Abstract
Kaposi's sarcoma (KS) and lymphoproliferative diseases induced by KS-associated herpesvirus (KSHV/human herpesvirus 8) cause substantial morbidity and mortality in human immunodeficiency virus-infected individuals. To understand KSHV biology it is useful to investigate closely related rhadinoviruses naturally occurring in nonhuman primates. Here we report evidence for a novel KSHV homolog in captive baboon species (Papio anubis and other). Using degenerate PCR we identified a novel rhadinovirus, PapRV2, that has substantial sequence identity to two essential KSHV genes, the viral polymerase and thymidylate synthase. A subset of animals exhibited detectable PapRV2 viral load in peripheral blood mononuclear cells. Extensive serological analysis of nearly 200 animals in the colony demonstrated that the majority carried cross-reacting antibodies that recognize KSHV or macaque rhadinovirus antigens. Seroreactivity increased with age, similar to the age-specific prevalence of KSHV in the human population. This establishes baboons as a novel resource to investigate rhadinovirus biology, which can be developed into an animal model system for KSHV-associated human diseases, vaccine development, and therapy evaluation.
Kaposi's sarcoma-associated herpesvirus (KSHV/human herpesvirus 8) is the causative agent of Kaposi's sarcoma (KS) and two lymphoproliferative diseases (primary effusion lymphoma and multicentric Castleman's disease) (reviewed in reference 5). While manifestations of KS are most severe in human immunodeficiency virus (HIV)-infected individuals (epidemic KS), KS also occurs in immunosuppressed organ transplant recipients (iatrogenic KS) and elderly Mediterranean men (classic KS) and is endemic in sub-Saharan Africa (endemic KS). In sub-Saharan African countries, where KS was already prevalent prior to the AIDS epidemic, KS has now become the most common malignancy (52).
Herpesviridae can be found in all vertebrates, from mice to elephants (47), including primates, birds, turtles, reptiles, and fish as well as invertebrates, such as oysters (19). Phylogenetic analysis shows that herpesvirus divergence closely paralleled mammalian and, in particular, primate speciation (1, 36). During this coevolution, the defining characteristics of herpesvirus biology, such as the ability to establish latency, host-polymerase-independent lytic replication and tropism were conserved. The Herpesviridae can be subdivided into the alpha, beta, and gamma branches, based upon sequence similarity and tissue tropism. Rhadinoviruses, of which KSHV is a member, belong to the gamma herpesviridae. These viruses establish latency in lymphocytes and are associated with lymphoproliferative diseases. Lymphocryptoviruses, including the prototypical member Epstein-Barr virus (EBV), display a similar tropism to rhadinoviruses and exhibit strong transforming abilities but are more distantly related members of the gamma herpesviridae. The strong evolutionary conservation among herpesviruses suggests that insights into fundamental mechanisms of herpesvirus biology can be gained from studying herpesviruses that infect nonhuman species.
Rhadinoviruses have been identified in both New World (platyrrhines) and Old World (catarrhines) primates (reviewed in reference 15). However, despite numerous attempts to date, only three rhadinoviruses have been isolated by in vitro culture techniques: herpesvirus saimiri (HVS) and herpesvirus ateles (HVA) from spider and squirrel monkeys, respectively, and rhesus rhadinovirus (RRV) from various macaque species (2, 16). Numerous other primate gammaherpesviruses have been identified either by testing animal sera for antibodies that cross-react with human herpesvirus glycoproteins or by amplifying sequences from conserved genes, such as the viral polymerase, using degenerate PCR (26, 28). These studies have yielded invaluable insights into rhadinovirus evolution. Hence, we applied a similar approach to search for rhadinoviruses endogenous to baboons. Investigations of homologous viruses constitute an important and attractive avenue to study virus-host interactions, the effect of targeted mutations in the context of the viral genome, antiviral agents and, ultimately, vaccine candidates.
Why investigate novel rhadinoviruses in baboons? Hunting for rhadinovirus homologs in Papio species offers a unique benefit: namely, the opportunity to develop a nonhuman primate animal model for the analysis of viral pathology and future development of vaccines. Baboons are among the very few primate species for which there are large and sustainable colonies available for biomedical research. Chimpanzees also harbor at least two rhadinovirus lineages (22, 26, 39), but they are not available for rhadinovirus or AIDS research. Additional rhadinoviruses have been found in many nonhuman primates (reviewed in reference 15), but these other species are mostly housed in zoos or captured in the wild and are not accessible for current biomedical research.
The many similarities between baboon physiology and human physiology have led to the use of baboons in a number of immunological and virological studies. (i) Human vaccines are routinely tested in baboons and appear to induce similar immune reactions (55). Hematological research in baboons is facilitated by the fact that human and baboon CD markers and immunoglobulin serotypes are sufficiently similar to allow the use of commercially available human reagents to study baboon hematology. (ii) Baboons are still under discussion as potential xenotransplant organ donors, since their organ sizes and major histocompatibility complex make-up are similar to those of humans (reviewed in reference 43). The potential for xenotransplantation has led to in-depth characterization of baboon immunology and virology, particularly baboon betaherpesviruses, which tend to reactivate upon transplantation and/or immunosuppression (4, 10, 11, 38). Several gammaherpesviruses, e.g., HVS (spider monkey) and MHV-68 (vole), are not known to cause disease in their natural host but may cause development of malignancies in other species (macaque and mouse, respectively). (iii) Finally, baboons have been investigated as models for AIDS and AIDS-associated infections. While baboon bone marrow and CD4 T cells are refractive to HIV type 1 (HIV-1) infection (7), HIV-2 and simian-human immunodeficiency virus readily infect baboons and cause AIDS-like symptoms (reviewed in reference 33). Importantly, in 1994 Levy and colleagues reported that HIV-2 inoculation was accompanied by the occurrence of retroperitoneal fibromatosis, which is considered a primate analog to KS, in some baboons (8). These factors prompted us to test the hypothesis that a KSHV homolog was present in Papio species.
