Abstract
In eubacterial translation, lack of a stop codon on the mRNA results in a defective, potentially toxic polypeptide stalled on the ribosome. Bacteria possess a specialized mRNA, called transfer messenger RNA (tmRNA), to rescue such a stalled system. tmRNA contains a transfer RNA (tRNA)-like domain (TLD), which enters the ribosome as a tRNA and places an ORF into the mRNA channel. This ORF codes for a signal marking the polypeptide for degradation and ends in a stop codon, leading to release of the faulty polypeptide and recycling of the ribosome. The binding of tmRNA to the stalled ribosome is mediated by small protein B (SmpB). By means of cryo-EM, we obtained a density map for the preaccommodated state of the tmRNA·SmpB·EF-Tu·70S ribosome complex with much improved definition for the tmRNA–SmpB complex, showing two SmpB molecules bound per ribosome, one toward the A site on the 30S subunit side and the other bound to the 50S subunit near the GTPase-associated center. tmRNA is strongly attached to the 30S subunit head by multiple contact sites, involving most of its pseudoknots and helices. The map clarifies that the TLD is located near helix 34 and protein S19 of the 30S subunit, rather than in the A site as tRNA for normal translation, so that the TLD is oriented toward the ORF.
Keywords: elongation factor Tu, preaccommodated state, rescue mechanism, transtranslation, transfer RNA-like domain
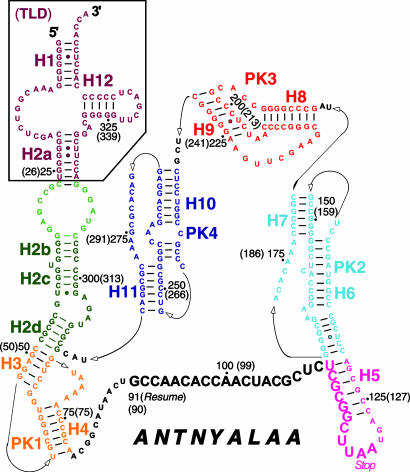
In bacterial cells, the normal process of translation is halted when the ribosome becomes trapped on incomplete mRNAs lacking a stop codon, resulting in a defective polypeptide attached to the P-site tRNA. To rescue such ribosomes and target the partially synthesized, potentially toxic peptide for degradation by proteases, bacteria maintain a quality-control system mediated by transfer messenger RNA (tmRNA), also called SsrA RNA or 10Sa RNA, and its associated protein, small protein B (SmpB) (1, 2). tmRNA functions as both transfer RNA (tRNA) and mRNA. Its tRNA-like domain (TLD) is charged at its 3′ CCA end with alanine (3, 4), whereas its mRNA-like domain contains an ORF, terminated with a stop codon, whose translation by the ribosome tags the defective peptide for recognition by proteases. In most tmRNAs, a string of pseudoknots (marked as PK1–PK4 in Fig. 1) is interspersed between the two functional domains. The switch of translation from the incomplete mRNA to the ORF of tmRNA is termed transtranslation. It has been shown in previous studies that tmRNA binds to ribosomes as a part of a complex with EF-Tu and GTP (5–8).
Fig. 1.
Secondary structure of tmRNA from T. thermophilus. The main three domains are TLD, MLD (mRNA-like domain), and PK (a succession of four pseudoknots). The MLD contains the nucleotide sequence known as ORF shown in bold letters for T. thermophilus, with the corresponding E. coli in parentheses, with resume and stop codons visible. The four PK domains (PK1 to PK4) along with 12 helices are shown in colors that are used consistently for the corresponding domains in all figures. The lines and dots represent the Watson–Crick and Wobble basepairings. The figure is adapted from the diagram for E. coli on the tmRDB web site (http://psyche.uthct.edu/dbs/tmRDB/tmRDB.html) (31).
This complex binds to a stalled ribosome in the presence of SmpB, allowing the initiation of transtranslation to occur (9, 10). Like tmRNA, the SmpB protein is well conserved among prokaryotic species (11) and is believed to be essential for the transtranslation process as a cofactor of tmRNA (9), enhancing both aminoacylation of tmRNA (7) and the interaction between tmRNA and the ribosome (12). During this process SmpB is likely to play a role on both the large and the small subunits of the ribosome (13).
A previous cryo-EM study of a preaccommodated tmRNA entering the stalled ribosome along with SmpB led to several findings (8). First, the TLD was found to be associated with EF-Tu in the same manner as an aminoacyl tRNA. Second, the remainder of the tmRNA was found to be highly structured and organized into a large, low-pitched spiral encircling the beak of the 30S subunit, placing the ORF into the vicinity of the mRNA entrance channel. Third, the density map showed one SmpB (in the following termed SmpB-1), forming a close interaction with both the TLD and the 50S ribosomal subunit.
