Abstract
To locate the transmissible gastroenteritis coronavirus (TGEV) packaging signal, the incorporation of TGEV subgenomic mRNAs (sgmRNAs) into virions was first addressed. TGEV virions were purified by three different techniques, including an immunopurification using an M protein-specific monoclonal antibody. Detection of sgmRNAs in virions by specific reverse transcription-PCRs (RT-PCRs) was related to the purity of virus preparations. Interestingly, virus mRNAs were detected in partially purified virus but not in virus immunopurified using stringent conditions. Analyses by quantitative RT-PCR confirmed that virus mRNAs were not present in highly purified preparations. Lack of sgmRNA encapsidation was probably due to the absence of a packaging signal (Ψ) within these mRNAs. This information plus that from the encapsidation of a collection of TGEV-derived minigenomes suggested that Ψ is located at the 5′ end of the genome. To confirm that this was the case, a set of minigenomes was expressed that included an expression cassette for an mRNA including the β-glucuronidase gene (GUS) plus variable sequence fragments from the 5′ end of the virus genome potentially including Ψ. Insertion of the first 649 nucleotides (nt) of the TGEV genome led to the specific encapsidation of the mRNA, indicating that a Ψ was located within this region which was absent from all of the other virus mRNAs. The presence of this packaging signal was further confirmed by showing the expression and rescue of the mRNA including the first 649 nt of the TGEV genome under control of the cytomegalovirus promoter in TGEV-infected cells. This mRNA was successfully amplified and encapsidated, indicating that the first 649 nt of TGEV genome also contained the 5′ cis-acting replication signals. The encapsidation efficiency of this mRNA was about 30-fold higher than the genome encapsidation efficiency, as estimated by quantitative RT-PCR. In contrast, viral mRNAs presented significantly lower encapsidation efficiencies (about 100-fold) than those of the virus genome, strongly suggesting that TGEV mRNAs in fact lacked an alternative TGEV Ψ.
Transmissible gastroenteritis coronavirus (TGEV) is a member of Coronaviridae, a family of positive-strand RNA viruses that infect birds and mammals and cause a variety of diseases, most frequently of the enteric and respiratory tract (15, 37). TGEV is an enveloped virus containing an internal core made of a positive-sense RNA genome of 28.5 kb, the nucleoprotein (N), and the carboxy terminus of the membrane (M) protein (16, 17, 54).
The genome and a nested set of subgenomic mRNAs are produced in the cytoplasm of TGEV-infected cells. A leader sequence at the 5′ end and open reading frame (ORF) 7 followed by the 3′ untranslated region (UTR) are present in all these mRNAs (Fig. 1A) (4, 28, 62). The incorporation of the viral RNA genome is a specific process based on the recognition of a set of sequences containing the packaging signal (Ψ) that directs the encapsidation of the genome RNA into virions (21, 29, 44, 45). Accordingly, a packaging signal has been characterized in the mouse hepatitis coronavirus (MHV) genome located within ORF 1b around 20 kb from the genome 5′ end in a sequence domain not present in the virus mRNAs (19, 47, 71). MHV Ψ has been narrowed down to a 190-nt RNA sequence of which a predicted 69-nt stem loop seems to be sufficient for RNA incorporation into virions (19). The homologous packaging signal in the bovine coronavirus (BCoV) genome has also been identified in a similar position (11). Studies of defective interfering (DI) RNA rescue (including the amplification and encapsidation of these minigenomes) have been carried out with coronaviruses of groups 1 and 3, TGEV, and the infectious bronchitis virus (IBV) (12, 27). However, the location and nature of the packaging signal in these viruses have not been precisely defined.
FIG. 1.
Detection of TGEV virus mRNAs in infected ST cells. (A) Scheme of the TGEV virus genome, represented as a bar in which the different ORFs are indicated as boxes. The virus mRNAs for each TGEV gene are represented as thin bars under the genome. L, leader sequence; UTR, 3′ UTR; An, polyadenylated sequence. Rep 1a, Rep 1b, S, 3a, 3b, E, M, N, and 7 indicate each of the TGEV ORFs. (B) Ethidium bromide-stained agarose gel of DNA products obtained by specific RT-PCRs to detect the virus mRNAs indicated above the panel. Mm, molecular size markers.
Encapsidation of coronavirus mRNAs in amounts 5- to 200-fold lower than those of the genome RNA has been reported for TGEV (63, 64), BCoV (25), and IBV (76). However, when mRNAs were detected in purified MHV virions, this detection was associated with contamination (6). Overall, these data indicate that the encapsidation of virus mRNAs in coronaviruses requires further studies.
In this study, we showed that TGEV mRNAs were absent from highly purified virions, most probably because these mRNAs do not contain an efficient Ψ. Interestingly, a TGEV RNA Ψ has been identified at the 5′ end of the TGEV genome in a sequence domain not present in the virus mRNAs. Furthermore, it has been shown that the presence of the first 649 nucleotides (nt) from the 5′ end of the TGEV genome (in addition to sequences from the 3′ end) was sufficient for minigenome RNA amplification and encapsidation.
MATERIALS AND METHODS
Cells and viruses.
Swine testicle (ST) cells (41) were grown in Dulbecco modified Eagle's medium supplemented with fetal calf serum. The TGEV PUR46-MAD strain was grown and titrated in ST cells as described previously (30).
Antibodies.
The murine monoclonal antibodies (MAbs) 3D.C10 and 25.22 (specific for the N and M proteins, respectively) have been described previously (9, 16, 17, 22, 30, 36, 58, 61).
Virus purification.
TGEV virions were purified by three techniques: (i) partial purification by concentration through a 15% sucrose cushion, (ii) purification in continuous sucrose gradients, and (iii) immunopurification using the M protein-specific MAb 25.22 (9, 16). For partial purification, virions from supernatants of TGEV-infected ST cells were sedimented through a 15% sucrose cushion in TEN buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 1 M NaCl) by ultracentrifugation at 27,000 rpm for 50 min at 4°C in a SW60Ti Beckman rotor and were recovered in TNE buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 0.1 M NaCl) to a final concentration of 1 μg/μl. In the second method, sedimented virions were further purified by ultracentrifugation through a linear sucrose gradient (30 to 42%) in TEN buffer as reported previously (30) and recovered in TNE to a final concentration of 1 μg/μl. TGEV immunopurification relied on the specific capture of sedimented virions with MAb 25.22 bound to enzyme-linked immunosorbent assay plates (Nunc) and subsequent washing in phosphate-buffered saline containing 0.1% bovine serum albumin as described previously (16). Sedimented virions were used instead of sucrose gradient-purified virions in immunopurifications to recover defective particles with lower densities than wild-type virions. When indicated, Tween 20 was added to washing buffer to achieve final concentrations of 0.05 or 0.5%. Virus purifications were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining in the case of the first two procedures and by fluorography using [35S]methionine-cysteine-labeled TGEV in the case of the immunopurifications, as described previously (16).
Cloning of minigenome M33-derived sequences in minigenome M39-GUS.
