Abstract
Members of the Hepadnaviridae family have been isolated from birds, rodents, and primates. A new hepadnavirus isolated from the woolly monkey, a New World primate, is phylogenetically distinct from other primate isolates. An animal model has been established for woolly monkey hepatitis B virus (WMHBV) by using spider monkeys, since woolly monkeys are endangered. In this study, a greater-than-genome length construct was prepared without amplification by using covalently closed circular DNA extracted from the liver of an infected woolly monkey. Transfection of the human liver cell line Huh7 with WMHBV DNA resulted in the production of viral transcripts, DNA replicative intermediates, and secreted virions at levels similar to those obtained with an infectious human HBV clone, demonstrating that the host range restriction of WMHBV is not at the level of genome replication. WMHBV particles from the medium of transfected cultures initiated an infection in a spider monkey similar to that obtained with virions derived from woolly monkey serum. In an attempt to adapt the virus for higher levels of replication in spider monkeys, immunosuppressed and newborn animals were inoculated. Neither procedure produced persistent infections, and the level of viral replication remained several logs lower than that observed in persistently infected woolly monkeys. These data demonstrate the production of an infectious clone for WMHBV and extend the characterization of the spider monkey animal model.
Hepatitis B virus (HBV) infections represent a worldwide health problem. Although acute HBV infection in adults is normally self-limiting, serious liver disease, including fulminant hepatitis, often develops. In addition, approximately 5 to 10% of the individuals infected as adults and 95% of those infected at birth become chronically infected. Persistently infected individuals are at risk for cirrhosis, end stage liver disease, and hepatocellular carcinoma (2). Most of our understanding of the pathogenesis and replication of HBV has been obtained by the use of related hepadnaviruses and their animal models, including duck HBV (12), woodchuck hepatitis virus (17), ground squirrel hepatitis virus (11), arctic ground squirrel hepatitis virus (20), heron hepatitis virus (16), and stork hepatitis virus (14). Infection of chimpanzees with human HBV is the only primate model for the analysis of HBV infections. Recently, a number of hepadnaviruses have been isolated from nonhuman primates. Most of the nonhuman primate isolates were from apes and clustered phylogenetically with human HBV isolates. Viruses have been isolated from gibbons (8, 13), chimpanzees (4, 5, 10, 19, 21), and orangutans (22). The origins of these viruses remain controversial. Although cross-species transmission from humans cannot be ruled out, chimpanzees from different geographical regions appear to be infected with distinct isolates (4), lending support to the concept that these viruses may have evolved with their hosts. However, the hepadnavirus isolated from woolly monkeys, woolly monkey HBV (WMHBV), clearly represents a new member of the Hepadnaviridae family (7). The ability to infect spider monkeys with this virus provided a new nonhuman primate model for the analysis of hepadnavirus infections and pathogenicity.
The WMHBV genome has the same genetic organization as human HBV. The core open reading frame (ORF) is the most conserved region of the genome, with approximately 15 and 24% divergence at the amino acid and nucleotide levels, respectively, in comparison to the human HBV core. The X region exhibits approximately 35% divergence between WMHBV and human HBV at the amino acid level, and the region of overlap between the pre-S1 and polymerase spacer regions also exhibits a high degree of divergence (30 to 32%). Nonetheless, recent studies have suggested that the WMHBV core and polymerase functions can substitute for the human HBV functions in genome replication (9) and that the WMHBV X region possesses transcriptional transactivating properties similar to those of the human HBV X protein (15).
In this report, we describe the construction of an infectious clone of WMHBV. The clone was derived by direct cloning from liver DNA of an infected woolly monkey without PCR amplification. Following transfection, the clone initiated replication in the human liver cell line Huh7 at levels similar to those of human HBV, and the virus secreted into the culture medium was infectious in spider monkeys. Attempts to improve the animal model by adaptation of the virus for better replication in spider monkeys, by either immunosuppression or inoculation of newborn animals, was not successful despite the induction of viremia of greater duration.
MATERIALS AND METHODS
Animals.
