Abstract
The viral bZIP transcription factor Zta (BZLF1, EB1, ZEBRA) mediates the switch between the latent and lytic cycles of Epstein-Barr virus (EBV). In part, its activity requires the formation of homodimers and interaction with specific DNA sequence elements (ZREs). Zta has an atypical zipper motif that has a lower stability than do typical bZIP proteins. Here we show that a synthetic peptide directed against the zipper can disrupt the DNA-binding function of Zta. This highlights the relevance of this region for the function of Zta and demonstrates that the zipper region is a potential target for therapeutic agents. We also unmask the relevance of a region adjacent to the zipper (CT region), which is required to direct the interaction of Zta with DNA and to transactivate ZRE-dependent promoters in vivo.
Epstein-Barr virus (EBV) is associated with several pathologies, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, and lymphoproliferative diseases in immunocompromised populations (6). EBV infects the host during early childhood and persists for the life of the individual with periodic bursts of replication and virus production (6).
The viral protein encoded by BZLF1, Zta (EB1, BZLF1, ZEBRA), is a key component of the induction of the lytic replicative cycle of EBV (8). Increased expression of Zta is one of the first events that can be detected following the induction of the lytic cycle in EBV-harboring B lymphocytes, and the enforced expression of Zta is sufficient to induce the lytic cycle in a cell containing latent EBV genomes (reviewed in references 21 and 24). Zta acts in part as a transcription factor; it activates its own expression and the expression of a subset of EBV genes through sequence-specific Zta response elements (ZREs) within their respective promoters (reviewed in references 30, 31, and 32). Zta also acts as a replication factor later in the lytic cycle; Zta interacts with the lytic origin of replication, again through specific ZREs (24, 28). Furthermore, Zta also reprograms the host cell cycle control machinery, since enforced expression of Zta induces cell cycle arrest in several cell lineages (2, 3, 11, 19, 20, 25, 26), although the effect is not universal (20). Interestingly the effect on cell cycle does not require a direct interaction with ZREs but occurs through the activation of a cellular transcription factor, C/EBPα (36). Zta also interacts with a number of other cellular factors (reviewed in reference 30) that may extend its ability to regulate the expression of genes that do not contain ZREs.
Zta is a member of the family of bZIP transcription factors (5, 7, 9, 14, 16, 23, 31, 34); it contains adjacent DNA contact (approximately amino acids 175 to 195) and multimerization domains (approximately amino acids 196 to 245) and can interact directly with specific DNA sequence elements, i.e., ZREs (5, 7, 10, 16, 23, 35) as a multimer (reviewed in references 24 and 31). By analogy with other members of the bZIP family, the multimerization interface of Zta has been predicted to fold through a coiled-coil structure (10, 14, 31). Biophysical evidence that this prediction holds true was recently provided (12). However, the thermal stability of the resulting structure is much lower than that of the coiled coil domains of canonical members of the bZIP family (12). This suggests that either the Zta dimerization interface is relatively weak or that elements outside the coiled coil act to stabilize dimer formation and thus the DNA-binding function of Zta.
We present an exploration of the dimerization region of Zta. By using a short synthetic peptide, homologous to the coiled coil region, we demonstrate that the coiled coil interface of Zta is relevant for the function of Zta as a protein, despite its low thermal stability. In addition, we explore the impact of the amino acid sequence variation found within the zipper region of Zta in natural isolates of EBV on DNA-binding dependent functions. These studies suggest that the zipper region of Zta is a suitable target for therapeutic agents designed to prevent viral lytic cycle reactivation. Furthermore, we unmask the function of a novel region (CT) of Zta, adjacent to the coiled coil, which is required for both DNA-binding and transactivation functions. The impact of this on the present model of Zta structure is discussed.
In order to probe the relevance of the coiled coil region of Zta, we attempted to disrupt the formation and/or stability of the coiled coil within the context of the full-length Zta protein. The principle behind this has been exploited previously to generate reagents that block the function of the coiled coil containing proteins APC and human immunodeficiency virus gp41 (4, 15, 29, 37) and is illustrated in Fig. 1A. To evaluate whether this approach is viable for Zta, a synthetic peptide (ZEDpep) corresponding to residues spanning the coiled coil region of Zta (amino acids 196 to 227 of the B95-8 sequence) was used. Initial experiments revealed that the addition of ZEDpep reduced DNA complex formation by 76% without altering the stability of Zta under the reaction conditions (Fig. 1B).