Degenerate PCR identified a novel rhadinovirus in baboons.
Captive baboons (Papio anubis, Papio cyano/anubis, Papio hamadryas, Papio chacma) were housed at the University of Oklahoma Health Sciences Center (OUHSC). Initial consensus PCR studies were carried out on peripheral blood mononuclear cells (PBMCs) from eight animals either directly or following culture and stimulation in vitro for 5 days with 20 ng of 12-O-tetradecanoylphorbol-13-acetate (TPA)/ml as described elsewhere for human primary effusion lymphoma (37, 45). Subsequent studies on serological and viral DNA prevalence in the colony were conducted on samples collected during biannual tuberculosis screening from 178 animals in the colony.
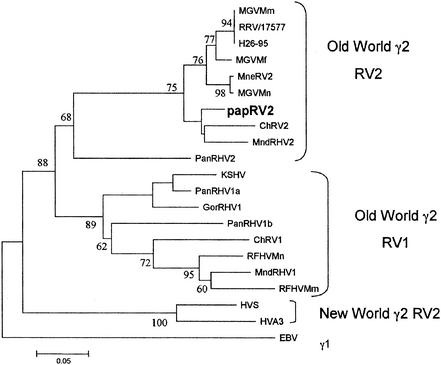
In order to identify a novel herpesvirus in the Papio species, we employed degenerate PCR (48). We targeted regions in the viral polymerase gene because these are highly conserved among all herpesviruses (alpha, beta, and gamma). Initially, a 172-bp pol sequence was amplified from TPA-stimulated PBMCs and found to have 83% identity to RRV and 64% identity to KSHV. A baboon-specific nested PCR primer was designed to extend the pol sequence (Table 1). Using a combination of degenerate and specific primers, a second pol sequence of 326 bp was amplified and found to have 84% identity to RRV and 70% identity to KSHV. The two overlapping pol sequences were then combined to construct a 475-bp fragment, the sequence of which shows 86% identity to RRV and 69% identity to KSHV. This 475-bp nucleotide sequence was translated into amino acids and aligned with herpesvirus pol sequences from other species using Clustal X1.81. Phylogenetic analysis included the protein distance, neighbor-joining, and maximum parsimony methods in MEGA 2.1. Bootstrap values were computed from 100 replicate samplings. A phylogenetic tree was constructed based upon amino acid similarity and is shown in Fig. 1. The baboon sequence clearly clusters with the RV2 group. It is most closely related to the RV2 viruses previously identified in mandrills and African green monkeys (23, 28). Of note, using the same pol primers described above, we also amplified a 179-bp EBV-like sequence from two of the baboon PBMC DNA samples. These sequences have 89% identity to human EBV, 69% identity to RRV, and 53% identity to KSHV.
TABLE 1.
PCR primers used in this study
| Primer | Gene region | 5′-3′ sequencea | Orientation |
|---|---|---|---|
| GDTD1B | Polymerase | CGGCATGCGACAAACACGGAGTCNGTRTCNCCRTA | − outer |
| DFASA | Polymerase | GTGTTCGACTTYGCNAGYYTNTAYCC | + outer |
| VYGA | Polymerase | ACGTGCAACGCGGTGTAYGGNKTNACNGG | + inner |
| PapRV2 S1 | Polymerase | GTCACGGTTTCCGCTATGTT | − inner |
| PapRV2 S2 | Polymerase | ACGGTTTCCGCTATGTTGAT | − inner |
| TS1A | TS | GAGYTGCTKTGGTTTMTSARGGG | + outer |
| TS1B | TS | CCCATRTCDSCBGAYCTCTG | − outer |
| TS2A | TS | GGNTTCCAGTGGAGRCAYTTYGG | + inner |
| TS2B | TS | TGRTACAGCTGRCAGGAMAGC | − inner |
| PapRV2 pol 1-f | Polymerase | GAGCATCATCCAGGCACACA | + real time |
| PapRV2 pol 1-r | Polymerase | GTGCAGGTGTAGGTCCCCC | − real time |
| PapRV2 pol 2-f | Polymerase | CTAGACAAACAGCAGCTGGCC | + real time |
| PapRV2 pol 2-r | Polymerase | ACCCCGGTGAACCCGT | − real time |
| PapRV2 TS-f | TS | TGACGATGCGCCGATC | + real time |
| PapRV2 TS-r | TS | AGCTGCGATACGTGATGGA | − real time |
| CMVpap gB-f | gB | CACAGAGCGGTAAGGTTGGC | + real time |
| CMVpap gB-r | gB | TACTCGTTCTCCTCCACCGG | − real time |
| SV40-f | Large T antigen | GTCTTCTACCTTTCTCTTCTTT | + real time |
| SV40-r | Large T antigen | GGAGCAGTGGTGGAA | − real time |
| Chymase pap-f | Chymase | CCTTCCCATCCCAATTCAACT | + real time |
| Chymase pap-r | Chymase | CTGTTCTTCCCCAGCCAG | − real time |
R = A/G, Y = C/T, M = A/C, K = G/T, S = C/G, B = C/G/T, N = A/C/G/T.
FIG. 1.