Black-handed spider monkeys (Ateles geoffroyi) were housed at the Southwest National Primate Research Center at the Southwest Foundation for Biomedical Research. Animals were cared for by members of the Department of Comparative Medicine in accordance with the Guide for the Care and Use of Laboratory Animals, and all protocols were approved by the Institutional Animal Care and Use Committee. FK506 (tacrolimus or Prograf) was purchased from Fujisawa Pharmaceutical Company. The animals were given FK506 orally twice daily at 100 μg/kg. The trough levels in blood were measured several times to ensure that therapeutic levels were being obtained. The enzyme-linked immunosorbent assays (ELISAs) for hepatitis B surface antigen (HBsAg) and antibodies to core protein (anti-hepatitis B core antigen [anti-HBcAg]) were purchased from Abbott Laboratories. The ELISA for antibodies to WMHBsAg was developed by using WMHBsAg purified from the medium of Huh7 cells transfected with a subgenomic clone of WMHBV expressing small, middle, and large envelope proteins (WM-SG pBS; see below). HBsAg from the medium of transfected Huh7 cells was concentrated by ultrafiltration (300,000 molecular weight cutoff) and ultracentrifugation through a 20% sucrose cushion. The partially purified material was used to coat microtiter wells (Immulon 2; Dynatech Laboratories) at 500 ng of HBsAg per well in borate-buffered saline (145 mM NaCl, 6 mM NaOH, 48 mM H3BO3, 50 mM KCl, pH 8.2). All incubations were for 1 h at 37°C, except for the final substrate incubation. Between incubation steps, wells were washed four times with phosphate-buffered saline (PBS)-0.05% Tween 20. Following coating, unoccupied protein binding sites were blocked with 5% nonfat dry milk in PBS. Serum samples were diluted 1:40 in antibody diluent (0.5% nonfat dry milk, PBS, 0.05% Tween 20) prior to incubation in coated wells. Bound antibody was detected with goat anti-human immunoglobulin G-horseradish peroxidase conjugate diluted 1:1,000 in antibody diluent. The substrate, 100 μl of 1-mg/ml 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS; Sigma), was incubated at room temperature until color development was stopped by the addition of 50 μl of 1% sodium dodecyl sulfate (SDS). Plates were read at 405 nm.
Quantitative PCR.
WMHBV DNA was isolated from serum by disruption of particles in TENS buffer (20 mM Tris-HCl [pH 7.4], 20 mM NaCl, 20 mM EDTA, 0.5% SDS) and digestion with proteinase K (1 mg/ml) for 2 h at 65°C. DNA was extracted with phenol-chloroform-isoamyl alcohol, and then chloroform and was ethanol precipitated. WMHBV DNA was then quantified by a real-time 5′ exonuclease PCR (TaqMan) assay with a primer-probe combination that recognizes a portion of the WMHBV surface gene. The primers (forward [nucleotides 589 to 609], 5′-GGACGGAAACTGCACTTGT-3′; reverse [nucleotides 659 to 639], 5′-GCCCAGTCCCATAGGAACTTT-3′) and probe (nucleotides 611 to 634; 5′-TTCCCATCCCCTCATCATGGGCTT-3′) were selected with the Primer Express software (Applied Biosystems, Foster City, Calif.). The fluorogenic probe was labeled with 6-carboxyfluorescein and 6-carboxytetramethylrhodamine and was obtained from Synthegen (Houston, Tex.). The primers and probe were used at 10 pmol/50-μl reaction mixture. The reactions were performed with the TaqMan PCR Core Reagent Kit (Applied Biosystems) and included a denaturation step of 10 min at 95°C and then 40 cycles of amplification under the universal TaqMan PCR standardized conditions, 15 s at 95°C for denaturation, and 1 min at 60°C for annealing and extension. Standards to establish genome equivalents were derived from cloned WMHBV DNA.
Plasmids.
The WMHBV greater-than-genome length (GGL) construct (WM GGL) was prepared from covalently closed circular DNA (cccDNA) isolated from liver tissue taken at autopsy from the woolly monkey that was the source of the original isolate of WMHBV. This animal died of fulminant liver failure. Approximately 200 mg of tissue was ground in 200 μl of TEN buffer (20 mM Tris-HCl [pH 7.4], 20 mM NaCl, 20 mM EDTA) and adjusted to contain 1% SDS and 1 M NaCl. Chromosomal DNA was allowed to precipitate overnight at 4°C, and a clarified supernatant containing the cccDNA was extracted three times with phenol-chloroform-isoamyl alcohol and then ethanol precipitated. The precipitate was suspended, digested with 40 μg of RNase A per ml for 45 min at 37°C, extracted with phenol-chloroform, and ethanol precipitated. The resulting DNA was digested with HindIII (the WMHBV genome contains a single HindIII site) and ligated into HindIII-digested pZero (Invitrogen). Thus, the clone was produced without the use of PCR amplification. The sequence of this clone was designated WMHBV-2 to distinguish it from the original WMHBV sequence. From the full-length WMHBV-2 clone, a GGL construct was produced (WM GGL) in a two-step cloning procedure that involved the cloning of an Eco52I (nucleotide position 1575)-to-HindIII (nucleotide position 123) fragment into NotI-HindIII-digested pBluescript SK+ (Invitrogen) and then addition of a HindIII-to-BsiWI (nucleotide position 1980) fragment into a HindIII-ApaI (T4 DNA polymerase blunted) intermediate vector. This yielded the WM GGL construct, spanning nucleotide positions 1575 to 1980. The human HBV GGL clone has been previously described (1, 3,18) and was derived from the ayw3 infectious clone. Both GGL clones were constructed in two different plasmid backgrounds, pBluescript SK+ and pZero. The nucleotide numbering convention used for both WMHBV and HBV corresponds to the HBV sequence numbering used by Galibert et al. (3). Subgenomic constructs were prepared to express the small, middle, and large envelope proteins in both pBluescript and pZero. For the WMHBV subgenomic construct cloned into pBluescript (WM-SG pBS), an SspI (blunted with Klenow)-to-KpnI (pBluescript site) fragment spanning nucleotide positions 2669 to 1980 was inserted into pBluescript cleaved at EcoRV and KpnI. This fragment was then moved to pZero as an EcoRV-to-KpnI fragment to produce WM-SG pZ. For Hu-SG pZ and Hu-SG pBS, a BglII-to-BglII fragment spanning nucleotide positions 2425 to 1987 was inserted into a BamHI-digested vector.
