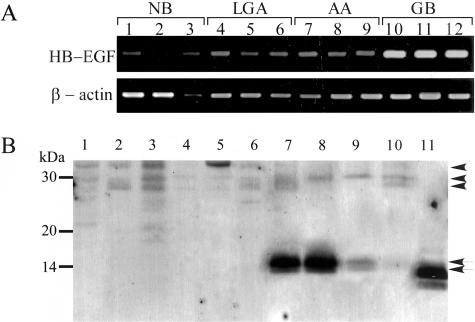
Because ADAM12m is implicated in the ectodomain shedding of proHB-EGF in mouse cardiomyocytes13 and embryonal fibroblasts,25 we studied the processing of proHB-EGF in glioblastoma tissues. When the mRNA expression of HB-EGF was examined in nonneoplastic brain tissues (n = 3), low-grade astrocytomas (n = 3), anaplastic astrocytomas (n = 3), and glioblastomas (n = 3) by RT-PCR, all of the tumors and nonneoplastic brain tissues expressed the mRNA of HB-EGF (Figure 7A). The expression was almost constitutive, although the level appeared to be higher in glioblastomas than in other tissues.
Figure 7.
The mRNA expression and immunoblotting of HB-EGF. A: mRNA expression of HB-EGF and β-actin in nonneoplastic brain tissues (lanes 1 to 3, NB), low-grade astrocytomas (lanes 4 to 6, LGA), anaplastic astrocytomas (lanes 7 to 9, AA), and glioblastomas (lanes 10 to 12, GB) was examined by RT-PCR as described in Materials and Methods. B: Immunoblotting analyses of HB-EGF in representative 10 cases. Tissue homogenates (20 μg/lane) of nonneoplastic brain (lanes 1 and 2), low-grade astrocytoma (lanes 3 and 4), anaplastic astrocytoma (lanes 5 and 6), or glioblastomas with ADAM12m expression at high (lanes 7 and 8), medium (lane 9), or low level (lane 10) were subjected to immunoblotting for HB-EGF as described in Materials and Methods. Recombinant HB-EGF (20 ng/lane) was used as for a positive control (lane 11). Note that the protein bands of 13.0 and 14.0 kd corresponding to processed HB-EGF (arrows) are found in the glioblastoma samples (lanes 7 to 10) showing a correlation with the ADAM12m expression, whereas the bands corresponding to membrane-anchored proHB-EGF of 27.0, 30.0, and 35.5 kd (arrowheads) are detected mainly in the nonneoplastic brain (lanes 1 and 2), low-grade astrocytoma (lanes 3 and 4), and anaplastic astrocytoma (lanes 5 and 6). Recombinant soluble HB-EGF, which lacks juxtamembrane, transmembrane, and cytoplasmic domains, shows the bands of 12.0, 13.5, and 14.5 kd.
By immunoblotting, nonneoplastic brain (n = 2), low-grade astrocytoma (n = 2), and anaplastic astrocytoma samples (n = 4), all of which had negligible or no ADAM12m expression, showed protein bands of 27.0, 30.0, and 35.5 kd, which correspond to membrane-anchored proHB-EGF (Figure 7B, lanes 1 to 6). In contrast, protein bands of 13.0 and 14.0 kd consistent with the soluble form of cleaved HB-EGF were identified in the tissue homogenates of glioblastoma samples (n = 6), which showed high, medium, and low levels of ADAM12m expression by real-time quantitative PCR (Figure 7B, lanes 7 to 10). Importantly, intensity of the processed forms appeared to correlate with the ADAM12m expression levels (Figure 7B).