FIG. 1.
A short synthetic peptide is able to disrupt the interaction of Zta with DNA. The schematic diagram of the rationale for this approach is shown in panel A. The bZIP region of Zta with the basic, DNA contact domain shown in black and the coiled coil region in grey are indicated. On the right, the short synthetic peptide (white box) interacts with the coiled coil region and prevents homodimerization and thus DNA binding. The ability of 10 nmol of ZEDpep to reduce the ability of full-length Zta to interact with a ZRE was evaluated by EMSA at 20°C. The reaction components were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and EMSA, and the relative amounts of Zta protein were calculated.
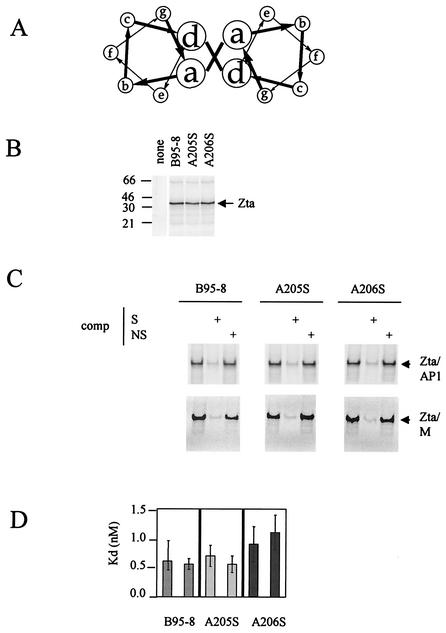
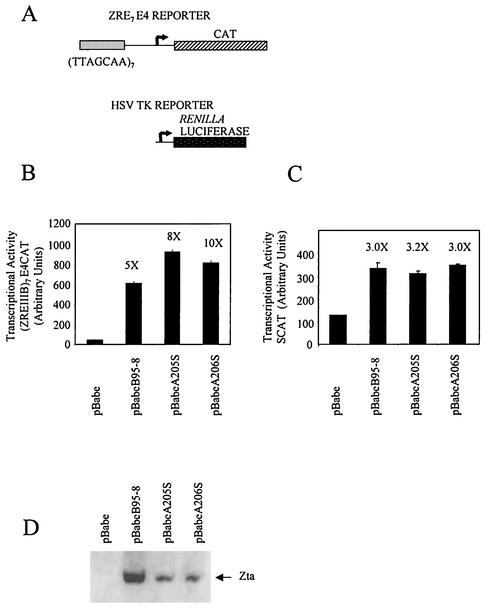
We previously demonstrated that the three natural sequence variants within the dimerization domain of Zta fold as dimeric coiled coils but that these structures have different stabilities (the Tm ranges from 17 to 25°C) (12). This was surprising, since the variant amino acids fall at the b and c positions of the heptad repeat, distant from the critical hydrophobic core formed by the a and d residues (Fig. 2A). It is therefore relevant to ask whether the different stabilities of the coiled coil motifs have an impact on dimerization-dependent functions of Zta, namely, the ability to interact with DNA in vitro and to transactivate ZRE-dependent promoters in vivo. In order to establish whether the three natural variants of the coiled coil sequence confer different properties to Zta, we reconstructed each variant within the context of the B95-8 Zta sequence, which generated vectors capable of expressing three forms of Zta termed as follows: B95-8, A205, and A206 (Fig. 2B). All three forms of Zta interacted specifically with two different ZREs (AP1 and M) (Fig. 2C). Further analysis of the strength of the interaction revealed similar Kd values for each Zta variant (Fig. 2D). The ability of each of the three variants to transactivate through ZREs in vivo was also compared. Using a reporter vector bearing seven ZREs upstream from a minimal promoter (1), together with a TK promoter as an internal control (Fig. 3A), we observed that all three Zta proteins transactivate well through the ZREs with A206S, effecting a marginally higher activation (Fig. 3B). Given that both A205S and A206S were expressed at lower levels than was B95-8 Zta in this experiment, it is clear that neither is compromised in function. Furthermore, we assessed the ability of these variants to up regulate the natural BSLF2+BMLF1 EBV promoter (27) in B cells (Fig. 3C). All three natural variants directed the up regulation of this promoter to a similar degree. This demonstrates that, despite the differences in the stability of the coiled coil structure within the zipper region of these variants, all have similar properties with respect to the ability to bind to ZREs and to transactivate gene expression through them.