Cells and transfections.
The human hepatoma cell line Huh7 was maintained in Dulbecco modified Eagle medium (DMEM)-F12 medium (1:1) containing 10% fetal bovine serum, 2 mM glutamine, and 50 μg of gentamicin per ml. Huh7 cells were approximately 80% confluent prior to transfection. TransIT-LT1 (Mirus; PanVera, Madison, Wis.) was added to 3 ml of serum-free DMEM-F12, and the mixture was incubated for 15 min at 23°C. DNA was added to the lipid-medium mixture at a ratio of 1 μg of DNA per 2 μg of lipid, and the mixture was incubated for 15 min at 23°C. Cultures were washed twice with serum-free medium and exposed to the lipid-DNA mixture for 6 h at 37°C. Following transfection, the lipid-DNA mixture was replaced with DMEM-F12 supplemented with 10% fetal bovine serum, 2 mM glutamine, and 50 μg of gentamicin per ml.
Immunoprecipitation.
Cells were harvested at various time points posttransfection by being washed twice with PBS and extracted with EB buffer (50 mM Tris-HCl [pH 9.0], 100 mM NaCl, 1% Igepal [Sigma]) containing protease inhibitors (100 μM leupeptin, 1 mM Pefabloc, 10 μM aprotinin, 10 μg of pepstatin per ml, and 1 mM EDTA). The medium was supplemented with 1% Igepal to remove the envelope prior to immunoprecipitation. Core particles were immunoprecipitated for 16 h at 4°C with rabbit anti-core antibody bound to protein A agarose beads. Following immunoprecipitation, the beads were washed twice with EB buffer and treated with 1.5 U of micrococcal nuclease (USB, Cleveland, Ohio) for 30 min at 37°C.
DNA extraction and Southern hybridization.
Core particles were eluted from the beads and disrupted by incubation with TENS buffer for 10 min at 23°C. The extract was clarified to remove the beads, and viral DNA was purified by digestion with proteinase K, extraction with phenol-chloroform, and precipitation with ethanol. DNA was analyzed by electrophoresis on 1% agarose gels in TAE buffer (0.04 M Tris-acetate, 2 mM EDTA) for 5 h at 100 V, followed by downward capillary transfer to GeneScreen Plus membranes (Perkin-Elmer) in 0.1 N NaOH for 1 h at 23°C. Membranes were hybridized with a probe consisting of an equal mixture of the 32P-labeled, full-length human HBV and WMHBV DNAs. Hybridization was performed with 50% formamide-7% SDS-0.25 M NaPO4 (pH 7.2)-0.25 M NaCl-1 mM EDTA for 16 h at 37°C, followed by three washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS and three washes in 0.2× SSC-0.1% SDS at 42°C. Membranes were dried and exposed to film.
RNA extraction and Northern hybridization.
Cultures were harvested at various times posttransfection in RNAzol B (Leedo, Houston, Tex.), and RNA was extracted and purified in accordance with the manufacturer's protocol. RNA was heated for 10 min at 70°C in RNA sample buffer containing 50% formamide and 2.2 M formaldehyde and resolved by electrophoresis in a 1% agarose gel containing 2.2 M formaldehyde for 16 h at 40 V. The gel was washed four times with water and once with 10× SSC, and RNA was transferred to a GeneScreen Plus membrane for 16 h in 20× SSC. RNA was UV cross-linked to the membrane, prehybridized for 5 h at 37°C in hybridization buffer (6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA at pH 7.7], 10% SDS, 200 μg of salmon sperm DNA per ml, 50% formamide), and hybridized in the same buffer for 16 h at 42°C with a probe consisting of an equal mixture of the 32P-labeled, full-length human HBV and WMHBV DNAs. Membranes were dried and exposed to film.
Endogenous polymerase assay.
For analysis by the endogenous polymerase assay, core particles were immunoprecipitated as described above. The beads were washed twice with endogenous polymerase (EP) buffer (50 mM Tris-HCl [pH 7.4], 75 mM NH4Cl, 1.0 mM EDTA, 20 mM MgCl2, 0.1 mM β-mercaptoethanol, 0.5% Tween 20), and the endogenous polymerase assay (6) was performed with the core particles still bound to the beads. The beads were suspended in 50 μl of EP buffer containing 100 μM unlabeled deoxynucleoside triphosphates (dATP, dGTP, and dCTP) and 5 μCi of [α-32P]TTP (3,000 Ci/mmol; NEN) and incubated at 37°C for 6 h. The beads were washed twice with EB buffer, and viral DNA was purified by proteinase K digestion and phenol extraction as described above. DNA was electrophoresed on a 1% agarose gel in 1× TAE buffer at 100 V for 4 h. The gels were fixed in 7.5% trichloroacetic acid, dried, and exposed to film.