M33 was the smallest efficiently encapsidated and replicated minigenome in TGEV-infected ST cells (27). The M33 5′ sequence is made from the first 2,144 nt of the genome (A region) and 568 nt from the overlapping sequence between ORFs 1a and 1b (nt 12195 to 12763) (B region). The A region was divided into four 550-nt fragments overlapping by 50 nt: A1, A2, A3, and A4. A- and B-derived sequences were amplified by PCR using primers that introduced SalI restriction sequences at both the 5′ and 3′ ends (Table 1). PCR products were digested with SalI and cloned into pCMV-M39-GUS plasmid (3) restricted with SalI. The M33-derived fragments (A1, A2, A3, A4, and B) were inserted upstream of the GUS gene under the control of the M39-GUS transcription-regulating sequence, generating minigenomes M39-GUS-A1, -A2, -A3, -A4, and -B under the control of the cytomegalovirus (CMV) promoter as described previously (3). The structures of all constructs were confirmed by sequencing.
TABLE 1.
Primers for cloning M33 sequences into pCMV-M39-GUS
| Region | Virus-sense primer (5′-3′) | Reverse-sense primer (5′-3′) | Amplified genome sequence (nt) |
|---|---|---|---|
| A1 | GGGTCGACGAAATATTTGTCTTTCTATGAAATC | CCGTCGACATGGCACCTCTGACAGTGCGAGC | 100-649 |
| A2 | GGGTCGACCGTTCTTGAGGACTTTGACCTTAAAATTG | CCGTCGACCATCACCAGGCTTAATATCACCC | 599-1149 |
| A3 | GGGTCGACTTTCTGGCAAAGTTAAGGGTGTC | CCGTCGACACGATTGTCTGGAACCACAAATGTTGGC | 1099-1649 |
| A4 | GGGTCGACGCTTTTACGATTGTAAACTACAAGCC | CCGTCGACTTCAAATGATGAACCAAGTTTTGC | 1599-2144 |
| B | GGGTCGACCAAATACCAACTGGCACACAAGATCC | CCGTCGACAATTCTTCAGTGCAAGCACCTACTGTC | 12195-12763 |
Cloning of mRNAs containing M33-derived sequences under the control of the CMV promoter.
To clone the mRNA-GUS cDNA in an expression plasmid under the control of the CMV promoter, cytoplasmic RNA was purified from ST cells infected with TGEV stocks containing minigenome M39-GUS. The mRNA-GUS 5′ fragment was amplified by reverse transcription-PCR (RT-PCR) using primers GUS 297 (Table 2) and CMV-5′TGEV (virus sense primer; 5′-GCTCGTTTGTGAACCGTACTTTTAAAGTAAAGTGAGTG-3′). This RT-PCR product included part of the CMV promoter fused to the mRNA-GUS 5′ end. Another PCR fragment was produced which contained most of the CMV promoter and 5′ end of the TGEV genome. In this PCR, the plasmid pCMV-M33NGUS (27) was used as a template with primers CMV-5′TGEV RS (reverse-sense primer; 5′-CTTTAAAAGTACGGTTCACTAAACGAGCTCTGCTT ATATAGACC-3′) and ApaI-CMV (virus sense primer; 5′-TATATAGGGCCCTTGACATTGATTATTGACTAG-3′). Using primers ApaI-CMV and GUS 297, the two PCR fragments were fused by overlap extension PCR. The resulting PCR product was restricted with NdeI and SalI and cloned into the pCMV-M39-GUS plasmid restricted with NdeI and SalI, generating the plasmid pCMV-mRNA-GUS.
TABLE 2.
Primers used for RT-PCR amplifications
| ORF | Reverse-sense primer (5′-3′) | Virus-sense primer (5′-3′) | Expected size (nt) |
|---|---|---|---|
| 1 | CTTGATGCACTAACTTCTG | CAGGATCCTGTAGACAAGTGTGTG | 1,200 |
| CTTGATGCACTAACTTCTG | GGCATGCTTGCTACTAGCTTGGTTGGTGC | 1,100 | |
| S | TAACCTGCACTCACTACCCC | Leader (AGATTTTGTCTTCGGACACCAACTCG) | 537 |
| 3a/3ba | TCAGCATGAGCTAAGCCACG | Leader | 615 |
| M | CCAAAACAACGGGCCATAATAGCC | Leader | 667 |
| N | TAGATTGAGAGCGTGACCTTG | Leader | 537 |
| Eb | GCGCATGCAATCACACGC | Leader | 625 |
| 7 | TCTGGTTTCTGCTAAACTCC | Leader | 191 |
| M39-GUS | GUS 297 (GACCCACACTTTGCCGTAATGAG) | 19949 (CTTGGTGGATCTGTTGCC) | 536 |
| mRNA-GUS | GUS 297 | Leader | 374 |
| β-actin mRNA | AGCACCGTGTTGGCGTAGAG | CGGGAGATCGTGCGGGACAT | 300 |
Primer hybridizes within the 3b gene to detect both mRNA 3a/3b and mRNA 3b.
Primer hybridizes with the 5′ end of the M gene.
To clone the mRNAs derived from minigenomes M39-GUS-A1, -A2, -A3, -A4, and -B under the control of the CMV promoter, the A- and B-derived sequences were amplified and restricted with SalI as described above and cloned in the plasmid pCMV-mRNA-GUS restricted with SalI.
Rescue of minigenomes and mRNAs in TGEV-infected ST cells.
Briefly, all constructs were transcribed from the CMV promoter in ST cells transfecting 5 μg of the engineered plasmids with 15 μl of Lipofectin as described by the manufacturer (GIBCO). Transfected cells were incubated overnight at 37°C and infected with TGEV at a multiplicity of infection (MOI) of 5. Supernatants were recovered at 24 h postinfection, and virions were passaged to confluent ST cells from two to four times as described previously (3).
RNA isolation and RT-PCR analysis.
Nontransfected or transfected ST cells (on 60-mm-diameter culture plates) were infected with TGEV at a MOI of 5. Cellular RNA was extracted at 20 h postinfection by using ULTRASPEC RNA extraction reagent and purified as described by the manufacturer (Biotecx). When RNA was extracted from transfected cells, contaminating plasmid DNA was digested with 0.3 U of DNase I (Roche)/μl by incubation for 1 h at 37°C. Then, RNA was repurified by the same procedure as described above to remove active DNase I. Absence of plasmid DNA was confirmed by standard and quantitative PCR before performing RT reactions. RT reactions were carried out using from 50 to 100 ng of RNA as described previously (23).
Using ULTRASPEC reagent as described by the manufacturer, RNA was extracted from 5 μg of virus purified in sucrose gradients or from 50 ng of immunopurified virus. Total Saccharomyces cerevisiae RNA (40 μg) or glycogen (Sigma) was added as a carrier during RNA precipitation. For detection of the genome, virus mRNAs, β-actin mRNA, M39-GUS-derived minigenomes, and mRNAs derived from minigenomes, RT-PCRs were performed using specific primers (Table 2).
DNA fragments from RT-PCRs were separated in 1% agarose gels in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0) containing 0.1% ethidium bromide and detected by UV light exposure. When indicated, RT-PCR products were purified using a High Pure PCR product purification kit (Qiagen). All purified DNA bands were sequenced.