The recent x-ray structure of SmpB in complex with the TLD of tmRNA (14) showed that the TLDs form an angle of 120° between the anticodon loop and the aminoacyl-acceptor stem (CCA), unlike the model by Valle et al. (8), in which an angle of 110° was assumed based on a prediction from transient electric birefringe measurements. In the x-ray structure, the SmpB is found at the elbow region of the TLD, which places the protein at the decoding site of the small subunit. This finding would imply the existence of two different sites for SmpB, one bound to the 50S subunit, the other to the 30S subunit. Indeed, various biochemical experiments have led to the conclusion that two or more copies of SmpB are bound to the ribosome during transtranslation (13, 15, 16). However, to date neither the precise number nor locations of the molecules have been ascertained. Another unsolved problem is the fate of tmRNA in the accommodated state. Wower et al. (17) concluded from biochemical data that the tmRNA is partially unstructured after release of EF-Tu from the ribosome and accommodation of TLD, after insertion of the ORF into the mRNA channel. Again, a confirmation of this result by structural methods has thus far been missing, and it is as yet unclear how the binding or release of the SmpBs is coordinated with the unraveling of tmRNA.
To address these questions, we obtained cryo-EM maps for complexes in two relevant states during transtranslation. The first is the preaccommodated state, with improved resolution particularly in the tmRNA–SmpB complex. The resulting map established the occurrence of two SmpBs in the complex, one binding to the 50S subunit, at the GTPase-associated center (GAC) near the site where it was previously found (SmpB-1) (8), the other (SmpB-2) to the 30S subunit close to the decoding site, but also in close vicinity to SmpB-1. In addition, the now-improved definition of the spiral-shaped density formed by the tmRNA pseudoknots allowed both the modeling of the tmRNA and the mapping of the contact sites between each of the pseudoknots of tmRNA and the 30S subunit with improved certainty. Significantly, the map shows that the 5′ end of the ORF is clearly positioned at the tip of the beak of the 30S subunit, indicating a novel path of the codons into the ribosome. The second state is the state after the release of EF-Tu, in which tmRNA has moved into the A site. The resulting density map, at 12-Å resolution, shows the tmRNA to be largely unstructured, in agreement with predictions from recent biochemical findings of Wower et al. (17).
Results
Biochemical Characterization of the tmRNA–Ribosome Complexes.
Two different tmRNA-ribosome entry complexes, corresponding to the preaccommodated and accommodated states of tmRNA, were reconstituted in vitro. For the preaccommodated state, the stalled 70S ribosome, containing no stop codon, with an initiator tRNA in the P site, reacts with alanylated tmRNA, SmpB, ribosomal protein S1, EF-Tu, and GTP in the presence of antibiotic kirromycin (details in Experimental Procedures). The presence of kirromycin prevents EF-Tu from leaving the ribosome after GTP hydrolysis, thus allowing the ternary complex of tmRNA·EF-Tu·GDP to be stabilized on the ribosome for visualization [preaccommodated state (8, 18–20)]. On the other hand, the accommodated state was reached in the absence of kirromycin and the presence of a deacylated tRNA in the P site. It was trapped by the absence of both EF-G and the tRNAs. In a first series of experiments the use of an aminoacylated P-site tRNA prevented tmRNA to stably occupy the A site, probably because of the high dissociation rate of accommodated tmRNA after transpeptidylation (occupancy close to zero as measured by incorporation of radioactive labeled tmRNA; data not shown). This result prompted us to use a deacylated tRNA, which produces an increase in the kinetic stability of the accommodated tmRNA in the A site, as described for tRNAs during canonical translation (21).
After reconstitution, stalled ribosomes in the absence of tmRNA (control), and the two complexes described above, were purified by gel filtration on a Superdex 200 chromatography column (GE Healthcare, Orsay, France). 70S ribosomes were eluted in the void volume of the column (see fractions 8 and 9 in Fig. 5a, which is published as supporting information on the PNAS web site; relative molecular mass >600,000). These fractions were pooled and submitted to SDS/PAGE analysis. After silver staining the bands corresponding to EF-Tu (44.6 kDa) and S1 (60 kDa) were separated from the ribosomal proteins (Fig. 5b, arrows and brace; compare control programmed ribosomes and tmRNA complexes). As expected, EF-Tu is present mainly when kirromycin is used (preaccommodated state). However, it was also detectable in the postaccommodated state, albeit at a very lower level. Exogenous protein S1 was detected to the same extent in the two complexes.
Western blot using anti-SmpB antibodies confirmed that the protein is present in both complexes but not on the programmed control ribosomes (Fig. 5c). Computer analysis of digital images of the Western blots was used to detect differences in relative band intensity of the immunologically detected SmpB within the preaccommodated and postaccommodated complexes. Quantitative measurements of SmpB within the various complexes indicated that ≈2 molecules of SmpB are associated per ribosome during the preaccommodated step, whereas ≈1 SmpB was recovered within the postaccommodated one. However, this result has to be interpreted in the light of the rates of occupancy of tmRNA within the two complexes. Based on the measurement of 32P radio-labeled tmRNA yields present in fractions 8 and 9, occupancy can indeed be evaluated to >95% in the preaccomodated complex versus ≈45% in the postaccommodated one (Fig. 5c). We cannot exclude the possibility that the lower SmpB/ribosome ratio recovered in the accommodated fraction is directly linked to the lower occupancy of tmRNA (see Discussion).