The deduced 158-amino-acid sequence of the baboon herpesvirus DNA polymerase gene was aligned with the sequences listed below using ClustalX version 1.81 and then analyzed using the protein distance and neighbor-joining methods in MEGA 2.1. Shown is the consensus dendrogram tree computed from 100 bootstrap iterations. Similar results were seen using maximum parsimony analysis including 100 bootstrap replicates. The sequences used were KSHV U75698, PanRHV1a AF250881, PanRHV1b AF250881, MndRHV2, AF282937, HVS M31122, HVA3 AF083424, GorRHV1 AF250886, MndRHV1, AF282943, ChRV1 AF029302, ChRV2 AJ251574, RFHVMn AF005478, RFHVMm AF005479, RRV/17577 AF083501, MneRV2 AF204167, H26-95 AF029302, MGVMf AF159032, MGVMm AF159033, MGVMn AF159031, PanRHV2 AAL06318, and EBV V01555.
The region covered by the pol primers will yield different length amplicons for different herpesvirus subgroups, e.g., all cytomegaloviruses (CMVs) have a large insert in that region of the polymerase gene which, by size of the amplified fragment, sets them apart from the gammaherpesviruses. In our strategy, we selected PCR products corresponding to the predicted size for gammaherpesviruses. Furthermore, the original pol primers were geared towards gamma viruses by choosing gammaherpesvirus-specific nucleotides at the 3′ end (48). To confirm the finding of a new rhadinovirus when using pol primers, degenerate thymidylate synthase (TS) primers were used to amplify a 230-bp sequence. We chose the TS as a second target, since this gene is not present in alpha, beta, or gamma 1 herpesviruses and, therefore, is specific for rhadinoviruses. Sequences obtained from two baboons proved to be 99% identical to each other. The consensus sequences from both animals had 88% identity to RRV and 71% identity to KSHV at the nucleotide level and 93 and 76% identity at the amino acid level. It is of note that of the five amplified products for pol and TS, four were obtained from short-term TPA-stimulated PBMCs, suggesting that this step sufficiently increased the viral load to enable detection.
Prevalence of PapRV2 sequences in baboons.
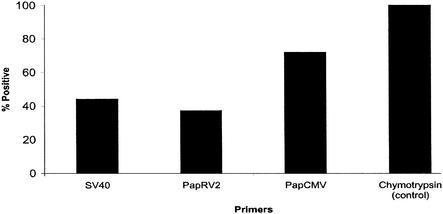
To confirm our degenerate PCR and to identify animals with detectable viral load, we designed a highly specific quantitative real-time PCR assay. We were able to amplify PapRV2 sequences from 35 out of 100 animals by using the primers PapRV2 TS-f and PapRV2 TS-r. Similar data were obtained using two independent sets of pol-specific primers (see Table 1 for primer sequences) (data not shown). All animals tested positive for baboon chymotrypsin, a housekeeping gene. A summary of the prevalence of viral sequences by real-time PCR is shown in Fig. 2. The real-time, quantitative DNA PCR conditions used an annealing temperature of 60°C, ensuring high specificity of the assay. All data presented here were derived using SYBR green as the method of detection, as previously described (18). Under these conditions, the PapRV2 primers did not yield any signal from nontemplate control, human CMV, or KSHV-positive BCBL-1 cell DNA (data not shown).
FIG. 2.
Bar graph showing the prevalence of PapRV2 in PBMCs from 100 baboons by using TaqMan screening with the indicated primers. Animals were counted positive if the real-time quantitative DNA-PCR signal of 106 cells was significant and a band was visible on an agarose gel. For comparison, the prevalence of SV40 and BaCMV is also reported. All animals were PCR positive for baboon chymotrypsin.
Baboons also harbor an endogenous cytomegalovirus (BaCMV) as well as simian virus 40 (SV40) (51). To investigate a possible correlation between gamma- and betaherpesvirus reactivation and SV40 prevalence, we determined the relative viral load in PBMCs for each virus by real-time quantitative PCR (see Table 1 for primers). In this assay total DNA was isolated directly from freshly drawn blood, without TPA induction and culture. The signal of the baboon-specific chymotrypsin gene was used to normalize input DNA, and viral load was calculated as the log10 relative differences (dCT) as previously described (18). This allows for a more robust statistical comparison at the lower limit of assay sensitivity. Detectable levels of PapRV2 correlated with PCR positivity for BaCMV (Pearson coefficient r = 0.90; n = 78), suggesting that both herpesviruses reactivated in response to a common stressor. In contrast, levels of PapRV2 did not correlate with SV40 PCR positivity (Pearson coefficient r = 0.25; n = 36).
Several independent PapRV2 PCR products were cloned and sequenced. All showed 100% identity to the target PapRV2 sequence identified by consensus PCR. Using either consensus or real-time PCR, we have now independently amplified, cloned, and sequenced the PapRV2 polymerase or the PapRV2 TS fragment from 14 animals.
Serology of PapRV2 in baboons.