Nucleotide sequence accession numbers.
The WMHBV-1 and WMHBV-2 nucleotide sequences have been submitted to the GenBank database and assigned accession no. AF046996 and AY226578, respectively.
RESULTS AND DISCUSSION
Construction of a WM GGL clone.
The initial WMHBV sequence was deduced by sequencing a series of PCR fragments amplified from viral DNA purified from the serum of an infected woolly monkey (7). This sequence was not assembled into a full-length clone, since it was found to contain a number of nucleotide changes in comparison to a clone derived without PCR amplification. The original sequence will be referred to as WMHBV-1. To avoid potential errors introduced by PCR, we decided to derive a clone directly from cccDNA extracted from the liver of an infected woolly monkey that died of fulminant hepatitis. Full-length WMHBV DNA was cloned by digestion at the unique HindIII site, and then a GGL clone was assembled as described in Materials and Methods. This clone was designated WMHBV-2 and differed from WMHBV-1 at 16 nucleotide and 15 amino acid positions.
Analysis of transcription of WMHBV-2 in Huh7 cells.
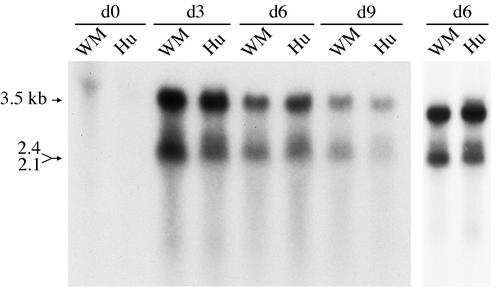
Since no woolly monkey or spider monkey liver cell lines were available, the replication potential of WMHBV-2 was analyzed with the human liver cell line Huh7 and directly compared to that of an HBV infectious clone of the ayw serotype (18). Initially, the transcriptional patterns of the two viruses were compared. Huh7 cells were transfected with WMHBV-2 GGL (WM GGL) or HBV ayw GGL (Hu GGL), and cultures were harvested at various times over a 9-day period. A typical transcript pattern was observed by Northern hybridization for both viruses, including the 3.5-kb pregenomic RNA and the 2.1- to 2.4-kb transcripts for the small, middle, and large envelope proteins (Fig. 1). The WMHBV genome appeared to produce transcript levels similar to those of HBV in Huh7 cells. The maximum level of viral RNA was observed at day 3 for both viruses, and the levels gradually declined thereafter but were still detectable at day 9. The 2.1- to 2.4-kb transcripts contain two distinctive mRNA species, one for the large envelope protein and one for the middle and small envelope proteins, which are poorly resolved on most gels. However, the level of the 2.4-kb transcript appeared to be reduced in the WM GGL-transfected cells. Analysis of a separate transfection at day 6 clearly indicated that the 2.4-kb transcript was reduced or absent in the WM GGL-transfected cells (Fig. 1, far right lanes).
FIG. 1.
Transcription profile of WMHBV-2 in Huh7 cells. The human liver cell line Huh7 was transfected with GGL constructs for both WMHBV-2 (WM) and HBV (Hu). Total cellular RNA was harvested from cultures at day 0 (d0), d3, d6, and d9 posttransfection. Viral RNA was analyzed by denaturing agarose gel electrophoresis and Northern hybridization with a mixed probe containing both WMHBV and HBV sequences. The migration of the subgenomic 2.1- to 2.4-kb and pregenomic 3.5-kb transcripts is indicated. A second preparation harvested on d6 was examined to confirm the reduced detection of the 2.4-kb transcript in the WMHBV-transfected cells.
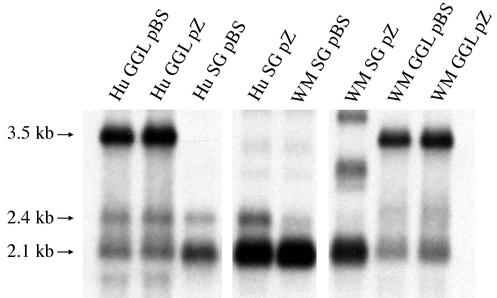
We further examined this difference in transcript patterns by preparing GGL constructs of both WMHBV and HBV in the vectors pBluescript SK+ (GGL pBS) and pZero (GGL pZ) and by preparing subgenomic clones that encode the envelope proteins with their promoters and the poly(A) site (SG pBS and SG pZ). Transfection of these constructs into Huh7 cells and analysis by Northern hybridization confirmed that the 2.4-kb transcript for the WMHBV large envelope protein was not as apparent as the equivalent transcript from HBV (Fig. 2). Whether this difference was due to reduced transcription from the WM pre-S promoter in Huh7 cells or comigration of the transcript with the 2.1-kb transcript was not pursued, since it was determined that the WMHBV-2 construct was infectious in spider monkeys (see below).