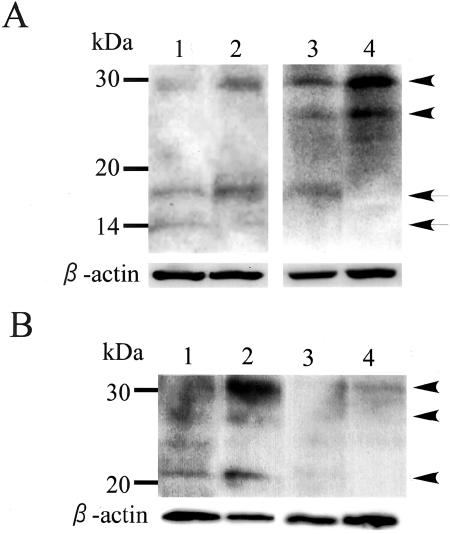
Inhibition of HB-EGF Processing by ADAM Inhibitor
To further study the involvement of ADAM12m in the processing of proHB-EGF in glioblastomas, glioblastoma tissues (n = 4) that expressed ADAM12m were cultured in the presence and absence of an ADAM inhibitor, KB-R7785, and HB-EGF species in the tumors were analyzed by immunoblotting. In the samples treated with KB-R7785, proHB-EGF and processed soluble form of HB-EGF appeared to increase and decrease, respectively, as compared with the control tissue (Figure 8A). In addition, we examined the effect of KB-R7785 on the processing of proHB-EGF from U251 glioblastoma cells that highly express ADAM12m. Using cell surface biotinylation and immunoprecipitation, faint bands for cell surface proHB-EGF of 20 to 30 kd were detected in the control cells without inhibitor treatment, but density of the bands remarkably increased in the cells treated with KB-R7785 (Figure 8B, lanes 1 and 2). Immunoprecipitation with nonimmune IgG showed only background signals (Figure 8B, lanes 3 and 4). All these data suggest that ADAM12m is implicated in the ectodomain shedding of proHB-EGF in glioblastoma cells.
Figure 8.
Inhibition of HB-EGF processing by ADAM inhibitor in glioblastoma tissues and U251 glioblastoma cells. A: Glioblastoma tissues were cultured in the presence (lanes 2 and 4) and absence (lanes 1 and 3) of KB-R7785, and HB-EGF species were analyzed by immunoblotting as described in Materials and Methods. ProHB-EGF and active HB-EGF species are indicated by arrowheads and arrows, respectively. The membrane was reprobed with anti-β-actin antibody. Two representative cases are shown. B: U251 glioblastoma cells were cultured in the presence (lanes 2 and 4) and absence (lanes 1 and 3) of KB-R7785, and biotinylated proHB-EGF species (arrowheads) were immunoprecipitated with anti-HB-EGF antibody (lanes 1 and 2) or nonimmune IgG (lanes 3 and 4) as described in Materials and Methods. As for a control, β-actin was immunoblotted as described in Materials and Methods.
Discussion
The present study provides the first analysis of ADAM expression in human malignant astrocytic tumors, and demonstrates that among the 13 different ADAM species with putative metalloproteinase activity, ADAM12m is selectively expressed in the glioblastoma tissues. This was first shown by RT-PCR of the glioma and nonneoplastic control brain tissues, and further demonstrated by real-time quantitative PCR, in situ hybridization, immunostaining, and immunoblotting. The mRNA expression patterns of the 13 different ADAM species in our samples could be divided into three groups; constitutive expression (ADAM9, ADAM10, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28s, and ADAM28m), negligible or no expression (ADAM8, ADAM12s, and ADAM30), and glioblastoma-selective expression (ADAM12m). In contrast to the selective expression of ADAM12m in the glioblastomas, ADAM12s, another splice variant of ADAM12, was not expressed in any samples examined. Because ADAM12s is highly expressed in term placenta but not in tumor cell lines,15 the tissue expression of these isoforms is differently regulated by alternate splicing. Iba and colleagues16 have previously reported that ADAM12m is immunolocalized to carcinoma cells in more than 50% of the human carcinoma samples of the breast (infiltrating ductal carcinoma), colon (adenocarcinoma), stomach (adenocarcinoma), and lung (squamous cell carcinoma) with little or no immunostaining in the corresponding normal epithelia. ADAM10 and ADAM17 are expressed in human cancer tissues,2,30 but they are also often expressed in normal tissues.30,31 Thus, the data in the present and previous studies16 suggest that ADAM12m is one of the ADAM species selectively expressed by human malignant tumor tissues.