Analysis of Cryo-EM Maps for Preaccommodated State.
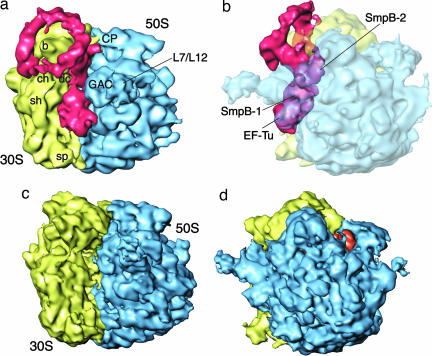
By means of cryo-EM and single-particle reconstruction techniques (22), the preaccommodated state complex was visualized at a resolution of 13.6 Å (Fig. 2a and b). For an additional comparison, a cryo-EM map (control map) of the stalled 70S ribosome in the absence of both tmRNA and SmpB has been produced at a resolution of 10 Å (Fig. 2 c and d). Although the previous data (8) led to a resolution of 14 Å for the overall map, both tmRNA and SmpB were poorly defined in their structural features. In the current study, the larger number of particles and higher degree of scrutiny in the selection of particles (see Experimental Procedures) resulted in a much clearer definition of the density attributable to tmRNA and SmpBs.
Fig. 2.
Reconstruction of tmRNA–SmpB complex bound with the 70S ribosome and EF-Tu. (a and b) Cryo-EM map generated for 70S·mRNA·tmRNA·EF-Tu·SmpB·S1 in the presence of GTP and kirromycin, in different orientations related by a rotation around the vertical axis. In a, the tmRNA density expands along finger-like protrusions to make contact with the 30S subunit when the density threshold is lowered (data not shown). In b, density for the ribosome is shown semitransparent; density for EF-Tu·tmRNA·SmpB is in red. (c and d) Cryo-EM map of 70S·mRNA·tRNA control. The 50S subunit is in blue, the 30S subunit is in yellow, and the E-site tRNA is in orange. Landmarks on the 50S subunit are as follows: CP, central protuberance; L7/L12, stalk formed by proteins L7/L12. Landmarks on 30S subunit are: sh, shoulder; b, beak; dc, decoding center; ch, entrance of mRNA channel; sp, spur.
The improvement in the resolution of the relevant features in the map led to a reexamination of the preaccommodated state. Overall, the density map closely resembles that obtained by Valle et al. (8), characterized by showing the combined density of EF-Tu, SmpB, and TLD at the entrance of the ribosomal intersubunit space, and tmRNA as a large spiraling thread of density surrounding the beak of the small ribosomal subunit. However, the two maps differ in the definition of the mass attributed to tmRNA and SmpB, the new map allowing a more extensive and precise interpretation by fitting an atomic model. (Protein S1 is not visible on the 30S subunit probably because of its loose binding; see ref. 23.)
We used the recently published x-ray structure of the complex formed by TLD and SmpB by Gutmann et al. (14) in the fitting of the present map. This structure includes two SmpB molecules and two partially fragmented copies of the TLD in one unit cell: a larger one, including nucleotides 13–57, and a smaller one, including nucleotides 16–39. We chose to use the larger copy of the TLD and tried different combinations of this TLD with the two SmpBs for the map fitting. When the first SmpB (according to the order in which the coordinates appear in the Protein Data Bank) and the chosen TLD were jointly fitted into the cryo-EM map, a unique fitting position was found in the map, with SmpB [henceforth named SmpB-2, reserving the term SmpB-1 for the position in which SmpB was previously observed (8)], oriented toward the A site of the 30S subunit (Fig. 6, which is published as supporting information on the PNAS web site) so the TLD is positioned toward the beak of the 30S subunit. Moreover, only this SmpB binds the elbow region of the TLD, as expected from previous chemical footprints (7, 24). In contrast, the combination of the TLD with the other copy of the SmpB in the x-ray structure does not match the density map. This result differs from the previous model by Valle et al. (8), where the density for the present SmpB-2 was attributed to the TLD, and no SmpB was found near the A site.
Unlike normal tRNA, the TLD does not have a closed anticodon loop (ASL) at the junction with the rest portion of tmRNA. An artificial tetraloop was added at the end of H2a in the x-ray structure to close the ASL equivalent (H2a) for the purpose of facilitating the crystallization. By contrast, in the real sequence of tmRNA, H2a is open and linked to H2b through unstructured loops C28–G35 and G319–G324. At the resolution of the present cryo-EM map (13.6 Å), it is impossible to determine these details conclusively.