FIG. 2.
Transcription profile of WMHBV subgenomic constructs. Constructs of WMHBV-2 (WM) and HBV (Hu) were prepared with two vectors, pBluescript (pBS) and pZero (pZ), to compare transcription profiles from GGL and subgenomic (SG) vectors that only express the envelope transcripts. Huh7 cells were transfected with each vector and harvested on day 3. Total cell RNA was analyzed by denaturing agarose gel electrophoresis and Northern hybridization with a mixed probe containing both WMHBV and HBV sequences. The migration of the subgenomic 2.1- to 2.4-kb and pregenomic 3.5-kb transcripts is indicated.
Analysis of replication of WMHBV-2 in Huh7 cells.
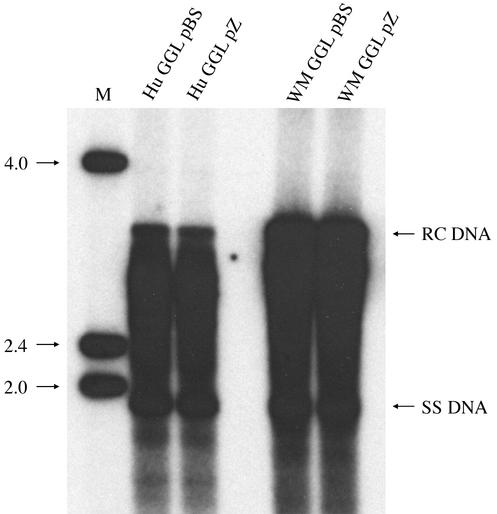
To examine the replication competence of the WMHBV-2 construct, Huh7 cells were transfected with WM GGL and Hu GGL and DNA replicative intermediates were examined in secreted virions. Medium was harvested 6 days posttransfection and treated with detergent to remove the envelope from particles. Core particles were immunoprecipitated with rabbit anti-core antibodies, and DNA extracted from the core particles was analyzed by Southern blot hybridization. Typical patterns for replicative intermediates were observed with both viruses including prominent bands representing single-stranded (SS) and relaxed-circular (RC) DNAs. Identical patterns were observed, independently of the GGL vector used (pBS or pZero), but minor differences in the abundance of RC DNA between WMHBV and HBV were observed (Fig. 3).
FIG. 3.
Replication of WMHBV in Huh7 cells. Huh7 cells were transfected with the WMHBV-2 (WM) and HBV (Hu) GGL constructs, and medium was harvested 6 days posttransfection. Core particles were immunoprecipitated from detergent-treated medium with anti-core antibodies. Viral DNA was purified from the core particles and analyzed by Southern blot hybridization with a mixed probe containing both HBV and WMHBV sequences. The migration of single-stranded (SS) and RC DNAs is indicated. The values on the left are the molecular sizes, in kilobases, of the markers in lane M.
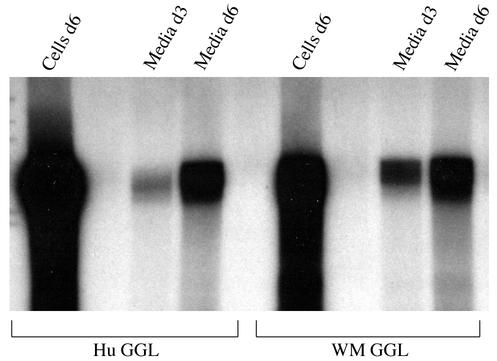
To further examine the in vitro replication competence of WMHBV-2 in Huh7 cells, endogenous polymerase assays were conducted in which the encapsidated reverse transcriptase extends the partially completed positive-strand DNA, allowing the incorporation of radiolabeled nucleotides. Huh7 cells were transfected with the GGL constructs, core particles were immunoprecipitated from the cells or detergent-treated medium, and core particles still bound to the antibody beads were used in the endogenous polymerase assays. Particles harvested from the medium on days 3 and 6 and from cell lysates on day 6 were examined. Radiolabeled DNA migrating at the position of RC DNA was observed for cells transfected with WM GGL and HBV GGL, and no difference was observed in the activity of the particles from the two viruses (Fig. 4). Collectively, these data indicate that WMHBV replicates in Huh7 cells at levels similar to those of HBV; thus, no host range restriction occurred at the level of genome replication.
FIG. 4.
Endogenous polymerase reaction of WMHBV particles. Huh7 cells were transfected with the WMHBV-2 (WM) and HBV (Hu) GGL constructs, and medium was harvested on day 3 (d3) and d6, and cells were harvested on d6. Core particles were immunoprecipitated from the cell lysates and detergent-treated medium with anti-core antibodies, and endogenous polymerase reactions were conducted with the core particles still bound to the immunobeads. Viral DNA was purified from the core particles and analyzed by gel electrophoresis and autoradiography.