Antibodies to KSHV antigens were not detected in any animal using immunofluorescence assay (IFA) methods (25), even though most baboon sera cross-reacted with human EBV-infected cells in a commercial EBV IFA (data not shown). Therefore, we developed rhadinovirus-specific enzyme-linked immunosorbent assays (ELISAs). RRV peptides for ELISAs were designed from the published amino acid sequences of RRV open reading frame (ORF) 65 (small capsomer interacting protein [SCIP]; PSDATTLDTRRSSQNKKSK; at 0.5 μg/ml), ORF 73 (latency-associated nuclear antigen [LANA], MWGSRQHRSGIVSGHGLRSSCRGHC and LGSPTPSPSGSAPVLASGLSPQNTS; at 0.5 μg/ml) and R8.1 (R-gp8.1, PSVRGPHAIPSSPFAIGTRSRP and FKPDPRLTVRVYPLRPERWQDTPSREQY; at 0.5 μg/ml) using antigenicity plots performed with Seqweb. These genes have been shown to be the most important KSHV antigens. KSHV peptides that had been previously reported (40, 41, 53) from corresponding gene regions were also evaluated for comparison. A panel of sera from 100 captive rhesus macaques was used to determine which peptides were most useful for larger-scale screening and to identify suitable positive and negative controls (data not shown). Further optimization of selected peptides was carried out to determine optimal peptide coating concentrations and assay cutoffs. KSHV ELISAs were performed using recombinant SCIP/ORF 65, K-gp8.1, and LANA/ORF 73. The SCIP/ORF 65 and K-gp8.1 assays have been previously described (17). The LANA/ORF 73 ELISA was performed using full-length baculovirus-expressed protein using essentially the same protocol. Sera were diluted 1:20 for the SCIP/ORF 65 and K-gp8.1 assays and 1:100 for the LANA/ORF 73 assays. Sera were considered to be positive for antibodies to RRV if they reacted with any of the peptides used. Similarly, sera were considered to be positive for antibodies to KSHV if they reacted to any of the KSHV peptides or recombinant proteins.
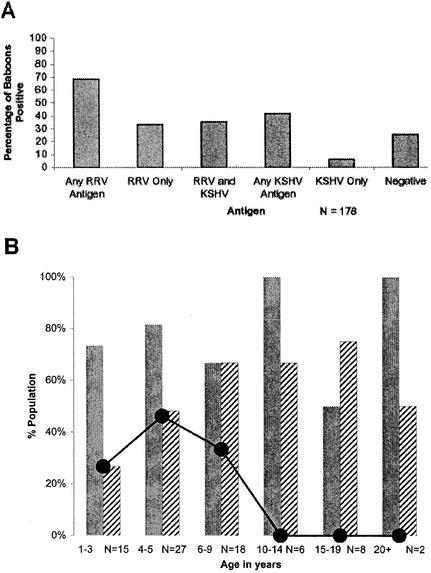
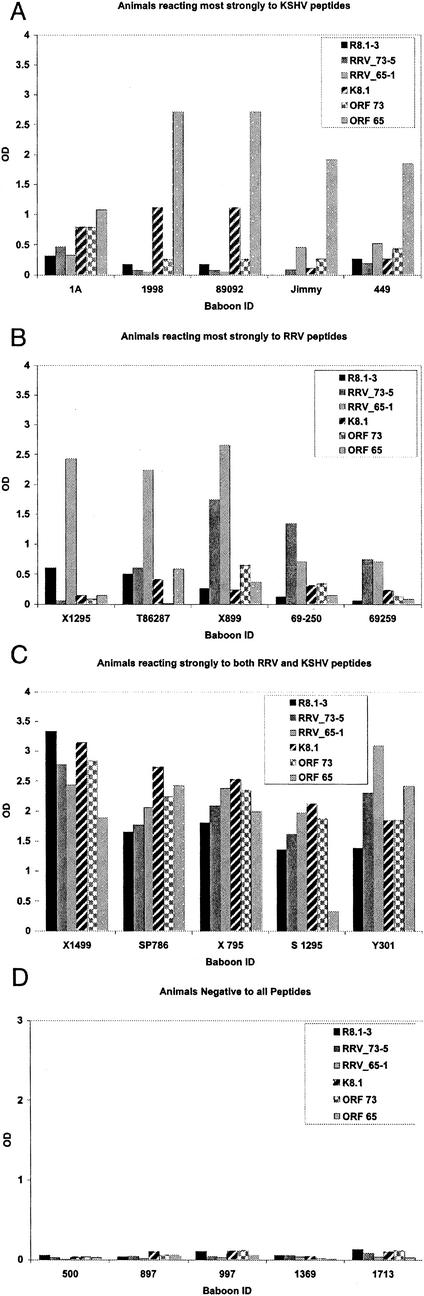
Using these novel ELISAs, antibodies to RRV antigens were detected more frequently than were antibodies to KSHV antigens (68.5% compared to 41.5%), and this difference was statistically significant (P = 0.001). Of those animals with antibodies to RRV antigens, 52% also had antibodies to KSHV antigens, while 48% had antibodies only to RRV antigens. Animals with antibodies to only KSHV antigens were rarely found: 85% of animals with antibodies to KSHV antigens also exhibited cross-reactivity to RRV antigens. A chi-square test of independence was run, and it indicated that KSHV seroreactivity is significantly more likely for those animals with RRV seroreactivity than for those not seroreactive for RRV (P < 0.001). A summary of antibody prevalence data is shown in Fig. 3A. The prevalence of antibodies to both RRV antigens and KSHV antigens increased with age, and there were no animals more than 10 years old who were seronegative for both (Fig. 3B). Logistic regression analyses were run, and the results indicated that the probability of reactivity to KSHV peptides increased significantly with age (P = 0.044), but the probability of reactivity to RRV peptides did not increase significantly by age (P = 0.85). This conclusion, however, must be viewed with caution, since accurate age was available for only 43% of animals. Figure 4 shows peptide reactivity profiles for individual baboons that reacted most strongly to KSHV peptides (Fig. 4A) or RRV peptides (Fig. 4B), reacted strongly to peptides from both viruses (Fig. 4C), or showed no reactivity to any of the peptides tested (Fig. 4D).
FIG. 3.