Calculation of encapsidation efficiencies by quantitative RT-PCR.
Confluent ST cells grown in 10 90-mm-diameter culture dishes were infected when indicated at a MOI of 10 with virus stocks containing mRNA-A1-GUS defective particles (passage 3). Cytoplasmic and purified virus RNAs were extracted at 12 h postinfection as described above. All PCRs were performed in PCR master mix buffer (Applied Biosystems), following the manufacturer's instructions. Real-time detection of amplicons was carried out using an ABI PRISM 7000 PCR detection system, and data processing was performed with an ABI PRISM 7000 SDS program. For quantitation of genome RNA, mRNAs for N and E proteins, and β-actin mRNA, primers for RT-PCRs specifically hybridizing within each of these RNAs (Table 3) were chosen attending to the following criteria: (i) melting temperature higher than 60°C, (ii) amplicon size of about 100 nt, and (iii) synthesizing of a single RT-PCR product. Concentration standards were generated by 10-fold dilutions of quantified amplicons (0.01, 0.001, and 0.0001 ng/μl). cDNA copies from RT reactions were estimated for infected cells and for virions immunopurified by using standard concentration lines with correlation coefficients (R2) higher than 0.99. Each sample was quantified in triplicate.
TABLE 3.
Primers used for real-time RT-PCR analyses
| Target | Reverse-sense primer (5′-3′) | Virus-sense primer (5′-3′) |
|---|---|---|
| Genome | TGCAAGGCATGCTGGCATTTTATAC | ACTCATTGAATTTAGGCAGCAAAGC |
| mRNA-N | CCTGGTTGGCCATTTAGAAGTTTAG | Leader primer |
| mRNA-E | TTCTTCTTTTAAGTCAATTTCGTTTAG | Leader primer |
| β-actin mRNA | CAGAGTCCATGACAATGCCAGTGGT | ATGTTTGAGACCTTCAACACGCC |
| mRNA-A1 | TCGCGATCCAGACTGAATGC | ATGGTCCGTCCTGTAGAAACCC |
Relative mRNA-to-genome encapsidation efficiencies were calculated as the mRNA/genome molar ratio in purified virions in relation to the ratio in infected ST cells. Five independent experiments were analyzed, and encapsidation efficiencies for the TGEV genome and all mRNAs were compared using a Wilcoxon nonparametric t test (43).
RESULTS
Detection of TGEV mRNAs in infected ST cells and in purified virions.
To determine whether TGEV mRNAs are specifically encapsidated, an RNA detection system based on RT-PCR was set up in TGEV-infected ST cells. Each virus mRNA was detected by RT-PCR using a leader-specific positive-sense primer (leader primer; Table 2) and reverse-sense primers specific for virus genes to amplify in each reaction a single member of the nested set of mRNAs. The presence of virus mRNAs was evaluated by RT-PCR at 20 h postinfection (Fig. 1A). DNA amplicons were separated in agarose gels containing 0.1% ethidium bromide, excised from gels, and sequenced. Bands with the expected size were detected in TGEV-infected cells (Fig. 1B), and their sequences corresponded to those of the viral mRNAs (data not shown).
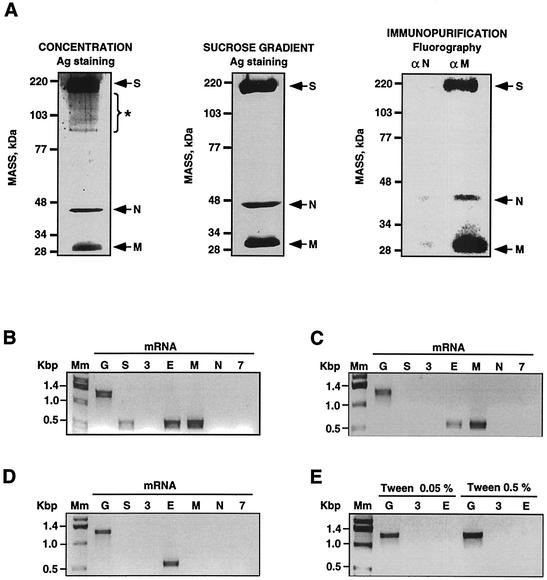
To determine whether viral mRNAs were encapsidated, TGEV virions were purified by three techniques: (i) partial purification by concentration through a 15% sucrose cushion, (ii) ultracentrifugation in continuous sucrose gradients, and (iii) immunopurification using the M protein-specific MAb 25.22 (specific for the amino terminus of the M protein). Purified viruses were analyzed by SDS-PAGE and silver staining in the case of partially and sucrose gradient-purified virus and by fluorography in the case of immunopurifications using 35S-labeled TGEV, as described previously (16). Fluorography was used instead of silver staining in the case of immunopurifications to avoid the partial overlap of the virus structural proteins with the immunoglobulins, bovine serum albumin, and protein A used in the immunopurification. All virus preparations contained the major structural proteins S (spike), N, and M (Fig. 2A). Partially purified virus also contained protein contaminants of high molecular mass. In contrast, sucrose gradient-purified and immunopurified virions did not contain significant protein contaminants (Fig. 2A). As a control, using the N protein-specific MAb 3D.C10, immunopurification of 35S-labeled virions was performed. As expected, when the N protein-specific MAb was used, only background levels of N and M proteins were detected, probably corresponding to very low levels of unassembled protein complexes (Fig. 2A). However, it cannot be concluded that immunopurified virions are totally free of protein contaminants, since only radioactively labeled proteins can be detected by fluorography. Additionally, in the conditions used for protein electrophoresis, proteins of molecular masses below 25 kDa could not be detected. Therefore, the presence of low-molecular-mass protein contaminants cannot be excluded.
FIG. 2.
Detection of virus mRNAs in purified TGEV virions. (A) SDS-PAGE and silver staining of virus proteins from partially purified TGEV virions (left panel) or sucrose gradient purified virus (central panel). Using the M protein-specific MAb 25.22 or the N protein-specific MAb 3D.C10 as a control, SDS-PAGE and fluorography of virus proteins from immunopurified TGEV virions were performed (right panel). TGEV major structural proteins are indicated with arrows. *, contaminant proteins. (B to E) Detection of the virus mRNAs (indicated by letters at the top of the gels) in purified virions by specific RT-PCRs in ethidium bromide-stained agarose gels from partially purified virus (B), sucrose gradient-purified virus (C), immunopurified virus (D) and immunopurified virus in the presence of the indicated concentrations of Tween 20 (E). Mm, molecular size markers.
The presence of virus mRNAs in these virus preparations was analyzed by RT-PCR (Fig. 2B). Partially purified TGEV contained mRNAs S, envelope (E), and M. Virus mRNAs E and M were also detected when the partially purified virions were further purified in sucrose gradients. Only mRNA E was detected after immunopurification and moderate washing. Interestingly, when Tween 20 at two concentrations (0.05 and 0.5%) was included during washing, no virus mRNAs were detected (Fig. 2B). These results showed that detection of TGEV mRNAs in purified virus was related to their homogeneity, strongly suggesting that virus mRNAs were not specifically encapsidated. For subsequent experiments, Tween 20 at 0.5% was included during immunopurifications.