Infectivity of WMHBV-2 in spider monkeys.
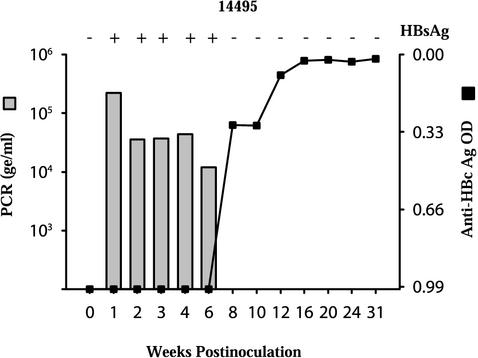
To confirm the infectivity of the WMHBV-2 clone, virus produced in Huh7 cells following transfection with WM GGL was inoculated into a spider monkey (Fig. 5). A large-scale transfection was performed with ten 100-mm dishes, and the medium was concentrated with ultrafiltration. A membrane with a 300,000 molecular weight cutoff was used to eliminate most of the serum and cellular proteins present in the medium. Spider monkey 14495 was inoculated intravenously with approximately 1012 genome equivalents (ge) of WMHBV as estimated by real-time, quantitative TaqMan PCR. Within 1 week postinoculation, viremia peaked at 2.2 × 105 ge/ml of serum. Viremia remained elevated until week 8, at which time viral clearance occurred. The HBsAg ELISA values exhibited a profile similar to that of viremia with high reactivity until week 6 and then negative values thereafter. Since baboons, rhesus monkeys, and tamarins inoculated with WMHBV were PCR negative by week 1 (data not shown), it can be concluded that the 6 weeks of viremia observed in spider monkey 14495 represents viral replication. In addition, seroconversion for antibodies to HBcAg and WMHBsAg occurred at week 8, at the time of viral clearance, which is another indicator of active replication. No significant rise in alanine transaminase occurred, indicating the absence of overt liver disease (data not shown). These data demonstrate that the WMHBV-2 clone is infectious in spider monkeys and induces an infection profile similar to that observed with woolly monkey serum containing WMHBV (7).
FIG. 5.
Infection of a spider monkey with WMHBV-2-derived particles. WMHBV particles were produced by transfection of Huh7 cells with WMHBV-2 GGL, and particles secreted into the medium were concentrated by ultrafiltration and ultracentrifugation. A spider monkey (14495) was inoculated intravenously with 1012 particles and monitored by TaqMan PCR for the level of viremia (bar graph) and by ELISA for the levels of HBsAg (plus and minus signs at the top of the graph) and anti-HBcAg (line graph). OD, optical density.
Effects of immunosuppression and inoculation at birth on the duration and level of WMHBV viremia in spider monkeys.
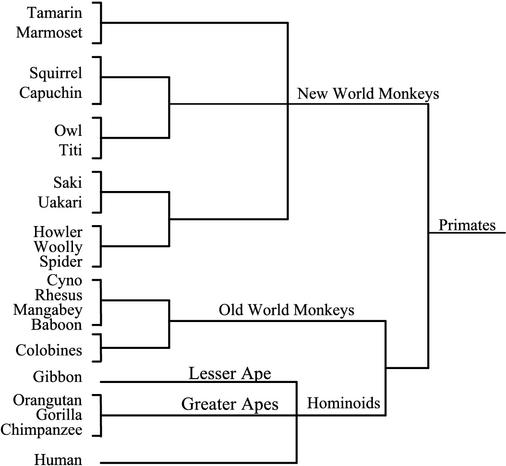
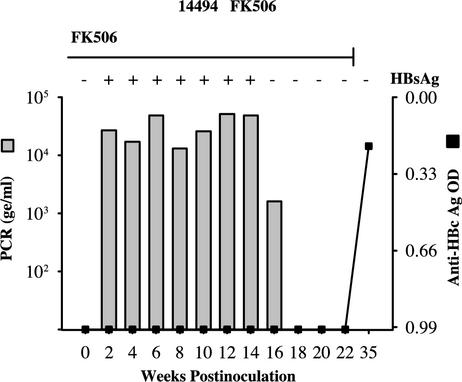
The permissiveness of spider monkeys for infection with WMHBV has provided an animal model for this new hepadnavirus. We have not observed significant replication in other New World (tamarins) or Old World (baboons, rhesus macaques, and chimpanzees) primates. The spider monkey is a close relative of the woolly monkey (Fig. 6); thus, the data suggest that, similar to those of other hepadnaviruses, the host range of WMHBV is very narrow. The level of viremia in chronically infected woolly monkeys is very high. Quantification by TaqMan assay revealed that the mean viral load was 1.8 × 109 ge/ml for the seven carriers for which serum was available. No acute resolving infections of woolly monkeys have been monitored. This level of viremia is 3 to 4 logs higher than the peak viremia in spider monkeys inoculated with virus from the infectious clone (2.2 × 105 ge/ml) or with woolly monkey serum (1.1 × 105 to 1.3 × 106 ge/ml) (7). We reasoned that a virus more fit for replication in spider monkeys would emerge because of random mutations if viremia could be prolonged or if chronic infection could be induced. We also reasoned that if viral levels were being suppressed by the immune system, immunosuppression might increase the level of viremia and enhance the potential for adaptation. Therefore, in an attempt to increase the duration and level of viremia, a spider monkey was immunosuppressed during infection with WMHBV. Animal 14494 was maintained on FK506 beginning 3 weeks prior to inoculation and for 22 weeks postinoculation, when viral clearance was documented by two consecutive blood samples negative for WMHBV DNA and HBsAg reactivity. Viremia was detectable until week 18, with peak viremia at 5.1 × 104 ge/ml (Fig. 7). HBsAg reactivity was present in the serum until week 14, and seroconversion for both anti-HBcAg and anti-WMHBsAg antibodies occurred between weeks 22 and 35. Unfortunately, serum samples were not