(A) Bar graph showing prevalence of antibodies to KSHV and RRV in 178 baboons. Sera from baboons were collected at a single time point and screened by ELISA. (B) Bar graph showing age-specific prevalence of antibodies to KSHV and RRV in 78 baboons for which accurate age information was available. Solid bars indicate percent reactivity to any RRV-derived peptide, and hatched bars indicate percent reactivity to any KSHV peptide. The dotted line represents the percentage of animals which had no reactivity to peptides derived from either virus. By age 10 to 14, all animals seroconverted.
FIG. 4.
Peptide ELISA reactivity profiles for 20 individual baboons. The optical density is shown on the y axes, the animal identi-fication number or name is shown on the x axes, and the reactivities to specific peptides are as shown in each corresponding legend. (A) Profiles of five animals with reactivity primarily to KSHV peptides; (B) profiles of five animals with reactivity mostly to RRV peptides; (C) profiles of five animals that reacted strongly against both KSHV and RRV peptides; (D) five animals that did not react to any of the KSHV or RRV peptides tested.
We were unable to plaque purify any baboon rhadinovirus by using the methods described. However, we routinely isolated various foamy viruses, BaCMV (betaherpesvirus), and even a trypanozoan from one animal. Presumably, those species grew much faster on fibroblasts and may have overgrown any potential gammaherpesvirus that could have been isolated otherwise. We also established several intermediate-term (4-week) and one permanent lymphoblastoid cell line from our baboon colony, but while the permanent cell line carried a baboon lymphocryptovirus (as expected from earlier reports [14]), it was PCR negative for rhadinovirus sequences.
Discussion.
In this study, we report the identification of a novel rhadinovirus in baboon species. Using degenerate PCR and, subsequently, specific real-time quantitative PCR, sequence information was obtained for fragments of two viral genes, pol and TS. Importantly, the same viral sequence was obtained from several baboons. Phylogenetic analysis showed that this novel baboon rhadinovirus belonged to the RV2 lineage, so we consequently named this virus PapRV2. Since only 750 discontinuous nucleotides were available for sequence comparison, this cautions against too-far-reaching interpretations of our phylogenetic analysis. We assume that both pol and TS sequences stem from the same RV2 virus species. However, until we have amplified the intervening sequence between the TS and pol sequences, we cannot exclude the theoretical possibility that there may be two RV2 isolates present in the same animal (one that amplifies with pol primers but not TS primers, and one which amplifies with TS but not pol primers). Our serological survey suggests the majority of animals in the OUHSC Baboon Facility may be infected with PapRV2. The serological data are corroborated by real-time quantitative PCR results, since almost one-third of the animals had detectable viral load in PBMCs, which indicated viral reactivation. However, no overt symptoms were associated with this level of viremia.
A comparison with other rhadinoviruses sets our results in perspective. Based on sequence comparison, two divergent lineages of primate rhadinoviruses, RV1 and RV2, have been identified in macaques, African green monkeys, and mandrills (49, 54). In addition, RV1 viruses have been identified in chimpanzees and gorillas (21-23). In chimpanzees, two distinct RV1 viruses and a more distantly related lineage that may represent an RV2 virus have been reported (27). RV1 and RV2 sequences have been discovered in every macaque species tested, and two isolates, RV2 mmu2695 (16) and RV2 mmu17577 (50), have been sequenced in entirety and replicate to high titers in culture. Thus far, no macaque RV1 isolate has been grown in culture. RV2mac viruses can infect related macaque species upon experimental inoculation (35, 57), although limited sequence comparison showed that RV2mac clusters by species of origin rather than facility (3, 6, 49, 54) This suggests that cross-species transmission of rhadinoviruses is rare even in captive colonies. The Oklahoma Baboon Research Resource houses three species of baboons, presently P. anubis (93 animals) and P. cyano/anubis (14 animals), with a few animals of the species P. chacma (7 animals) and P. hemandyas (3 animals). Currently, we do not have enough sequence information to distinguish rhadinoviruses of different baboon species, since all of our sequence data were derived from Papio anubis. It is not clear, however, whether such an endeavor would be informative, since most of the baboon species interbreed and produce fertile offspring, indicating that they may represent only subspecies.
To assess the prevalence of rhadinoviruses in the OUHSC baboon colony, we used serological assays based on RRV, a macaque RV2 virus, and KSHV, a human RV1 virus, to detect cross-reactive antibodies in baboons. Assuming that antibodies to RRV antigens indicate infection with a baboon RV2 virus and that antibodies to KSHV antigens indicate infection with a baboon RV1 virus, we can speculate on the existence of a second RV1-like virus in baboons. Alternatively, we may not fully appreciate the complex nature of antibody cross-reactivity to antigens of these closely related viruses. Cross-reactivity is likely to occur between RV1 and RV2 antigens, especially for highly homologous antigens such as SCIP/ORF 65. The development of specific antibody assays for the baboon viruses will be needed for more-definitive studies. Prevalence estimates using the RRV and KSHV assays are likely to underestimate the true prevalence of either agent in the baboon colony, since the sensitivity of these assays for the detection of cross-reactive antibodies in another species is undoubtedly lower than those for baboon RV1- and RV2-specific assays would be.
Over 90% of the animals at the New England Regional Primate Research Center and the Oregon Regional Primate Research Center are seropositive for RRV, the macaque RV2 virus (16, 35, 57). Transmission is thought to occur by close contact between adults and infants, since hand-rearing of juveniles prevents seroconversion (35). Mother-to-child transmission of KSHV, likewise, is implicated in areas where KS is endemic (13, 42). Studies on macaque RV1 prevalence and transmission are hampered by lack of a good serological assay and the insensitivity of available PCR assays (12, 49, 54; M. L. Bosch, K. B. Strand, and T. M. Rose, Letter, J. Virol. 72:8458-8459, 1998); thus, the seroprevalence and routes of transmission for this virus are largely unknown.