Helix 1 of the TLD is equivalent to the CCA stem of tRNA for normal translation, but coordinates of this helix were not solved in the crystal structure. Thus, another x-ray structure including the CCA stem from the Cys-tRNACys·EF-Tu·GDPNP complex (25) was used to complete the model of the TLD for the purpose of fitting the present map. The coordinates of EF-Tu from the same crystal (1B23) were also used for the present fitting. The relative orientation of EF-Tu to the CCA stem in the x-ray structure (1B23) was retained. Placement of both the CCA stem and EF-Tu resulted in an overall satisfying match to the density.
After the fitting of the TLD·SmpB and EF-Tu·CCA-stem moieties, a portion of the density remained unexplained, which was found of sufficient size and suitable shape to accommodate another SmpB (henceforth called SmpB-1), in a position where the protein contacts both the TLD and EF-Tu, pointing toward the 50S subunit (Fig. 6b). This finding indicates that there are two SmpB molecules (SmpB-1 and SmpB-2) that coexist on the ribosome in the preaccommodated state of tmRNA. Our observation is consistent with the biochemical findings indicating that more than one SmpB molecule is associated with the complex formed by the tmRNA and the stalled ribosome (13, 15, 16).
After the docking of EF-Tu, TLD, SmpB-1, and SmpB-2, the remaining spiraling density must account for helices and pseudoknot-rich domains of tmRNA. The improved resolution of the cryo-EM map in this region allowed us to confirm that the ORF is highly structured in the preaccommodated state. Based on the positions of helix 5 and PK1 at the two ends of the ORF, an atomic model for fitting the spiraling density was built by using components of the model by Zwieb et al. (26); see Fig. 6 c and d. The coordinates that were determined previously (8) are now replaced by our model.
The x-ray structure of SmpB includes several residues forming random coils at the two terminals. Based on the present fitting positions, we surmise that these terminals would be actively involved in the interactions among SmpB and the TLD, and the two subunits of the ribosome, but it is impossible to determine such details at the present level of structural resolution.
Interactions of SmpBs and tmRNA with 30S and 50S Ribosomal Subunits.
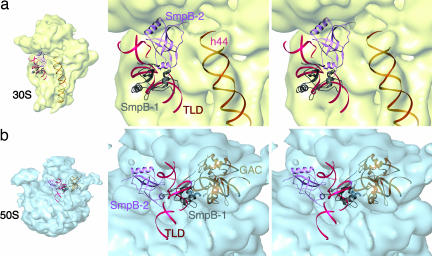
The current study shows the presence of two molecules of protein SmpB bound to the tmRNA, namely SmpB-2 at the decoding center of 30S subunit, and SmpB-1 contacting the GAC of the 50S subunit (see Fig. 7, which is published as supporting information on the PNAS web site). The interactions among the two SmpBs, the TLD, and the ribosomal subunits can be characterized as follows:
SmpB-2.
This copy of the molecule overlaps with the distal portion of the anticodon loop of the preaccommodated aminoacyl-tRNA as seen in normal translation (20). SmpB-2 possibly interacts with the nucleotides involved in codon–anticodon recognition, namely A1492-A1493 in h44 and nucleotides around G530 in h18 of 16S rRNA (Fig. 3a). Specifically, in the current fitting, the C-terminal of SmpB-2, which may be quite flexible under physiological conditions, points toward the position of the codon in the A site observed for normal translation. The N-terminal of SmpB-2 is positioned near the intersection of H1 (the equivalent of the CCA of normal tRNA) and H12 (the equivalent of the T loop of normal tRNA). Thus, SmpB-2 forms a tight network with the surrounding components.
Fig. 3.
Ribosomal subunits interacting with SmpBs and tmRNA. Different views of the 70S ribosome are used to explain the interactions of SmpB-1·SmpB-2·TLD complex with the 30S subunit (a) and the 50S subunit (b). Components of tmRNA are the same colors as in Fig. 2c.
SmpB-2 remains at the same position in both the absence (R. Gillet, S.K., W.L., M. Hallier, B. Felden, and J.F., unpublished work) and the presence of tmRNA, but it cannot be definitely decided whether the SmpB remains in the same orientation, because of the globular shape of SmpB and the resolution limitation of the current map
TLD.
The TLD interacts with each of the components forming the complex of tmRNA·SmpBs·EF-Tu and the ribosome. Helix 2a (the equivalent of the anticodon loop) possibly contacts h34 alongside SmpB-2. The T-loop equivalent in the TLD interacts with SmpB-2, whereas the D-loop equivalent interacts with SmpB-1. Relative to EF-Tu, helix 1 (the equivalent of the CCA stem) is positioned in the orientation of the CCA stem in the A/T-site tRNA for normal translation.
SmpB-1.