available at times between weeks 22 and 35. Although chronic infection was not attained in this animal, the duration of viremia was greater than that previously observed in immunocompetent animals (7). The number of animals that could be devoted to this approach was limited. Spider monkeys are not being bred for biomedical research at any primate center in the United States. For our studies, we obtained the few remaining animals from a colony previously housed at another facility. We are breeding additional animals for these studies, and several of these were used in the studies involving inoculation of newborn spider monkeys (see below). Nonetheless, in a single animal, immunosuppression yielded 18 weeks of viremia, in comparison to the 4 to 6 weeks of viremia observed in all of the previously inoculated immunocompetent spider monkeys. Although the extended duration of viremia may have been due to the genetic background of this animal, it was most likely a result of the immunosuppression. Rather than pursue this approach with additional animals, alternative strategies for the induction of chronicity were chosen.
FIG. 6.
Classification of primates. The classification scheme for primates is illustrated. The primates are divided into New World primates, Old World primates, and hominoids, which include the lesser and greater apes. The common names of many of the primate genera are shown. Note the close relationship between spider and woolly monkeys. Hepadnaviruses have been isolated from woolly monkeys, gibbons, orangutans, and chimpanzees.
FIG. 7.
Infection of an immunosuppressed spider monkey with WMHBV. A spider monkey (14494) was immunosuppressed with FK506 (200 μg/kg/day) for 3 weeks prior to and 22 weeks after inoculation with WMHBV and monitored by TaqMan PCR for the level of viremia (bar graph) and by ELISA for the levels of HBsAg (plus and minus signs at the top of the graph) and anti-HBcAg (line graph). OD, optical density.
One alternative strategy by which to achieve persistent infection is inoculation of animals at birth, since most humans exposed at birth develop chronic infections while adults normally clear the virus. Two spider monkeys (14333 and 16275) were inoculated at birth. Blood samples were infrequently obtained from these animals, since they were being carried by their mothers. The serum of the first animal (14333) was positive for viral DNA at months 2, 5, 9, and 10, but viral clearance occurred by month 14. Peak viremia was 2.9 × 104 ge/ml. The second animal (16275) was PCR positive at month 2 but negative by month 5. Thus, in neither case did inoculation at birth result in chronic infection or higher levels of viremia. However, since the duration of viremia was greater with both immunosuppression and inoculation at birth, additional efforts are warranted to improve the animal model for WMHBV.
The factors limiting the level of WMHBV viremia in spider monkeys are not apparent. Presumably, proper interaction with an essential cellular protein is lacking or reduced in spider monkeys. One possible explanation for the lack of adaptation to a more “fit” virus is that an overlapping essential function prevents the emergence of a virus with higher replication competence. The overlapping nature of the polymerase ORF with other ORFs and cis functions certainly constrains the evolution of hepadnaviruses. Surprisingly, the WMHBV infectious clone replicates in the human cell line Huh7 at levels similar to those of HBV. Although receptor interactions cannot be examined in Huh7 cells, it is unlikely that receptor interactions account for the reduced viremia. The lack of interaction between the receptor and WMHBV in spider monkeys would presumably completely block infection, while a reduced affinity between the virus and the receptor would presumably still result in spread of the infection to all susceptible hepatocytes. Another possible factor that limits viremia is greater suppression of WMHBV replication by the innate immune system of spider monkeys. Further evaluation of this virus-host interaction may provide additional information about the factors that regulate hepadnavirus infections.
Acknowledgments
This work was supported by grants RO1 AI46609 and P51 RR13986 from the National Institutes of Health.