Our in situ hybridization study demonstrated that neoplastic astrocytic cells are responsible for the gene expression of ADAM12m in glioblastoma tissues. Immunostaining using monoclonal antibody specific to the cytoplasmic tail domain of ADAM12m further confirmed the expression by glioblastoma cells but not by stromal cells such as endothelial cells. Importantly, ADAM12m was immunolocalized predominantly on the cell membranes of glioblastoma cells, although a few glioblastoma cells showed weak cytoplasmic staining. These findings indicate that ADAM12m exists mainly on the cell membranes of glioblastoma cells, and suggest that some glioblastoma cells accumulate it within the cytoplasm because of the overproduction.
Like other ADAM species, ADAM12m is synthesized in an inactive proform and activated within the cells by being processed to an active form by the action of furin-like proprotein convertases(s).32,33 Previous immunoblotting studies showed that COS-7 cells transiently transfected with expression vectors encoding ADAM12m produce its precursor form of ∼110 to 120 kd and active form of ∼90 kd in ∼1:1 ratio.32,33 In the present study, however, immunoblotting identified only a protein band of ∼90 kd in both homogenates of glioblastoma tissues and lysates of U251 glioblastoma cells, and demonstrated that the protein band represents glycoprotein generated by N-linked protein glycosylation of ∼75-kd protein. Because the antibody used for immunoblotting recognizes the cytoplasmic tail domain of ADAM12m and the molecular weights of glycosylated and unglycosylated proteins correspond to the expected sizes of active glycosylated and unglycosylated forms of ADAM12m shown by other studies32,33 and estimated by the amino acid sequences,15 it is conceivable that ADAM12m is readily activated within the glioblastoma cells and presented on their cell surfaces in the active form.
One of the important findings in the present study is that the expression levels of ADAM12m directly correlated with MIB1-positive cell index of the malignant astrocytic tumors. This suggests the possible involvement of ADAM12m in the proliferation of glioblastomas. The marked proliferative activity of glioblastomas is explained mainly by enhanced EGF signaling through EGFR. The EGFR gene is amplified in 30 to 40% of human glioblastoma cases.18 In approximately half of the glioblastomas with EGFR gene amplification, the amplification is accompanied by specific deletion of a portion of the extracellular domain of EGFR through gene rearrangement.19–21 This truncated EGFR is constitutively active without ligand binding, and thus continuously transduces its signals to promote glioblastoma cell growth.22,23 On the other hand, glioblastomas expressing the wild-type EGFR with or without amplification proliferate in paracrine and/or autocrine pathway through the ligand-dependent EGF signaling.23,24 Astrocytic tumors are known to express the EGFR ligands, ie, EGF, TGF-α, and HB-EGF, as well as EGFR,23,24 and HB-EGF plays a key role in the EGFR signals in glioma cells.24 Because HB-EGF is synthesized as a membrane-anchored proform and processed to soluble active forms that function by binding to EGFR, ectodomain shedding is one of the key regulation steps for the activity of HB-EGF.13 Recent studies revealed that ADAM12m processes proHB-EGF to the soluble active forms in mouse myocardiocytes13 and embryonal fibroblasts.25 In the present study, we have demonstrated that although HB-EGF is constitutively expressed in both astrocytic tumors and nonneoplastic brain tissues, production of soluble HB-EGF forms from its precursor correlates with the expression of active ADAM12m in glioblastomas and the processing in glioblastomas and U251 glioblastoma cells is inhibited by ADAM inhibitor. Previous studies have shown that ADAM912 and MMP-334 are also implicated for ectodomain shedding of HB-EGF. However, the present study indicated that ADAM9 is constitutively expressed in both nonneoplastic brain tissues and astrocytic tumors without correlation to the HB-EGF processing. Although MMP-3 was immunolocalized in 40% of the glioblastoma cases,35 our quantitative analysis by sandwich enzyme immunoassay demonstrated that the production level in the glioblastoma tissues is negligible.36 Altogether, these data suggest the possibility that ADAM12m selectively expressed in glioblastomas contributes to the prominent proliferation of glioblastoma cells through shedding of HB-EGF.