Besides the already noted interactions of SmpB-1 with the TLD and SmpB-2, SmpB-1 forms a close contact with the GAC, including the A1067 loop in 23S rRNA (Fig. 3b), which interacts with the D loop of tRNA in normal translation (20). The region of interaction between SmpB-1 and the TLD is in agreement with available data from footprinting and mutational analysis (7, 27). The random coil at the C terminal of SmpB-1 that appears in the x-ray structure is, according to the fitting, near the groove between the A1067 and A1095 loops, leading to the conclusion that the region around the loops is involved in the interaction between SmpB-1 and the GAC. Thus, the binding between SmpB-1 and the GAC appears to be stronger than what was found in the case of normal translation between the GAC and aminoacyl tRNA.
Another important aspect of our results is the way SmpB-1 and SmpB-2 interact with each other and with the TLD portion of tmRNA (Fig. 7). SmpB-2 and SmpB-1 loosely form a cross with the TLD, sitting at the nexus of the two domains of TLD. One role of SmpB-2 is evidently to stabilize the unusual shape of the TLD, which differs strongly from the 90° L shape of the normal tRNA. Thus, SmpB-1 and SmpB-2 appear to create a stable scaffold that allows the stretched shape of the TLD to reach the two remotely located, functionally essential elements of the ribosome: the decoding site on the small subunit and the GAC on the large subunit.
tmRNA.
Analysis of our map delineates the contact sites between tmRNA and the 30S subunit, along most of the spiral density formed by tmRNA (see Fig. 8, which is published as supporting information on the PNAS web site). First, the density of helix 2 is tightly fused with the 30S subunit head, at the position S19 and helix 34, seemingly through the involvement of multiple residues. Second, the single strand connecting the ORF and PK1 at the 3′ end contacts helix 33c, and the 5′ of the ORF contacts helix 34 of the 30S subunit. Third, helix 5 is largely fused with protein S3 of the 30S subunit. Fourth, PK2 contacts helix 41 of the 30S subunit. Fifth, the single strand in PK3 forms a weak contact with helix 33c of the 30S subunit. PK4 seems to be the only structural element that does not directly interact with the ribosome, which may be the reason PK4 is the most unstructured portion in the accommodated state, the state that will be discussed in the next section.
Accommodation of the TLD into the A Site.
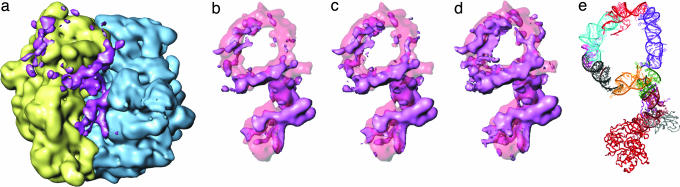
In addition to the study of the preaccommodated state of the tmRNA–ribosome complex, we carried out a cryo-EM study of its accommodated state. In normal translation, EF-Tu is promptly released from the ribosome after GTP hydrolysis, the tRNA is accommodated into the A site, and its aminoacyl end reaches the peptidyl-transfer center. Our study was designed to observe the corresponding accommodated state of the TLD and the disposition of the remainder of the tmRNA in this state. We visualized the complex in this state at a resolution of 12 Å (Fig. 4). We can summarize the findings as follows.
SmpB-2 is observed in the same position and with the same high occupancy as in the preaccommodated state. At a low density threshold, which is much lower than the level appropriate for visualizing the ribosome (≈50%), helix 5, PK2, PK3, and PK1 are seen to form a spiral-shaped density disconnected at PK4 (Fig. 4e). Although, overall, the density is weak compared with the preaccommodated state, the density attributable to PK1 is relatively strong. The disappearance of portions of tmRNA is interpreted as a consequence of the unstructured nature and high mobility of tmRNA in these regions. At a high threshold, the remaining density is only for PK1 (Fig. 4c), which is consistent with its high compact size and stability as recently shown by NMR (28).
Helix 2, including H2b, H2c, and H2d, remains in contact with PK1. Even at a very high density threshold, at which the spiral-shaped density mostly disappears, helix 2 including the TLD shows up as the strongest portion in the remaining mass of the tmRNA (Fig. 4c).
There is also an additional density occupying a small portion of the mass of EF-Tu, but a major part of the density for EF-Tu is not present in the current state, even at a significantly decreased density threshold. These data are in agreement with the very weak signal by EF-Tu within the complex as detected by silver-staining gels (Fig. 5b).
Fig. 4.
Accommodated tmRNA–SmpB complex bound to 70S ribosome in the presence of ribosomal protein S1. 30S subunit is in yellow and 50S subunit is in blue. (a) Cryo-EM map of tmRNA-SmpB shown with low density threshold, along with 70S ribosome shown with normal threshold. (b–d) tmRNA–SmpB complex displayed with different density thresholds, from normal (b) to very low (d). (e) Atomic model of tmRNA complex shown in Fig. 6d, shown for comparison.
Discussion
Mechanisms of Transtranslation.