REFERENCES
- 1.Beames, B., and R. E. Lanford. 1993. Carboxy-terminal truncations of the HBV core protein affect capsid formation and size of the encapsidated HBV RNA. Virology 194:597-607. [DOI] [PubMed] [Google Scholar]
- 2.Beasley, R. P. 1988. Hepatitis B virus: the major etiology of hepatocellular carcinoma. Cancer 61:1942-1956. [DOI] [PubMed] [Google Scholar]
- 3.Galibert, F., E. Mandart, F. Fitoussi, P. Tiollais, and P. Charnay. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646-650. [DOI] [PubMed] [Google Scholar]
- 4.Hu, X. L., A. Javadian, P. Gagneux, and B. H. Robertson. 2001. Paired chimpanzee hepatitis B virus (ChHBV) and mtDNA sequences suggest different ChHBV genetic variants are found in geographically distinct chimpanzee subspecies. Virus Res. 79:103-108. [DOI] [PubMed] [Google Scholar]
- 5.Hu, X. L., H. S. Margolis, R. H. Purcell, J. Ebert, and B. H. Robertson. 2000. Identification of hepatitis B virus indigenous to chimpanzees. Proc. Natl. Acad. Sci. USA 97:1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan, P. M., R. L. Greenman, J. L. Gerin, R. H. Purcell, and W. S. Robinson. 1973. DNA polymerase associated with human hepatitis B antigen. J. Virol. 12:995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanford, R. E., D. Chavez, K. M. Brasky, R. B. Burns III, and R. Rico-Hesse. 1998. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc. Natl. Acad. Sci. USA 95:5757-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanford, R. E., D. Chavez, R. Rico-Hesse, and A. Mootnick. 2000. Hepadnavirus infection in captive gibbons. J. Virol. 74:2955-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lott, L., L. Notvall, and R. E. Lanford. 2003. Transcomplementation of core and polymerase functions of the woolly monkey and human hepatitis B viruses. Virology 308:330-339. [DOI] [PubMed]
- 10.MacDonald, D. M., E. C. Holmes, J. C. Lewis, and P. Simmonds. 2000. Detection of hepatitis B virus infection in wild-born chimpanzees (Pan troglodytes verus): Phylogenetic relationships with human and other primate genotypes. J. Virol. 74:4253-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marion, P. L., L. S. Oshiro, D. C. Regnery, G. H. Scullard, and W. S. Robinson. 1980. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc. Natl. Acad. Sci. USA 77:2941-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason, W. S., G. Seal, and J. Summers. 1980. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J. Virol. 36:829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norder, H., J. W. Ebert, H. A. Fields, I. K. Mushahwar, and L. O. Magnius. 1996. Complete sequencing of a Gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology 218:214-223. [DOI] [PubMed] [Google Scholar]
- 14.Pult, I., H. J. Netter, M. Bruns, A. Prassolov, H. Sirma, H. Hohenberg, S. F. Chang, K. Frolich, O. Krone, E. F. Kaleta, and H. Will. 2001. Identification and analysis of a new hepadnavirus in white storks. Virology 289:114-128. [DOI] [PubMed] [Google Scholar]
- 15.Schuster, R., E. Hildt, S.-F. Chang, O. Terradillos, T. Pollicino, R. Lanford, W. H. Gerlich, H. Will, and S. Schaefer. 2002. Conserved transactivating and pro-apoptotic functions of hepadnaviral X protein in ortho- and avihepadnaviruses. Oncogene 21:6606-6613. [DOI] [PubMed] [Google Scholar]
- 16.Sprengel, R., E. F. Kaleta, and H. Will. 1988. Isolation and characterization of a hepatitis B virus endemic in herons. J. Virol. 62:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summers, J., J. M. Smolec, and R. Snyder. 1978. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc. Natl. Acad. Sci. USA 75:4533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sureau, C., J. W. Eichberg, G. B. Hubbard, J. L. Romet-Lemonne, and M. Essex. 1988. A molecularly cloned hepatitis B virus produced in vitro is infectious in a chimpanzee. J. Virol. 62:3064-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi, K., S. Mishiro, and A. M. Prince. 2001. Novel hepatitis B virus strain from a chimpanzee of Central Africa (Pan troglodytes troglodytes) with an unusual antigenicity of the core protein. Intervirology 44:321-326. [DOI] [PubMed] [Google Scholar]
- 20.Testut, P., C.-A. Renard, O. Terradillos, L. Vitvitski-Trepo, F. Tekaia, C. Degott, J. Blake, B. Boyer, and M. A. Buendia. 1996. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J. Virol. 70:4210-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaudin, M., A. J. Wolstenholme, K. N. Tsiquaye, A. J. Zuckerman, and T. J. Harrison. 1988. The complete nucleotide sequence of the genome of a hepatitis B virus isolated from a naturally infected chimpanzee. J. Gen. Virol. 69:1383-1389. [DOI] [PubMed] [Google Scholar]
- 22.Warren, K. S., J. L. Heeney, R. A. Swan, Heriyanto, and E. J. Verschoor. 1999. A new group of hepadnaviruses naturally infecting orangutans (Pongo pygmaeus). J. Virol. 73:7860-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]