Relative encapsidation efficiencies of viral mRNAs.
To confirm that virus mRNAs were not encapsidated, the efficiency of incorporation of mRNAs N and E to virions in relation to TGEV genome encapsidation was estimated by quantitative RT-PCR. Viral mRNAs N and E were chosen since mRNA N was one of the most abundant viral mRNAs in infected cells, while mRNA E remained after extensive virus washing (Fig. 2B), although there was a random variability from experiment to experiment regarding the contaminant virus mRNAs that remained after immunopurification. However, both mRNAs N and E were the contaminants detected most frequently on a regular basis. As a nonviral mRNA control, encapsidation efficiency for cellular β-actin mRNA was also estimated. When sucrose gradient-purified TGEV was used, the apparent encapsidation efficiencies of mRNAs N and E (in relation to genome encapsidation) were variable and high, at up to fivefold higher in the case of mRNA E (Table 4). In contrast, cellular β-actin mRNA levels (unrelated to TGEV mRNAs) were low in purified virions. These results suggested that either virus mRNAs were encapsidated or there were abundant contaminating mRNAs, as observed in previous experiments (Fig. 2B). Nevertheless, when immunopurified virions were used, encapsidation efficiencies for virus mRNAs significantly (P < 0.05) dropped to very low levels (comparable to that of β-actin mRNA) of about 100- to 1,000-fold lower than genome encapsidation efficiencies, as determined by the nonparametric t test of Wilcoxon (Table 4). These results confirmed that virus mRNAs were not specifically encapsidated. The lower proportion of β-actin mRNA compared to virus mRNAs in sucrose gradient-purified viruses could have been due to the lesser presence of this mRNA in the cell compartment in which the virus was assembled.
TABLE 4.
Relative mRNA-to-genome encapsidation efficiencies determined by quantitative real-time RT-PCR
| Target | Encapsidation efficiency (severalfold) by:
|
|
|---|---|---|
| Sucrose gradient purification | Immunopurification | |
| Genome | 1 | 1 |
| mRNA Na | 0.27 ± 0.035 | 0.008 ± 0.0018 |
| mRNA Ea | 5 ± 3 | 0.0004 ± 0.0005 |
| β-actin mRNA (ns)b | 0.001 ± 0.0009 | 0.006 ± 0.0006 |
Significant differences (P < 0.05) in mRNA encapsidation efficiencies determined from virus purified by the two methods.
ns, no significant differences (P > 0.05) in mRNA encapsidation efficiencies determined from virus purified by the two methods.
Location of the TGEV genome region containing Ψ.
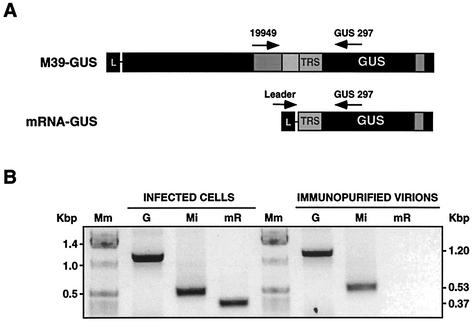
TGEV mRNAs were not incorporated into virions, probably because they lacked a packaging signal. To confirm whether this was the case, encapsidation of minigenome M39-GUS and of mRNA-GUS transcribed from an expression cassette inserted within this minigenome (Fig. 3A) was studied. Both M39-GUS and mRNA-GUS were detected at comparable levels by specific RT-PCRs in infected cells (Fig. 3B). In contrast, only M39-GUS was detected in immunopurified virions (Fig. 3B). Since both M39-GUS and mRNA-GUS contained the leader sequence and the same 3′ end (Fig. 3A), TGEV Ψ was probably located within the 5′ sequence of minigenome M39-GUS.
FIG. 3.
Specific encapsidation of TGEV minigenome M39-GUS. (A) Scheme representing the M39-GUS minigenome containing the expression cassette for the GUS protein (upper bar) and the mRNA-GUS derived from M39-GUS by transcription (lower bar). Oligonucleotides used for specific RT-PCRs are shown as arrows above the bars. L, leader; TRS, transcription-regulating sequence; GUS, β-glucuronidase gene. The arrows above the bars indicate the positions in the sequence where the primers for the RT-PCR amplification were located. The numbers indicate the sequence nucleotides at which the primer starts in the viral genome or within the GUS gene. (B) Ethidium bromide-stained agarose gel of RT-PCR products specific for the genome (G), minigenome M39-GUS (Mi), and mRNA-GUS (mR) in infected ST cells and immunopurified virions, as indicated in Materials and Methods. Mm, molecular size markers.
To locate the TGEV genome region that contained the packaging signal, sequences potentially containing Ψ were selected from the 5′ sequence of the M33 minigenome also included in minigenome M39 (Fig. 4A), since M33 was the smallest minigenome efficiently rescued in a previous study (27). The 5′ sequence of M33 is derived from two discontinuous regions from the TGEV genome: one (A region) of 2,144 nt and another (B region) of 568 nt from positions 12195 to 12763 of the TGEV genome (27, 54) (Fig. 4A). The A region was divided into four fragments of 550 nt, overlapping each other by 50 nt (Fig. 4A). At the 3′ end of minigenome M39, an expression cassette was inserted consisting of a TRS, each of the selected fragments potentially containing Ψ, and the GUS gene (Fig. 4B). Therefore, cells infected with a helper TGEV and transfected with a plasmid encoding this minigenome would produce mRNAs containing each of the inserted sequences plus GUS RNA, in addition to the standard helper virus RNAs. In principle, only the mRNA containing the Ψ would be encapsidated. Plasmids encoding these minigenomes were transfected to ST cells, and minigenomes were synthesized from the CMV promoter as described previously (27). Minigenomes were amplified through two passages in confluent ST cell cultures (Fig. 4B). The presence of minigenome and mRNAs in infected cells and in immunopurified virions was investigated by RT-PCR as described above (Fig. 3A). Minigenomes and mRNAs were detected in cells in comparable amounts independently of the inserted region (Fig. 4C). In contrast, only the mRNA containing the A1 region (mRNA-A1-GUS) was detected in immunopurified virions (Fig. 4C).
FIG. 4.
Structure of TGEV minigenome M33 and location of the genome region containing the TGEV Ψ. (A) Scheme representing the TGEV genome (upper bar) and the M33 minigenome (lower large bar). The sequence fragments forming M33 (derived from TGEV genome) are indicated (I, II, III, and IV); bar sections A and B represent discontinuous genome regions present in the 5′ sequence of M33. Thin bars below the M33 minigenome bar represent overlapping PCR-amplified sequences of 550 nt from the A and B regions that potentially contain the packaging signal (A1 to A4 and B). Numbers above and below the M33 bar represent nucleotide positions in the TGEV genome. L, leader sequence; An, polyadenylated sequence. Rep 1a, Rep 1b, S, 3a, 3b, E, M, N, and 7 indicate each of the TGEV ORFs. (B) A scheme of the experimental procedure used to locate the TGEV Ψ is shown. The M39 minigenome containing an expression cassette is represented by the top bar. The shorter bar below the M39 minigenome bar represents the insert containing M33-derived sequences. The minigenome transfected to helper TGEV-infected ST cells was amplified by successive virus passage in confluent ST cell monolayers (P0 to P2). TRS, transcription-regulating sequence; TGEV, helper virus; RNA 0 to RNA 2, cytoplasmic and virion RNAs. (C) Ethidium bromide-stained agarose gels of specific RT-PCR amplified cDNAs from the genome (G), minigenome (Mi), and mRNA-A1-GUS to mRNA-B-GUS from cells and immunopurified virus including Tween-20 in washing buffer, as indicated. mR, mRNA-GUS; Mm, molecular size markers.