Overall seroreactivity in the OUHSC baboon colony increased with age, suggesting a mode of transmission from adult to infant similar to that of KSHV. However, it should be noted that our analysis is based on a one-time cross-sectional sampling and that accurate age was available for only 43% of animals. Longitudinal observations on selected animals are currently in progress. Baboons also carry BaCMV (11) and another gammaherpesvirus, herpesvirus papio, an EBV homolog. A comparison between the seroconversion rates for PapRV2 (this study), BaCMV (11), and herpesvirus papio (24) suggests that the natural history of herpesviruses in baboons is similar to that of herpesviruses in humans, namely, that the likelihood for seroconversion in animals or humans increases as they reach sexual maturity and the viruses reactivate in response to stress and/or immunosuppression.
We did not find any pathology associated with PapRV2 in the OUHSC colony. This is not surprising, since none of the animals were immune compromised, a prerequisite for the development of most gammaherpesvirus diseases in their native hosts (29, 56). For instance, macaques infected with the RV1 virus RFHV only developed retroperitoneal fibromatosis when coinfected with the immunosuppressive retrovirus SRV2 (49), and experimental infections with RRV isolates requires SIVmac239 coinfection to elicit viral reactivation (12) and lymphoproliferative diseases (57). If macaques are experimentally infected with SIVmac, most animals will succumb to opportunistic infections and SIVmac-induced lymphadenopathy before the endogenous rhadinovirus can reactivate. Hence, lymphomas are rare in captive animals coinfected with SIV (9, 20, 30, 34, 35, 44, 46).
Barnett et al. reported that induction of KS-like lesions in baboons resulted from HIV-2 strain HIV-2UC2 infection (8). HIV-2 was used since, unlike SIV or HIV-1, HIV-2 isolates induce severe immunosuppression and persistent infection in baboons (31, 32). The KS-like lesions, therefore, likely resulted from reactivation of endogenous papio rhadinoviruses as a result of HIV-2-induced immunosuppression. We have identified baboons with high PapRV2 viral loads as well as animals with high antibody titers in the colony. This will allow us to conduct more-targeted molecular and longitudinal epidemiologic studies as well as infection studies. Thus far, only two animals in the OUHSC baboon colony tested seropositive for SIV (another two animals are seropositive for simian retrovirus), and these animals are being closely monitored. The finding of rhadinoviruses in baboons and the development of appropriate diagnostic tools indicate that baboons could be a promising new model for vaccine studies and AIDS-associated malignancies.
Nucleotide sequence accession numbers.
The 475-bp pol nucleotide sequence and the 230-bp TS nucleotide sequence of PapRV2 have been deposited in GenBank under accession numbers AY270026 and AY270027, respectively.
Acknowledgments
We thank Rebecca Hines-Boykin for invaluable technical help and Doug Powell for statistical expertise. We also thank Larry Arthur, Jeff Lifson, B. Damania, and E. Mocarski for providing reagents and B. Damania for critical reading.
This work was supported by NIH grants RR1555777, EB53309, and CA97951 to D.P.D. and RR12317-04 to G.W. and R.K. and a Pew Scholar Award to D.H.K. J.P. is supported by NIH training grant AI07364 to the OUHSC Department of Microbiology and Immunology. This project has been funded in part with federal funds from the National Cancer Institute and the National Institutes of Health under contract no. N01-CO-12400.
REFERENCES
- 1.Alba, M. M., R. Das, C. A. Orengo, and P. Kellam. 2001. Genomewide function conservation and phylogeny in the Herpesviridae. Genome Res. 11:43-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allan, J. S., S. R. Broussard, M. G. Michaels, T. E. Starzl, K. L. Leighton, E. M. Whitehead, A. G. Comuzzie, R. E. Lanford, M. M. Leland, W. M. Switzer, and W. Heneine. 1998. Amplification of simian retroviral sequences from human recipients of baboon liver transplants. AIDS Res. Hum. Retrovir. 14:821-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antman, K., and Y. Chang. 2000. Kaposi's sarcoma. N. Engl. J. Med. 342:1027-1038. [DOI] [PubMed] [Google Scholar]
- 6.Auerbach, M. R., S. C. Czajak, W. E. Johnson, R. C. Desrosiers, and L. Alexander. 2000. Species specificity of macaque rhadinovirus glycoprotein B sequences. J. Virol. 74:584-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker, R. 1995. FDA approves baboon bone marrow transplant for AIDS patient. Food and Drug Administration. BETA 1995:7-8. [PubMed]
- 8.Barnett, S. W., K. K. Murthy, B. G. Herndier, and J. A. Levy. 1994. An AIDS-like condition induced in baboons by HIV-2. Science 266:642-646. [DOI] [PubMed] [Google Scholar]
- 9.Baskin, G. B., K. J. Cremer, and L. S. Levy. 2001. Comparative pathobiology of HIV- and SIV-associated lymphoma. AIDS Res. Hum. Retrovir. 17:745-751. [DOI] [PubMed] [Google Scholar]