Transtranslation is a complex process involving the entering of tmRNA and two molecules of SmpB into the ribosome. The extended configuration in which tmRNA is present makes the complex highly variable. Previously, only a single state of the tmRNA–ribosome complex, equivalent to the preaccommodated state of the translating ribosome, had been visualized by cryo-EM (8), but its interpretation rested on a structural model of the TLD (assumption of tRNA mimicry) that proved to be incorrect.
Using the published x-ray coordinates of a re-engineered TLD–SmpB complex (14) in the analysis of the improved cryo-EM map and reanalysis of the existing map (8), we have been able to obtain a more complete picture of the arrangement of tmRNA components and SmpB on the ribosome in the preaccommodated state. Our results indicate that two molecules coexist in the preaccommodated state, consistent with biochemical studies that indicated the presence of more than one copy: one (SmpB-1) being identical to the one identified by Valle et al. (8), the other (SmpB-2) being positioned at the A site of the 30S subunit and complexed with the TLD in the same manner as shown by Gutmann et al. (14). According to a recent analysis of the interaction between tmRNA and SmpB from Thermus thermophilus (29) only one SmpB binds to tmRNA (at a location equivalent to the D loop and variable loop of tRNA) outside the framework of the ribosome. These data strengthen the idea that the formation of a SmpB scaffold around tmRNA occurs only in contact with the ribosome and that it guides the molecule into a productive conformation (R. Gillet, S.K., W.L., M. Hallier, B. Felden, and J.F., unpublished work).
From the arrangement of the two SmpB molecules in the complex with tmRNA and the stalled ribosome, we can deduce that they play two very important roles in the transtranslation. One is that the two molecules together form a scaffold to stabilize the TLD structure in the shape (angle between domains of 120°) required for its import into the ribosome. The other role is that the SmpBs establish key interactions with the decoding center and the GAC, which are both sites that the TLD cannot directly reach. Both interactions are required, in the course of normal translation, to productively engage the ribosome: at the decoding center, the nucleotides involved in stabilizing the codon–anticodon contact and in issuing a conformational signal are A1492 and A1493 of h44 and G530 of h18. At the GAC, the loop carrying A1067 may be on the receiving end of the signal (20). A plausible working hypothesis is that the TLD accommodation would use the same mechanism that is used in the accommodation of an aminoacylated tRNA, because the TLD, just as tRNA, enters with EF-Tu in the GTP state and requires GTP hydrolysis for EF-Tu to disengage and leave.
The visualization of the binding complex of tmRNA, SmpBs, and the ribosome still leaves open the question how these moieties might recognize the stalled ribosome. There must be a high degree of cooperativity in two pivotal processes: (i) between the binding and positioning of SmpBs on the ribosome and the entry of tmRNA into the ribosome, and (ii) between the release of EF-Tu and the accommodation of the TLD. Some light is shed on the first step of assembly by the discovery (R. Gillet, S.K., W.L., M. Hallier, B. Felden, and J.F., unpublished work) that in the absence of tmRNA, a single copy of SmpB binds to the stalled ribosome, at the same site as SmpB-2.
From the Preaccommodated State to the Accommodated State.
A recent study by Wower et al. (17) indicated that a portion of tmRNA, ranging from PK2 to PK4, is unstructured when the TLD of the tmRNA is accommodated. The present study of the complex in the accommodated state shows that several portions of density visible in the cryo-EM map of the preaccommodated state have disappeared, including those for EF-Tu and PK2–PK4 of the tmRNA. This disappearance must be attributed to the variability in the conformations of the corresponding portions after the accommodation, as they are averaged out in the reconstruction. Thus, the remaining portions with relatively strong density likely represent rather immobile structural elements. We made an attempt of modeling the TLD in the accommodated state by placing its equivalent of the CCA stem into the A site, in the vicinity of the peptidyl transferase center, while retaining SmpB-1 in the same relative position to the CCA stem as in the preaccommodated state. This placement, however, resulted in a steric clash between SmpB-1 and the tRNA in the P site (data not shown). Thus, it is unlikely that SmpB-1 will stay in the same relative orientation to the CCA stem once the TLD is accommodated. Relative quantification of SmpB within this complex shows that one molecule of SmpB remains associated to each ribosome after accommodation (Fig. 5c). Although we cannot rule out that this decrease in the SmpB/ribosome ratio is caused by the poor occupancy of the tmRNA ternary complex to the ribosome after accommodation, this observation, in agreement with biochemical data collected from Shpanchenko et al. (30), suggests as a possible reason the departure of SmpB-1, which was reported to contact the large subunit only transiently during transtranslation (13).
Experimental Procedures
Preparation of the 70S·tmRNA·EF-Tu·GDP·SmpB·S1·kir Complex.