From a biological and clinical point of view, glioblastomas are classified to primary glioblastomas developing in a rapid de novo manner and secondary glioblastomas evolved by progression from less malignant gliomas.17,37,38 Recent molecular genetic analyses of the glioblastomas have indicated that primary and secondary glioblastomas are characterized by the alterations of the EGFR and p53 genes, respectively.37,38 Therefore, it is interesting to know in which glioblastoma subsets ADAM12m is overexpressed to shed HB-EGF. However, we could not perform the genetic analyses of our samples because of the restrictions on the informed consent from the patients and limited amounts of the tissue samples. Thus, the issue remains to be answered by future work.
Recent accumulated evidence has indicated that ADAM12 plays an important role in supporting tumor cell adhesion, which is mediated through binding of the cysteine-rich domain of ADAM12 to syndecans, cell surface proteoglycans.6,16 Because syndecans become expressed in the reactive astrocytes surrounding the region of the brain necrosis induced by the cryo-injuries,39 it might be possible to speculate that ADAM12m facilitates glioblastoma cells to attach to the reactive astrocytes present near the leading invasive edge. This may explain the so-called “secondary structures” of glioblastoma invasive pattern, which represents the glioblastoma cell accumulation in the borders of the brain such as subpial zone of the cortex, subependymal, and perivascular regions.40 Alternatively, ADAM12m might be involved in mutual cell attachment and spreading of glioblastoma cells, which enables the glioblastoma cells to form tumor cell aggregates that are commonly observed in human glioblastomas during the invasive growth. Further studies are necessary to elucidate whether glioblastoma cells within the human glioblastoma tissues express syndecans and/or β1 integrin, which are used for the ADAM12-mediated cell attachment and spreading.6
Acknowledgments
We thank Michiko Uchiyama and Yuko Hashimoto for their technical assistance.
Footnotes
Address reprint requests to Yasunori Okada, M.D., Ph.D., Department of Pathology, School of Medicine, Keio University. 35 Shinanomachi, Shinjuku-ku, Tokyo 160-0016, Japan. E-mail: okada@sc.itc.keio.ac.jp.
Supported by the Ministry of Education, Science, and Culture of Japan (grant-in-aid B2-11240206 to Y.O.).
References
- Rooke J, Pan D, Xu T, Rubin GM. KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- Yavari R, Adida C, Bray-Ward P, Brines M, Xu T. Human metalloprotease-disintegrin Kuzbanian regulates sympathoadrenal cell fate in development and neoplasia. Hum Mol Genet. 1998;7:1161–1167. doi: 10.1093/hmg/7.7.1161. [DOI] [PubMed] [Google Scholar]
- Emi M, Katagiri T, Harada Y, Saito H, Inazawa J, Ito I, Kasumi F, Nakamura Y. A novel metalloprotease/disintegrin-like gene at 17q21.3 is somatically rearranged in two primary breast cancers. Nat Genet. 1993;5:151–157. doi: 10.1038/ng1093-151. [DOI] [PubMed] [Google Scholar]
- Arner EC, Pratta MA, Trzaskos JM, Decicco CP, Tortorella MD. Generation and characterization of aggrecanase. A soluble, cartilage-derived aggrecan-degrading activity. J Biol Chem. 1999;274:6594–6601. doi: 10.1074/jbc.274.10.6594. [DOI] [PubMed] [Google Scholar]
- Schlondorff J, Blobel CP. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J Cell Sci. 1999;112:3603–3617. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- Iba K, Albrechtsen R, Gilpin B, Frohlich C, Loechel F, Zolkiewska A, Ishiguro K, Kojima T, Liu W, Langford JK, Sanderson RD, Brakebusch C, Fassler R, Wewer UM. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to β1 integrin-dependent cell spreading. J Cell Biol. 2000;149:1143–1156. doi: 10.1083/jcb.149.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane MA, Marcinkiewicz C, Vijay-Kumar S, Wierzbicka-Patynowski I, Niewiarowski S. Viper venom disintegrins and related molecules. Proc Soc Exp Biol Med. 1998;219:109–119. doi: 10.3181/00379727-219-44322. [DOI] [PubMed] [Google Scholar]