These results indicated that a packaging signal was present within the first 649nt of the TGEV genome and that its insertion into mRNA-GUS (Fig. 3) led to the specific encapsidation of this mRNA (Fig. 4C). Interestingly, it was observed that mRNA-A1-GUS encapsidation apparently interfered with TGEV genome incorporation into virions, since the amount of genomic RNA was reduced when the mRNA included the A1 sequence but not when the mRNAs lacked this region (Fig. 4C).
Replication of mRNA-A1-GUS.
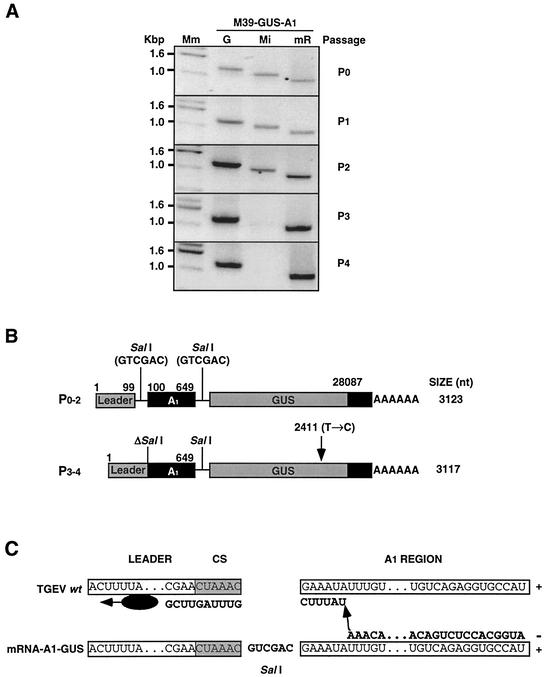
During M39-GUS-A1 rescue it was observed that this minigenome tended to disappear throughout cell passage (Fig. 5A). In contrast, mRNA-A1-GUS, launched by M39-GUS-A1 by transcription at passage 0, increased throughout virus passage, suggesting that mRNA-A1-GUS rescue was independent of the presence of M39-GUS-A1 (Fig. 5A). The increase of mRNA-A1-GUS levels in successive passages suggested that this mRNA was amplified by the helper virus, and the mRNA-A1-GUS from passage 0 to 3 was sequenced (Fig. 5B). From passage 0 to 2 its sequence was identical to that expected for the mRNA transcribed from minigenome M39-A1-GUS, containing two SalI restriction sites flanking the A1 region. In contrast, at passage 3 the SalI restriction site between the leader core sequence (CS) and the A1 region was lost (Fig. 5B). This reversion to the wild-type sequence probably occurred by recombination between the mRNA-A1-GUS and the helper virus genome (Fig. 5C) during the synthesis of the mRNA-A1-GUS negative or positive strand, indicating that the mRNA-A1-GUS corresponded to newly synthesized RNA and not to an mRNA transcribed from minigenome M39-GUS-A1.
FIG. 5.
Rescue of M39-GUS-A1 in ST cells. (A) Ethidium bromide-stained agarose gels of specific RT-PCR products for the genome (G), minigenome M39-GUS-A1 (Mi), and mRNA-A1-GUS (mR) from ST cells infected with the helper TGEV and transfected with minigenome M39-GUS-A1 at the indicated passages (P0 to P4). Mm, molecular size markers. (B) Scheme of the structure and sequence of mRNA-A1-GUS rescued in the indicated cell passages. The SalI restriction sites flanking the A1 fragment (present in the mRNA transcribed from minigenome M39-GUS-A1) are indicated above the bars. ΔSalI, deletion of the SalI restriction site. Numbers above the bars represent positions in the TGEV genome. The position of a nucleotide substitution within the GUS gene is indicated with an arrow. (C) Scheme of the potential recombination event between the mRNA-A1-GUS and TGEV genome that could have been produced anywhere in the A1 region during the synthesis of either the mRNA-A1-GUS positive strand or its complementary strand, leading to an mRNA with the 5′ end sequence identical to that of the genomic RNA. The SalI restriction site between the leader and the A1 region is shown in the mRNA-A1-GUS. CS, transcription-regulating core sequence located at the 3′ end of the leader. The viral replicase complex is represented as an ellipse, and the progress of RNA synthesis is indicated with arrows. +, Positive-stranded RNA; −, nascent negative-stranded RNA.
To determine whether this mRNA-A1-GUS was amplified by the helper virus, all mRNAs containing M33-derived sequences were cloned under the control of the CMV promoter and ST cells were transfected with these constructs. Transfected cells were infected with TGEV, and supernatants were passaged in confluent ST cell monolayers. Intracellular RNA was extracted in each passage, and mRNAs were analyzed by RT-PCR (Fig. 6A). Interestingly, only the mRNA-A1-GUS was detected in all passages, while the rest of the mRNAs were only detected at passage 0. This confirmed that mRNA-A1-GUS was encapsidated and also amplified; otherwise, its presence would have been drastically reduced at passage 4 by dilution. The mRNA-A1-GUS from passages 1 to 4 was sequenced. From passage 0 to 2 its sequence was identical to that of the transfected construct, containing the two SalI restriction sites flanking the A1 region. Interestingly, from passage 3 to 4, the SalI restriction site between the leader CS and the A1 region was also lost, as already observed for the mRNA-A1-GUS launched from minigenome M39-GUS-A1 (Fig. 5B and C). This reversion to the wild-type sequence probably occurred by recombination between the mRNA-A1-GUS and the helper virus genome (Fig. 5C), indicating that the mRNA-A1-GUS corresponded to newly synthesized RNA. In fact, at passage 5 the amounts of TGEV genome and mRNA-A1-GUS were undetectable during the first hour after infection and had clearly increased at 3 h postinfection (Fig. 6B). In contrast, β-actin mRNA levels were high from the beginning and were kept constant (Fig. 6B). These results confirmed that mRNA-A1-GUS most probably contained a packaging signal and that it was also amplified by the helper virus.
FIG. 6.
Rescue of mRNA-A1-GUS in ST cells. (A) Ethidium bromide-stained agarose gels of specific RT-PCR products for the genome (G) and mRNA-A1-GUS (mR) from ST cells in the P0 to P4 virus passages. The constructs transfected in passage 0 are indicated above the gels. G and mR, amplified DNAs from the genome and the mRNA, respectively. (B) Ethidium bromide-stained agarose gels of specific RT-PCR DNAs amplified from the genome, mRNA-A1-GUS, and β-actin mRNA in TGEV-infected ST cells containing the mRNA-A1-GUS (passage 5) at the postinfection times indicated above the gels. Mm, molecular size markers.