- 10.Blewett, E. L., D. H. Black, N. W. Lerche, G. White, and R. Eberle. 2000. Simian foamy virus infections in a baboon breeding colony. Virology 278:183-193. [DOI] [PubMed] [Google Scholar]
- 11.Blewett, E. L., G. White, J. T. Saliki, and R. Eberle. 2001. Isolation and characterization of an endogenous cytomegalovirus (BaCMV) from baboons. Arch. Virol. 146:1723-1738. [DOI] [PubMed] [Google Scholar]
- 12.Bosch, M. L., E. Harper, A. Schmidt, K. B. Strand, S. Thormahlen, M. E. Thouless, and Y. Wang. 1999. Activation in vivo of retroperitoneal fibromatosis-associated herpesvirus, a simian homologue of human herpesvirus-8. J. Gen. Virol. 80:467-475. [DOI] [PubMed] [Google Scholar]
- 13.Bourboulia, D., D. Whitby, C. Boshoff, R. Newton, V. Beral, H. Carrara, A. Lane, and F. Sitas. 1998. Serologic evidence for mother-to-child transmission of Kaposi sarcoma-associated herpesvirus infection. JAMA 280:31-32. [DOI] [PubMed] [Google Scholar]
- 14.Cho, Y. G., A. V. Gordadze, P. D. Ling, and F. Wang. 1999. Evolution of two types of rhesus lymphocryptovirus similar to type 1 and type 2 Epstein-Barr virus. J. Virol. 73:9206-9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damania, B., and R. C. Desrosiers. 2001. Simian homologues of human herpesvirus 8. Philos. Trans. R. Soc. Lond. B 356:535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engels, E. A., D. Whitby, P. B. Goebel, A. Stossel, D. Waters, A. Pintus, L. Contu, R. J. Biggar, and J. J. Goedert. 2000. Identifying human herpesvirus 8 infection: performance characteristics of serologic assays. J. Acquir. Immune Defic. Syndr. 23:346-354. [DOI] [PubMed] [Google Scholar]
- 18.Fakhari, F. D., and D. P. Dittmer. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 76:6213-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farley, C. A., W. G. Banfield, G. Kasnic, Jr., and W. S. Foster. 1972. Oyster herpes-type virus. Science 178:759-760. [DOI] [PubMed] [Google Scholar]
- 20.Fortgang, I. S., P. J. Didier, and L. S. Levy. 2000. B-cell leukemia in a rhesus macaque (Macaca mulatta) infected with simian immunodeficiency virus. Leuk. Lymphoma 37:657-662. [DOI] [PubMed] [Google Scholar]
- 21.Greensill, J., and T. F. Schulz. 2000. Rhadinoviruses (gamma2-herpesviruses) of Old World primates: models for KSHV/HHV8-associated disease? AIDS 14(Suppl. 3):S11-S19. [PubMed] [Google Scholar]
- 22.Greensill, J., J. A. Sheldon, K. K. Murthy, J. S. Bessonette, B. E. Beer, and T. F. Schulz. 2000. A chimpanzee rhadinovirus sequence related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8: increased detection after HIV-1 infection in the absence of disease. AIDS 14:F129-F135. [DOI] [PubMed] [Google Scholar]
- 23.Greensill, J., J. A. Sheldon, N. M. Renwick, B. E. Beer, S. Norley, J. Goudsmit, and T. F. Schulz. 2000. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among Old World primates? J. Virol. 74:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenson, H. B., Y. Ench, S. J. Gao, K. Rice, D. Carey, R. C. Kennedy, J. R. Arrand, and M. Mackett. 2000. Epidemiology of herpesvirus papio infection in a large captive baboon colony: similarities to Epstein-Barr virus infection in humans. J. Infect. Dis. 181:1462-1466. [DOI] [PubMed] [Google Scholar]
- 25.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. (Erratum, 2:1041.) [DOI] [PubMed]
- 26.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, and A. Gessain. 2000. KSHV-like herpesviruses in chimps and gorillas. Nature 407:151-152. [DOI] [PubMed] [Google Scholar]
- 27.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, and A. Gessain. 2001. A novel gamma 2-herpesvirus of the rhadinovirus 2 lineage in chimpanzees. Genome Res. 11:1511-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, J. Rigoulet, T. Petit, and A. Gessain. 2000. Simian homologues of human gamma-2 and betaherpesviruses in mandrill and drill monkeys. J. Virol. 74:11993-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lennette, E. T., D. J. Blackbourn, and J. A. Levy. 1996. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet 348:858-861. [DOI] [PubMed] [Google Scholar]
- 30.Li, S. L., E. E. Kaaya, C. Ordonez, M. Ekman, H. Feichtinger, P. Putkonen, D. Bottiger, G. Biberfeld, and P. Biberfeld. 1995. Thymic immunopathology and progression of SIVsm infection in cynomolgus monkeys. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:1-10. [PubMed] [Google Scholar]
- 31.Locher, C. P., S. W. Barnett, B. G. Herndier, D. J. Blackbourn, G. Reyes-Teran, K. K. Murthy, K. M. Brasky, G. B. Hubbard, T. A. Reinhart, A. T. Haase, and J. A. Levy. 1998. Human immunodeficiency virus-2 infection in baboons is an animal model for human immunodeficiency virus pathogenesis in humans. Arch. Pathol. Lab. Med. 122:523-533. [PubMed] [Google Scholar]
- 32.Locher, C. P., D. J. Blackbourn, B. G. Herndier, G. Reyes-Teran, S. W. Barnett, K. K. Murthy, and J. A. Levy. 1998. Transient virus infection and pathogenesis of a new HIV type 2 isolate, UC12, in baboons. AIDS Res. Hum. Retrovir. 14:79-82. [DOI] [PubMed] [Google Scholar]