Salt-washed T. thermophilus 70S ribosomes free of S1 were prepared as described (8). EF-Tu and S1 from T. thermophilus were overexpressed in Escherichia coli by using the T7 expression system and purified by using ion exchange and gel-filtration chromatography. C-terminal His-tagged SmpB from T. thermophilus was produced from a pET21a plasmid. After the overproduction, the mixture was heated at 70°C for 30 min, allowing the separation of free SmpB from the RNA-bound protein. The T. thermophilus tmRNA gene was cloned downstream of a T7 RNA polymerase promoter. Plasmids were linearized with BsmBI restriction enzyme before RNA was synthesized in vitro by using s MEGAscript T7 kit (Ambion, Austin, TX). Purification was performed by electrophoresis on denaturing gels. The tmRNA was heated at 80°C for 2 min in a folding buffer (5 mM MgCl2/20 mM NH4Cl/10 mM Hepes-KOH, pH 7.5) and cooled for 30 min at room temperature before being aminoacylated. The alanylation was performed in a 200-μl medium containing 25 mM Hepes-KOH (pH 7.5), 30 mM NH4Cl, 7 mM MgCl2, 2 mM ATP, 6 mM phosphoenolpyruvate, 10 g/ml PK, 30 μM alanine, 20 μM EF-Tu·GTP, 1 μM SmpB, 1 μM tmRNA, and 2 μM AlaRS. (The maximum rate of alanylation of T. thermophilus tmRNA by E. coli AlaRS was >60% as measured by trichloroacetic acid precipitation of radioactivity from labeled alanine.) After incubation for 30 min at 37°C, the sample was concentrated 10-fold after addition of 0.1 vol of 2 M ice-cold potassium acetate (pH 5.0). This EF-Tu·AlatmRNA·SmpB complex was used for binding to the ribosome. Ribosomal P-site tRNA complexes were obtained by incubating 70S ribosomes (1 μM) with 2 μM of mRNA consisting of the sequence GGCAAGGAGGUAAAAAUG and E. coli fMet-tRNAfMet (2 μM) for 30 min at 37°C. The concentrated solution of the EF-Tu·tmRNA·SmpB complex was directly added to an equal volume of P-site-occupied 70S ribosomes in the presence of 100 μM kirromycin and, for S1-containing samples, a 3-fold excess of S1 protein. The SmpB concentration was increased 2-fold before continuing the incubation for 30 min at 37°C.
For the preparation of the S1-containing accommodated complex, the same procedure was used except that 70S ribosomes (1 μM) were incubated in the presence of E. coli deacylated tRNAfMet (2 μM). Deacylated P-site tRNA was used to increase the kinetic stability of tmRNA within the A site after accommodation (21). No kirromycin was used during the following procedures.
Purification of Programmed 70S Complexes by Size Exclusion Chromatography.
After filtration through 0.22-μm filters, samples (100 μl) were applied to a Superdex 200 HR 10/30 column (GE Healthcare) equilibrated with 5 mM Hepes-KOH (pH 7.5), 10 mM NH4Cl, 10 mM MgOAc, and 50 mM KCl at 4°C. The fractions containing the purified 70S ribosomes were pooled, concentrated, and applied to an SDS/PAGE analysis with 15% polyacrylamide gel. Gels were stained by using the PlusOne Silver Staining kit (GE Healthcare) or subjected to SmpB immunodetection by Western blotting. Rabbit polyclonal antibodies used in the experiments were directed against E. coli SmpB protein (16) and strongly cross-reacted with T. thermophilus SmpB. Detection was performed by using chemiluminescence. tmRNA occupancy measurements were evaluated by using the same procedure, except that 32P-radiolabeled tmRNA was used and subsequently quantified within the purified 70S fractions by liquid scintillation counting on a Wallac 1409 (Perkin–Elmer, Wellesley, MA).
Cryo-EM and Image Processing.
Ribosomal samples were diluted to a final concentration of 32 nM and used directly for cryo-EM grid preparation following standard procedures. Micrographs at ×50,000 (±2%) magnification were taken on a Philips FEI (Eindhoven, The Netherlands) Tecnai F20 with field emission gun operated at 200 KV and low electron dose (≈15e Å−2). The micrographs were scanned with a pixel size corresponding to 2.82 Å on the object scale with a Zeiss Imaging scanner (Z/I Imaging Corporation, Huntsville, AL). The image processing was carried out with the SPIDER package (32) and included a reference-guided projection classification and alignment, contrast transfer function correction of segregated defocus groups, and correction of the high-frequency amplitudes by using low-angle x-ray scattering data (33). The numbers of particles used in the 3D reconstructions were 48,252, 52,829, and 19,472 for the 70S·tRNA control, 70S tRNA·tmRNA·EF-Tu·SmpB·S1·kir preaccommodated complex, and 70S·tRNA·tmRNA·EF-Tu·SmpB·S1 accommodated complex, respectively. In the same order, the 0.5 cutoff in the Fourier shell correlation was 10, 13.6, and 12 Å, respectively. The docking of atomic model of tmRNA into the cryo-EM maps was done by using O, and the visualization was performed by using IRIS Explorer (Numerical Algorithm Group, Downers Grove, IL), Ribbons (34), and Insight II (Accelrys, San Diego, CA).