- Bjarnason JB, Fox JW. Snake venom metalloendopeptidases: reprolysins. Methods Enzymol. 1995;248:345–368. doi: 10.1016/0076-6879(95)48023-4. [DOI] [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ, Black RA. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Shirakabe K, Wakatsuki S, Kurisaki T, Fujisawa-Sehara A. Roles of meltrin beta/ADAM19 in the processing of neuregulin. J Biol Chem. 2001;276:9352–9358. doi: 10.1074/jbc.M007913200. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Hirata M, Hasuwa H, Iwamoto R, Umata T, Miyado K, Tamai Y, Kurisaki T, Sehara-Fujisawa A, Ohno S, Mekada E. A metalloprotease-disintegrin, MDC9/meltrin-γ/ADAM9 and PKCdelta are involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. EMBO J. 1998;17:7260–7272. doi: 10.1093/emboj/17.24.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, Ohmoto H, Node K, Yoshino K, Ishiguro H, Asanuma H, Sanada S, Matsumura Y, Takeda H, Beppu S, Tada M, Hori M, Higashiyama S. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat Med. 2002;8:35–40. doi: 10.1038/nm0102-35. [DOI] [PubMed] [Google Scholar]
- Loechel F, Fox JW, Murphy G, Albrechtsen R, Wewer UM. ADAM 12-S cleaves IGFBP-3 and IGFBP-5 and is inhibited by TIMP-3. Biochem Biophys Res Commun. 2000;278:511–515. doi: 10.1006/bbrc.2000.3835. [DOI] [PubMed] [Google Scholar]
- Gilpin BJ, Loechel F, Mattei MG, Engvall E, Albrechtsen R, Wewer UM. A novel, secreted form of human ADAM 12 (meltrin α) provokes myogenesis in vivo. J Biol Chem. 1998;273:157–166. doi: 10.1074/jbc.273.1.157. [DOI] [PubMed] [Google Scholar]
- Iba K, Albrechtsen R, Gilpin BJ, Loechel F, Wewer UM. Cysteine-rich domain of human ADAM 12 (meltrin α) supports tumor cell adhesion. Am J Pathol. 1999;154:1489–1501. doi: 10.1016/s0002-9440(10)65403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavenee WK, Furnari FB, Nagane M, Huang H-JS, Newcomb EW, Bigner DD, Weller M, Berens ME, Plate KH, Israel MA, Noble MD, Kleihues P. Diffusely infiltrating astrocytomas. Kleihues P, Cavenee WK, editors. Lyon: IARC Press,; Pathology and Genetics of Tumors of the Nervous System. 2000:pp 10–21. [Google Scholar]
- Louis DN, Gusella JF. A tiger behind many doors: multiple genetic pathways to malignant glioma. Trends Genet. 1995;11:412–415. doi: 10.1016/s0168-9525(00)89125-8. [DOI] [PubMed] [Google Scholar]
- Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey PA, Wong AJ, Vogelstein B, Zalutsky MR, Fuller GN, Archer GE, Friedman HS, Kwatra MM, Bigner SH, Bigner DD. Anti-synthetic peptide antibody reacting at the fusion junction of deletion-mutant epidermal growth factor receptors in human glioblastoma. Proc Natl Acad Sci USA. 1990;87:4207–4211. doi: 10.1073/pnas.87.11.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci USA. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor α, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- Mishima K, Higashiyama S, Asai A, Yamaoka K, Nagashima Y, Taniguchi N, Kitanaka C, Kirino T, Kuchino Y. Heparin-binding epidermal growth factor-like growth factor stimulates mitogenic signaling and is highly expressed in human malignant gliomas. Acta Neuropathol (Berl) 1998;96:322–328. doi: 10.1007/s004010050901. [DOI] [PubMed] [Google Scholar]