- 33.Locher, C. P., S. A. Witt, B. G. Herndier, K. Tenner-Racz, P. Racz, and J. A. Levy. 2001. Baboons as an animal model for human immunodeficiency virus pathogenesis and vaccine development. Immunol. Rev. 183:127-140. [DOI] [PubMed] [Google Scholar]
- 34.Maggiorella, M. T., F. Monardo, M. L. Koanga-Mogtomo, L. Cioe, L. Sernicola, F. Corrias, C. D. Baroni, P. Verani, and F. Titti. 1998. Detection of infectious simian immunodeficiency virus in B- and T-cell lymphomas of experimentally infected macaques. Blood 91:3103-3111. [PubMed] [Google Scholar]
- 35.Mansfield, K. G., S. V. Westmoreland, C. D. DeBakker, S. Czajak, A. A. Lackner, and R. C. Desrosiers. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J. Virol. 73:10320-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mesri, E. A., E. Cesarman, L. Arvanitakis, S. Rafii, M. A. Moore, D. N. Posnett, D. M. Knowles, and A. S. Asch. 1996. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J. Exp. Med. 183:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaels, M. G., F. J. Jenkins, K. St. George, M. A. Nalesnik, T. E. Starzl, and C. R. Rinaldo, Jr. 2001. Detection of infectious baboon cytomegalovirus after baboon-to-human liver xenotransplantation. J. Virol. 75:2825-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nath, B. M., K. E. Schumann, and J. D. Boyer. 2000. The chimpanzee and other non-human-primate models in HIV-1 vaccine research. Trends Microbiol. 8:426-431. [DOI] [PubMed] [Google Scholar]
- 40.Olsen, S. J., R. Sarid, Y. Chang, and P. S. Moore. 2000. Evaluation of the latency-associated nuclear antigen (ORF73) of Kaposi's sarcoma-associated herpesvirus by peptide mapping and bacterially expressed recombinant Western blot assay. J. Infect. Dis. 182:306-310. [DOI] [PubMed] [Google Scholar]
- 41.Pau, C. P., L. L. Lam, T. J. Spira, J. B. Black, J. A. Stewart, P. E. Pellett, and R. A. Respess. 1998. Mapping and serodiagnostic application of a dominant epitope within the human herpesvirus 8 ORF 65-encoded protein. J. Clin. Microbiol. 36:1574-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plancoulaine, S., L. Abel, M. van Beveren, D. A. Tregouet, M. Joubert, P. Tortevoye, G. de The, and A. Gessain. 2000. Human herpesvirus 8 transmission from mother to child and between siblings in an endemic population. Lancet 356:1062-1065. [DOI] [PubMed] [Google Scholar]
- 43.Platt, J. L. (ed.). 2001. Xenotransplantation. ASM Press, Washington D.C.
- 44.Putkonen, P., E. E. Kaaya, D. Bottiger, S. L. Li, C. Nilsson, P. Biberfeld, and G. Biberfeld. 1992. Clinical features and predictive markers of disease progression in cynomolgus monkeys experimentally infected with simian immunodeficiency virus. AIDS 6:257-263. [DOI] [PubMed] [Google Scholar]
- 45.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 46.Rezikyan, S., E. E. Kaaya, M. Ekman, A. F. Voevodin, H. Feichtinger, P. Putkonen, E. Castanos-Velez, G. Biberfeld, and P. Biberfeld. 1995. B-cell lymphomagenesis in SIV-immunosuppressed cynomolgus monkeys. Int. J. Cancer 61:574-579. [DOI] [PubMed] [Google Scholar]
- 47.Richman, L. K., R. J. Montali, R. L. Garber, M. A. Kennedy, J. Lehnhardt, T. Hildebrandt, D. Schmitt, D. Hardy, D. J. Alcendor, and G. S. Hayward. 1999. Novel endotheliotropic herpesviruses fatal for Asian and African elephants. Science 283:1171-1176. [DOI] [PubMed] [Google Scholar]
- 48.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose, T. M., K. B. Strand, E. R. Schultz, G. Schaefer, G. W. Rankin, Jr., M. E. Thouless, C. C. Tsai, and M. L. Bosch. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 71:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi, L., J. Ho, L. A. Norling, M. Roy, and Y. Xu. 1999. A real time quantitative PCR-based method for the detection and quantification of simian virus 40. Biologicals 27:241-252. [DOI] [PubMed] [Google Scholar]
- 52.Sitas, F., and R. Newton. 2001. Kaposi's sarcoma in South Africa. J. Natl. Cancer Inst. Monogr. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 53.Spira, T. J., L. Lam, S. C. Dollard, Y. X. Meng, C. P. Pau, J. B. Black, D. Burns, B. Cooper, M. Hamid, J. Huong, K. Kite-Powell, and P. E. Pellett. 2000. Comparison of serologic assays and PCR for diagnosis of human herpesvirus 8 infection. J. Clin. Microbiol. 38:2174-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strand, K., E. Harper, S. Thormahlen, M. E. Thouless, C. Tsai, T. Rose, and M. L. Bosch. 2000. Two distinct lineages of macaque gamma herpesviruses related to the Kaposi's sarcoma associated herpesvirus. J. Clin. Virol. 16:253-269. [DOI] [PubMed] [Google Scholar]
- 55.Watts, A. M., J. R. Stanley, M. H. Shearer, P. S. Hefty, and R. C. Kennedy. 1999. Fetal immunization of baboons induces a fetal-specific antibody response. Nat. Med. 5:427-430. [DOI] [PubMed] [Google Scholar]
- 56.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, A. S. Denton, et al. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 57.Wong, S. W., E. P. Bergquam, R. M. Swanson, F. W. Lee, S. M. Shiigi, N. A. Avery, J. W. Fanton, and M. K. Axthelm. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J. Exp. Med. 190:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]