The increased, effectively doubled number of particles for the preaccommodated state (58,699) compared with the previous reconstruction (29,098), along with the use of a higher correlation threshold for rejection, led to a much improved definition of the density for tmRNA, even though the overall resolution of the entire ribosome–tmRNA complex (measured by Fourier shell correlation with 0.5 criterion) was only slightly higher.
Supplementary Material
Acknowledgments
We thank M. Watters for assistance with the illustrations; V. Ramakrishnan (Medical Research Council, Cambridge, U.K.) for providing the T. thermophilus ribosomes; and B. Felden for insightful comments on the manuscript. This work was supported by National Institutes of Health Grants R37 GM29169 and R01 GM55440 (to J.F.) and Rennes Métropole and the Région Bretagne Programme de Recherche d' Initiative Régionale no. 691 (to R. Gillet).
Abbreviations
- tRNA
transfer RNA
- TLD
tRNA-like domain
- SmpB
small protein B
- tmRNA
transfer messenger RNA
- GAC
GTPase-associated center.
Footnotes
The authors declare no conflict of interest.
References
- 1.Karzai AW, Roche ED, Sauer RT. Nat Struct Biol. 2000;6:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 2.Saguy M, Gillet R, Metzinger L, Felden B. Biochimie. 2005;87:897–903. doi: 10.1016/j.biochi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. Proc Natl Acad Sci USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ushida C, Himeno H, Watanabe T, Muto A. Nucleic Acids Res. 1994;22:33923396. doi: 10.1093/nar/22.16.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudinger-Thirion J, Giege R, Felden B. RNA. 1999;5:989–992. doi: 10.1017/s135583829999101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barends S, Wower J, Kraal B. Biochemistry. 2000;39:2652–2658. doi: 10.1021/bi992439d. [DOI] [PubMed] [Google Scholar]
- 7.Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. J Mol Biol. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 8.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Science. 2003a;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 9.Karzai AW, Susskind MM, Sauer RT. EMBO J. 1999;18:37933799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wower IK, Zwieb CW, Guven SA, Wower J. EMBO J. 2000;19:6612–6621. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keiler KC, Shapiro L, Williams KP. Proc Natl Acad Sci USA. 2000;97:7778–7783. doi: 10.1073/pnas.97.14.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu Y, Ueda T. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- 13.Hallier M, Desreac J, Felden B. Nucleic Acids Res. 2006;34:1935–1945. doi: 10.1093/nar/gkl097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 15.Wower J, Zwieb CW, Hoffman DW, Wower IK. Biochemistry. 2002;41:8826–8836. doi: 10.1021/bi0201365. [DOI] [PubMed] [Google Scholar]
- 16.Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B. J Biol Chem. 2004;279:5978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- 17.Wower IK, Zwieb C, Wower J. RNA. 2005;11:668–673. doi: 10.1261/rna.7269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valle M, Sengupta J, Swani NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M. Nat Struct Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 20.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Nat Struct Biol. 2003b;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 21.Semenkov YP, Rodnina MV, Wintermeyer W. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 22.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. New York: Oxford Univ Press; 2006. [Google Scholar]
- 23.Sengupta J, Agrawal RK, Frank J. Proc Natl Acad Sci USA. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzinger L, Hallier M, Felden B. Nucleic Acids Res. 2005;33:2384–2394. doi: 10.1093/nar/gki534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen P, Thirup S, Kjeldgaard M, Nyborg J. Struct Fold Des. 1999;7:143–156. [Google Scholar]
- 26.Zwieb C, Larsen N, Wower J. Nucleic Acids Res. 1998;26:166–167. doi: 10.1093/nar/26.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nonin-Lecomte S, Felden B, Dardel F. Nucleic Acids Res. 2006;34:1847–1853. doi: 10.1093/nar/gkl111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nameki N, Someya T, Okano S, Suemasa R, Kimoto M, Hanawa-Suetsugu K, Terada T, Shirouzu M, Hirao I, Takaku H, et al. J Biochem (Tokyo) 2005;138:729–739. doi: 10.1093/jb/mvi180. [DOI] [PubMed] [Google Scholar]
- 30.Shpanchenko OV, Zvereva MI, Ivanov PV, Bugaeva EY, Rozov AS, Bogdanov AA, Kalkum M, Isaksson LA, Nierhaus KH, Dontsova OA. J Biol Chem. 2005;280:18368–18374. doi: 10.1074/jbc.M409094200. [DOI] [PubMed] [Google Scholar]
- 31.Knudsen B, Wower J, Zwieb C, Gorodkin J. Nucleic Acids Res. 2001;29:171–172. doi: 10.1093/nar/29.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 33.Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 34.Carson M. J Appl Crystallgr. 1991;24:103–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.