- Kurisaki T, Masuda A, Sudo K, Sakagami J, Higashiyama S, Matsuda Y, Nagabukuro A, Tsuji A, Nabeshima Y, Asano M, Iwakura Y, Sehara-Fujisawa A. Phenotypic analysis of meltrin α (ADAM12)-deficient mice: involvement of meltrin α in adipogenesis and myogenesis. Mol Cell Biol. 2003;23:55–61. doi: 10.1128/MCB.23.1.55-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada M, Nakamura H, Ikeda E, Fujimoto N, Yamashita J, Sato H, Seiki M, Okada Y. Expression and tissue localization of membrane-type 1, 2, and 3 matrix metalloproteinases in human astrocytic tumors. Am J Pathol. 1999;154:417–428. doi: 10.1016/S0002-9440(10)65288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Nakamura H, Inoue M, Imai K, Noguchi M, Sato H, Seiki M, Okada Y. Expression and tissue localization of membrane-types 1, 2, and 3 matrix metalloproteinases in human invasive breast carcinomas. Cancer Res. 1997;57:2055–2060. [PubMed] [Google Scholar]
- Goishi K, Higashiyama S, Klagsbrun M, Nakano N, Umata T, Ishikawa M, Mekada E, Taniguchi N. Phorbol ester induces the rapid processing of cell surface heparin-binding EGF-like growth factor: conversion from juxtacrine to paracrine growth factor activity. Mol Biol Cell. 1995;6:967–980. doi: 10.1091/mbc.6.8.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger PC, Shibata T, Kleihues P. The use of the monoclonal antibody Ki-67 in the identification of proliferating cells: application to surgical neuropathology. Am J Surg Pathol. 1986;10:611–617. doi: 10.1097/00000478-198609000-00003. [DOI] [PubMed] [Google Scholar]
- Borrell-Pages M, Rojo F, Albanell J, Baselga J, Arribas J. TACE is required for the activation of the EGFR by TGF-α in tumors. EMBO J. 2003;22:1114–1124. doi: 10.1093/emboj/cdg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkkainen I, Rybnikova E, Pelto-Huikko M, Huovila AP. Metalloprotease-disintegrin (ADAM) genes are widely and differentially expressed in the adult CNS. Mol Cell Neurosci. 2000;15:547–560. doi: 10.1006/mcne.2000.0848. [DOI] [PubMed] [Google Scholar]
- Pabic HL, Bonnier D, Wewer UM, Coutand A, Musso O, Baffet G, Clement B, Theret N. ADAM12 in human liver cancers: TGF-β-regulated expression in stellate cells is associated with matrix remodeling. Hepatology. 2003;37:1056–1066. doi: 10.1053/jhep.2003.50205. [DOI] [PubMed] [Google Scholar]
- Cao Y, Kang Q, Zhao Z, Zolkiewska A. Intracellular processing of metalloprotease disintegrin ADAM12. J Biol Chem. 2002;277:26403–26411. doi: 10.1074/jbc.M110814200. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Raab G, Moses MA, Fernandez CA, Klagsbrun M. Matrix metalloproteinase-3 releases active heparin-binding EGF-like growth factor by cleavage at a specific juxtamembrane site. J Biol Chem. 1997;272:31730–31737. doi: 10.1074/jbc.272.50.31730. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kubota T, Kabuto M, Sato K, Kawano H, Hayakawa T, Okada Y. Production of matrix metalloproteinases and tissue inhibitor of metalloproteinases-1 by human brain tumors. J Neurosurg. 1994;81:69–77. doi: 10.3171/jns.1994.81.1.0069. [DOI] [PubMed] [Google Scholar]
- Shiomi T, Okada Y. MT1-MMP and MMP-7 in invasion and metastasis of human cancers. Cancer Metastasis Rev. 2003;22:145–152. doi: 10.1023/a:1023039230052. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Ohgaki H. Primary and secondary glioblastomas: from concept to clinical diagnosis. Neuro-oncology. 1999;1:44–51. doi: 10.1093/neuonc/1.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Tachibana O, Sata K, Yonekawa Y, Kleihues P, Ohgaki H. Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 1996;6:217–223. doi: 10.1111/j.1750-3639.1996.tb00848.x. [DOI] [PubMed] [Google Scholar]
- Iseki K, Hagino S, Mori T, Zhang Y, Yokoya S, Takaki H, Tase C, Murakawa M, Wanaka A. Increased syndecan expression by pleiotrophin and FGF receptor-expressing astrocytes in injured brain tissue. Glia. 2002;39:1–9. doi: 10.1002/glia.10078. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Burger PC, Collins VP, Newcomb EW, Ohgaki H, Cavenee WK. Glioblastoma. Kleihues P, Cavenee WK, editors. Lyon: IARC Press,; Pathology and Genetics of Tumors of the Nervous System. 2000:pp 29–39. [Google Scholar]