Estimation of mRNA-A1-GUS encapsidation efficiency by quantitative RT-PCR.
The relative mRNA-A1-GUS and TGEV genome encapsidation efficiencies were estimated in immunopurified virions by quantitative RT-PCR using specific primers (Table 3). All calculations were performed with molar ratios as described in Materials and Methods. The mRNA-A1-GUS encapsidation efficiency was 33- ± 2.6-fold (mean ± standard deviation; n = 5, coefficient of variation = 8%) higher than that of TGEV genome encapsidation and about 200-fold higher than that of virus mRNA N (0.16 ± 0.04, mean ± standard deviation; n = 5, coefficient of variation = 25%). These differences were statistically significant (P < 0.05) by the Wilcoxon t test. These results indicated that mRNA-A1-GUS was specifically and efficiently encapsidated and most probably contained the TGEV major packaging signal.
DISCUSSION
In this study, the TGEV packaging signal was located within the first 649 nt of the genome in a sequence domain absent from all TGEV mRNAs. In fact, detection of virus mRNAs in virions depended on the purity of virus preparations and viral mRNAs were not detected in highly immunopurified viruses, indicating that viral mRNAs were not specifically encapsidated. This observation was supported by the results of quantitative RT-PCR.
The findings obtained in this work supported those reported for MHV, for which mRNAs were detected in purified virus only when the Ψ was inserted into these RNAs (6), and also those of publications that concluded that the presence of mRNAs in virus preparations was associated with contamination (48, 49).
The engineering of infectious coronavirus cDNAs has shown that the full-length genome is sufficient to generate infectious coronaviruses either from cDNA or from in vitro-transcribed RNA molecules (2, 7, 70, 73, 74). However, although encapsidation of RNA genomes is a specific process required for virus propagation, incorporation into virions of tRNAs, rRNAs, mRNAs, and subgenomic RNAs has been reported for several viruses (1, 25, 38, 46, 55, 59, 60, 63, 76). Encapsidation of mRNAs has also been described for coronaviruses (25, 63, 76).
We have shown that the presence of viral mRNAs within the viral particles is a function of the degree of purity of the virions. Therefore, since it would be very difficult to have virions absolutely free of contaminant, we would expect the presence of minimal mRNA quantities within the viral particles to be detected. In fact, this occurred when highly sensitive techniques such as quantitative real time RT-PCR were used. Therefore, the conclusion that a given virus preparation is free of mRNA is a relative statement. However, it was found that while minigenomes are specifically encapsidated, both the viral and the cellular mRNAs were nonspecifically associated to the viral particle, since their presence in the virus preparations significantly decreased with the homogeneity of the preparation, in contrast to the presence of the viral genome or of Ψ-containing minigenomes that are encapsidated independently of the purification degree.
To locate the TGEV region that contained the Ψ, a positive approach was undertaken by introducing selected sequences from minigenome M33 into mRNA-GUS, which was not incorporated into virions. The location of TGEV Ψ in the leader and 3′ third of the TGEV genome was in principle discarded, since all virus mRNAs contain the leader and the 3′ end and were not specifically encapsidated. Nevertheless, a possible collaboration of the 3′ end with sequences located at the 5′ end of the genome cannot be ruled out. Since the mRNA-GUS containing the first 649 nt of TGEV genome (mRNA-A1-GUS) was specifically encapsidated, it was concluded that a major TGEV Ψ is located within the TGEV genome 5′ end. Surprisingly, the packaging signal of MHV and BCoV was located within ORF 1b about 20 kb from the genome 5′ end (Fig. 7). However, although the presence of this packaging signal located in ORF 1b was convincingly demonstrated for MHV and BCoV, it has also been reported that a defective minigenome (DI-RNA Drep), consisting of the BCoV genome 5′-terminal 498 nt, the N ORF (1,344 nt), and the 3′ genome end, was also efficiently rescued (8, 11). These results suggest that another cis-acting signal involved in BCoV RNA encapsidation must be located within the BCoV genome ends. Furthermore, DI-RNA sequences from either the 5′ or 3′ UTRs (in group 3 coronaviruses such as IBV) were required for minigenome incorporation into viral particles (12). The insertion of the MHV Ψ in nonreplicating RNAs led to encapsidation of these RNAs (6, 72). However, this encapsidation was estimated as inefficient (albeit it was never quantitatively determined), suggesting that other factors or sequences might be required for efficient encapsidation (6). It is, therefore, possible that group 2 coronavirus encapsidation could be mediated by two different RNA motifs, one located at the 5′ end of the genome and another at the end of ORF 1b. It would be interesting to quantify the extent of RNA encapsidation observed for MHV and BCoV mRNAs and DI-RNAs to determine which RNA motif represents the major packaging signal between the two potential encapsidation domains.
FIG. 7.
Location of coronavirus cis-acting packaging signals. A scheme of the TGEV genome indicating the viral ORFs (represented as boxes) is shown. The positions of TGEV, MHV, and BCoV packaging signals are indicated with arrows. Numbers below the genome bar indicate approximate virus genome nucleotide positions. Rep 1a, Rep 1b, S, 3a, 3b, E, M, N, and 7 indicate each of the TGEV ORFs.
When mRNA-A1-GUS was launched by transcription from the CMV promoter in the absence of a minigenome encoding this mRNA, it was rescued in TGEV-infected ST cells. These results implied that mRNA-A1-GUS was replicated by the TGEV helper virus, showing that the first 649 nt from the TGEV genome were sufficient for RNA replication and encapsidation. The requirement of the first 649 nt from the TGEV genome for minigenome replication is similar to that of the cis-acting RNA minimal replication sequences for MHV, BCoV, and IBV replication (12, 31, 40, 57). Since the region containing the TGEV Ψ has been located within the same sequence domain required for replication (27), it cannot be excluded at this time that RNA encapsidation in TGEV could be coupled to genome replication, as in the case of poliovirus (51) and flavivirus (32, 33). Additional work is being performed to determine whether TGEV RNA replication and encapsidation activities can be mapped within different sequence domains.
The encapsidation efficiency of mRNA-A1-GUS was around 30-fold higher than that of genome encapsidation, as determined by quantitative RT-PCR. The higher encapsidation efficiency of this mRNA could be due to its size, in that it is considerably smaller than the full-length genome. If both the full-length genome and the mRNA containing the A1 region are recognized by the same Ψ, mRNA-A1-GUS encapsidation might interfere with full-length genome encapsidation, explaining the reduced incorporation of genomic and minigenome RNAs into virions shown in Fig. 4.
Genome encapsidation in RNA viruses is a specific process that involves cis- and trans-acting factors. A packaging signal is usually required as a cis-acting signal that confers specificity for genome incorporation into virions. These signals have been identified in several RNA and DNA viruses (5, 13, 14, 18, 20, 24, 34, 42, 53, 56, 65, 75). Interestingly, independently of the virus species, most packaging signals have been located within genome ends (Table 5). In addition, trans-acting factors are also required for genome encapsidation, including binding of virus structural proteins and even host proteins to nucleocapsids. As an example, human immunodeficiency virus type 1 (HIV-1) genome encapsidation is mediated by interactions in cis and in trans of the nucleocapsid domain of Gag protein. In HIV-2 the packaging signal is located in the genome and also in mRNAs, but only genome RNA is encapsidated because encapsidation in HIV-2 is coupled to translation of the Gag protein (24). Furthermore, in hepatitis B virus, host cellular proteins binding to the Ψ modulate genome replication (67).
TABLE 5.
Locations of cis-acting packaging signals in virus species
| Virus species | Family | Genomea | Location of cis-acting packaging signals |
|---|---|---|---|
| Hepatitis B virus | Hepadnaviridae | dsDNA | 5′ End of pregenomic RNA |
| Adeno-associated virus | Parvoviridae | ssDNA | Inverted terminal repeats |
| Yeast L-A virus | Totiviridae | dsRNA | 3′ End of L-A (+) ssRNA strand |
| Influenza virus | Orthomyxoviridae | ssRNA (−) | 5′ Genome ends |
| Human parainfluenza virus-3 | Paramyxoviridae | ssRNA (−) | 5′ And 3′ extracistronic sequences |
| Rabies virus | Rhabdoviridae | ssRNA (−) | 3′ UTR |
| Vesicular stomatitis virus | Rhabdoviridae | ssRNA (−) | 5′ And 3′ UTRs |
| Bunyamwera virus | Bunyaviridae | ssRNA (−) | 5′ Genome terminal sequences |
| Hantavirus | Bunyaviridae | ssRNA (−) | 5′ Genome terminal sequences |
| HIV-1 and HIV-2 | Retroviridae | ssRNA (+) | 5′ Genome end |
| Poliovirus | Picornaviridae | ssRNA (+) | Several genome regions; coupled to replication |
| Flavivirus | Flaviviridae | ssRNA (+) | Several genome regions; coupled to replication |
| Brome mosaic virus | Bromoviridae | ssRNA (+) | 5′ And 3′ proximal regions of 3a ORF |
dsDNA, double-stranded DNA; ssDNA, single-stranded DNA; dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; −, negative strand; +, positive strand.
trans-acting factors could also be implicated in coronavirus encapsidation. The coronavirus genome is packed by the N protein, forming the nucleocapsid (10, 17, 50, 69). It has been shown that two-thirds of TGEV virion M protein specifically binds to the nucleocapsid by a 16-amino-acid peptide located within the carboxy terminus, leading to stabilization of the internal core (17). M protein-nucleocapsid binding has also been characterized in MHV-infected cells and could be responsible for nucleocapsid incorporation into budding virions (48, 49). Recently, genetic evidence revealed that MHV M protein binds to N protein, although a direct protein-protein interaction has not been shown (35). Whether coronavirus RNA encapsidation is mediated by interactions between M and N proteins or directly between the M protein and the Ψ is currently a subject of investigation. In addition, it has been demonstrated that host proteins specifically bind to the coronavirus genome (26, 39, 66, 68) and it is, therefore, also possible that host factors are also involved in coronavirus genome encapsidation.
In summary, a packaging signal has been located in the first 649 nt of TGEV genome in a domain absent from TGEV mRNAs. Identification and characterization of TGEV Ψ would allow the improvement in biosafety of a new generation of virus vectors based on the TGEV genome (52).
Acknowledgments
We thank Hubert Laude for providing M protein-specific MAb 25.22.
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT), La Consejería de Educación y Cultura de la Comunidad de Madrid, Fort Dodge Veterinaria, and the European Communities (Frame V, Key Action 2, Control of Infectious Disease Projects QLRT-1999-00002, QLRT-1999-30739, and QLRT-2000-00874). D.E. and A.I. received postdoctoral fellowships from the European Union (Frame V, Key Action 2, Control of Infectious Disease Projects). C.C. received a predoctoral fellowship from the Comunidad de Madrid.
REFERENCES
- 1.Adkins, B., and T. Hunter. 1981. Identification of a packaged cellular mRNA in virions of Rous sarcoma virus. J. Virol. 39:471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazán, F., J. M. González, Z. Pénzes, A. Izeta, E. Calvo, J. Plana-Durán, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An, S., A. Maeda, and S. Makino. 1998. Coronavirus transcription early in infection. J. Virol. 72:8517-8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bancroft, C. T., and T. G. Parslow. 2002. Evidence for segment-nonspecific packaging of the influenza A virus genome. J. Virol. 76:7133-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos, E. C. W., J. C. Dobbe, W. Luytjes, and W. J. M. Spaan. 1997. A subgenomic mRNA transcript of the coronavirus mouse hepatitis virus strain A59 defective interfering (DI) RNA is packaged when it contains the DI packaging signal. J. Virol. 71:5684-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, R. Y., and D. A. Brian. 1996. cis requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J. Virol. 70:2201-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charley, B., and H. Laude. 1988. Induction of alpha interferon by transmissible gastroenteritis coronavirus: role of transmembrane glycoprotein E1. J. Virol. 62:8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cologna, R., and B. Hogue. 1998. Coronavirus nucleocapsid protein: RNA interactions. Adv. Exp. Med. Biol. 440:355-360. [PubMed] [Google Scholar]
- 11.Cologna, R., and B. G. Hogue. 2000. Identification of a bovine coronavirus packaging signal. J. Virol. 74:580-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalton, K., R. Casais, K. Shaw, K. Stirrups, S. Evans, P. Britton, T. D. K. Brown, and D. Cavanagh. 2001. cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J. Virol. 75:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 2002. Positional effect of deletions on viability, especially on encapsidation, of Brome mosaic virus D-RNA in barley protoplasts. Virology 293:314-319. [DOI] [PubMed] [Google Scholar]
- 14.Dannull, J., A. Surovoy, G. Jung, and K. Moelling. 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 13:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enjuanes, L., D. Brian, D. Cavanagh, K. Holmes, M. M. C. Lai, H. Laude, P. Masters, P. Rottier, S. G. Siddell, W. J. M. Spaan, F. Taguchi, and P. Talbot. 2000. Coronaviridae, p. 835-849. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 16.Escors, D., E. Camafeita, J. Ortego, H. Laude, and L. Enjuanes. 2001. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. J. Virol. 75:12228-12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flodell, S., J. Cromsigt, J. Schleucher, K. Kidd-Ljunggren, and S. Wijmenga. 2002. Structure elucidation of the hepatitis B virus encapsidation signal by NMR on selectively labeled RNAs. J. Biomol. Struct. Dyn. 19:627-636. [DOI] [PubMed] [Google Scholar]
- 19.Fosmire, J. A., K. Hwang, and S. Makino. 1992. Identification and characterization of a coronavirus packaging signal. J. Virol. 66:3522-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimura, T., and R. Esteban. 2000. Recognition of RNA encapsidation signal by the yeast L-A double-strander RNA virus. J. Biol. Chem. 275:37118-37126. [DOI] [PubMed] [Google Scholar]
- 21.Fujimura, T., R. Esteban, L. M. Esteban, and R. B. Wickner. 1990. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell 62:819-828. [DOI] [PubMed] [Google Scholar]
- 22.Gebauer, F., W. A. P. Posthumus, I. Correa, C. Suñé, C. M. Sánchez, C. Smerdou, J. A. Lenstra, R. Meloen, and L. Enjuanes. 1991. Residues involved in the formation of the antigenic sites of the S protein of transmissible gastroenteritis coronavirus. Virology 183:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González, J. M., Z. Penzes, F. Almazán, E. Calvo, and L. Enjuanes. 2002. Stabilization of a full-length infectious cDNA clone of transmissible gastroenteritis coronavirus by the insertion of an intron. J. Virol. 76:4655-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin, S. D. C., J. F. Allen, and A. M. L. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann, M. A., P. B. Sethna, and D. A. Brian. 1990. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 64:4108-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, P., and M. M. C. Lai. 2001. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 75:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izeta, A., C. Smerdou, S. Alonso, Z. Penzes, A. Méndez, J. Plana-Durán, and L. Enjuanes. 1999. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 73:1535-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong, Y. S., and S. Makino. 1994. Evidence for coronavirus discontinuous transcription. J. Virol. 68:2615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jewell, N. A., and L. M. Mansky. 2000. In the beginning: genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 81:1889-1899. [DOI] [PubMed] [Google Scholar]
- 30.Jiménez, G., I. Correa, M. P. Melgosa, M. J. Bullido, and L. Enjuanes. 1986. Critical epitopes in transmissible gastroenteritis virus neutralization. J. Virol. 60:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joo, M., S. Banerjee, and S. Makino. 1996. Replication of murine coronavirus defective interfering RNA from negative-strand transcripts. J. Virol. 70:5769-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khromykh, A. A., A. N. Varnavski, P. L. Sedlak, and E. G. Westaway. 2001. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 75:4633-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouznetzoff, A., M. Buckle, and N. Tordo. 1998. Identification of a region of the rabies virus N protein involved in direct binding to the viral RNA. J. Gen. Virol. 79:1005-1013. [DOI] [PubMed] [Google Scholar]
- 35.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laude, H., J. Gelfi, L. Lavenant, and B. Charley. 1992. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J. Virol. 66:743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laude, H., K. Vanreeth, and M. Pensaert. 1993. Porcine respiratory coronavirus—molecular features and virus host interactions. Vet. Res. 24:125-150. [PubMed] [Google Scholar]
- 38.Levin, J. G., and J. G. Seidman. 1979. Selective packaging of host tRNA's by murine leukemia virus particles does not require genomic RNA. J. Virol. 29:328-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, H.-P., X. Zhang, R. Duncan, L. Comai, and M. M. C. Lai. 1997. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. USA 94:9544-9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luytjes, W., H. Gerritsma, and W. J. M. Spaan. 1996. Replication of synthetic defective interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology 216:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClurkin, A. W., and J. O. Norman. 1966. Studies on transmissible gastroenteritis of swine. II. Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can. J. Comp. Med. Vet. Sci. 30:190-198. [PMC free article] [PubMed] [Google Scholar]
- 42.McVoy, M. A., D. E. Nixon, J. K. Hur, and S. P. Adler. 2000. The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J. Virol. 74:1587-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motulsky, H. 1995. Comparing two paired groups: paired t and Wilcoxon tests, p. 225-229. Intuitive Biostatistics. Oxford University Press, New York, N.Y.
- 44.Mougel, M., and E. Barklis. 1997. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 71:8061-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mougel, M., Y. Zhang, and E. Barklis. 1996. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J. Virol. 70:5043-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narayanan, K., C.-J. Chen, J. Maeda, and S. Makino. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77:2922-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayanan, K., and S. Makino. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen, V.-P., and B. G. Hogue. 1997. Protein interactions during coronavirus assembly. J. Virol. 71:9278-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osborne, J. C., and R. M. Elliott. 2000. RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J. Virol. 74:9946-9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Penzes, Z., J. M. González, E. Calvo, A. Izeta, C. Smerdou, A. Mendez, C. M. Sánchez, I. Sola, F. Almazán, and L. Enjuanes. 2001. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes 23:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peters, G. G., and J. Hu. 1980. Reverse transcriptase as the major determinant for selective packaging of tRNA's into avian sarcoma virus particles. J. Virol. 36:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pirttimaa, M. J., and D. H. Bamford. 2000. RNA secondary structures of the bacteriophage phi6 packaging regions. RNA 6:880-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Repass, J. F., and S. Makino. 1998. Importance of the positive-strand RNA secondary structure of a murine coronavirus defective interfering RNA internal replication signal in positive-strand RNA synthesis. J. Virol. 72:7926-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Risco, C., I. M. Antón, L. Enjuanes, and J. L. Carrascosa. 1996. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 70:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rumenapf, T., D. T. Brown, E. G. Strauss, M. König, R. Ramirez-Mitchel, and J. H. Strauss. 1995. Aura alphavirus subgenomic RNA is packaged into virions of two sizes. J. Virol. 69:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rumenapf, T., E. G. Strauss, and J. H. Strauss. 1994. Subgenomic mRNA of Aura alphavirus is packaged into virions. J. Virol. 68:56-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sánchez, C. M., G. Jiménez, M. D. Laviada, I. Correa, C. Suñé, M. J. Bullido, F. Gebauer, C. Smerdou, P. Callebaut, J. M. Escribano, and L. Enjuanes. 1990. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology 174:410-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-220. [DOI] [PubMed] [Google Scholar]
- 63.Sethna, P. B., M. A. Hofmann, and D. A. Brian. 1991. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 65:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sethna, P. B., S.-L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Severson, W. E., X. Xu, and C. B. Jonsson. 2001. cis-acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi, S. T., P. Huang, H.-P. Li, and M. M. C. Lai. 2000. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 19:4701-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin, H. J., S. S. Kim, Y. H. Cho, S. G. Lee, and H. M. Rho. 2002. Host cell proteins binding to the encapsidation signal ɛ in hepatitis B virus RNA. Arch. Virol. 147:471-491. [DOI] [PubMed] [Google Scholar]
- 68.Spagnolo, J. F., and B. G. Hogue. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thiel, V., J. Herold, B. Schelle, and S. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 71.van der Most, R. G., P. J. Bredenbeek, and W. J. M. Spaan. 1991. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J. Virol. 65:3219-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woo, K., M. Joo, K. Narayanan, K. H. Kim, and S. Makino. 1997. Murine coronavirus packaging signal confers packaging to nonviral RNA. J. Virol. 71:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: the transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, C., C. S. Lee, and P. Guo. 1994. The proximate 5′ and 3′ ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage phi 29 DNA. Virology 201:77-85. [DOI] [PubMed] [Google Scholar]
- 76.Zhao, X., K. Shaw, and D. Cavanagh. 1993. Presence of subgenomic mRNAs in virions of coronavirus IBV. Virology 196:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]