FIG. 2.
All three natural variants within the dimerization domain of Zta interact equivalently with DNA. The locations of amino acid residues A205 and A206 occur at b and c positions of the heptad repeat in the predicted structure of the coiled coil as shown in panel A. The coding sequences for A205 and A206 were each altered by using site-directed mutagenesis to encode serine within the B95-8 Zta cDNA (27). The resulting plasmids, B95-8, A205S, and A206S, were transcribed and translated in a rabbit reticulocyte lysate system as for Fig. 1. The proteins were fractionated on a sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis gel (B). The input plasmid is indicated above each lane, and the migration of molecular weight markers is given on the left (in kilodaltons). Following exposure to a phosphorimager (Storm), the concentrations of the proteins were normalized and their ability to interact with a double-strand version of a canonical AP1 site and a ZRE from the BSLF2+BMLF1 promoter (M) was evaluated by EMSA at 20°C (C). The input proteins are indicated above with the radiolabeled probe (2 ng), and nonlabeled competitor (comp) DNA—either specific, S, (AP1) (500 ng), or nonspecific, N, (500 ng)—is shown above each track. (D) The Kd values of the interaction of the B95-8, A205S, and A206S Ztas were determined by EMSA with increasing probe concentration. The data were quantitated by using phosphorimaging, and the concentrations of bound and free probe were determined according to the method described by Stone et al. (33).
FIG. 3.
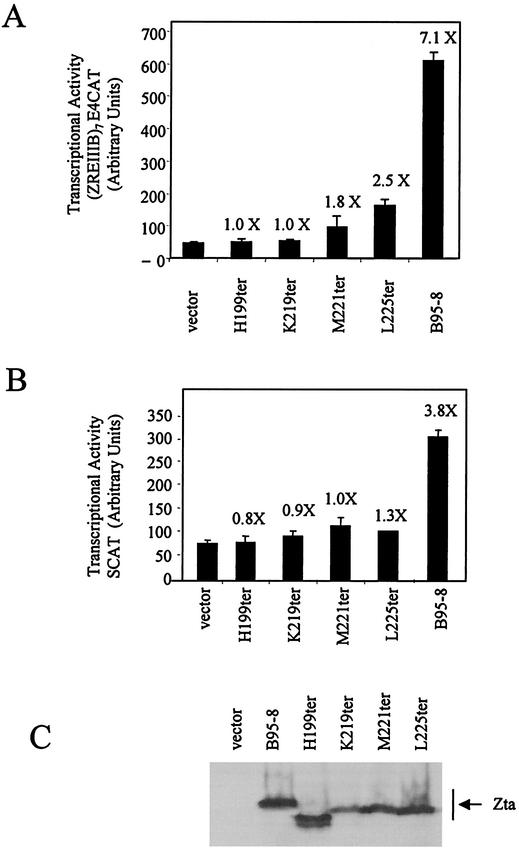
All three forms of Zta transactivate a synthetic ZRE containing promoter equivalently in vivo. HeLa cells (obtained through ECACC) were transfected by using Effectene (Qiagen) with a total of 2 μg of DNA containing the reporter vectors, ZRE7CAT (0.5 μg) and HSVTK luciferase (0.5 μg) (Promega) (A), and either a Zta expression vector containing the full-length Zta sequences (1.0 μg) or the respective “empty” vector pBabe (22) (1.0 μg) as a control. DG75 cells were transfected by using electroporation with a total of 20 μg of DNA containing the reporter vectors, SCAT (5.0 μg) (27) and HSVTK luciferase (5.0 μg) (Promega) (C), and either a Zta expression vector containing the full-length Zta sequences (10.0 μg) or the respective “empty” vector pBabe (22) (10.0 μg) as a control. Forty-eight hours later the chloramphenicol acetyltransferase and luciferase activities were determined (Promega). The relative activation (n-fold) after correction for different transfection efficiencies is shown above the chloramphenicol acetyltransferase activity data in panels B and C. Western blot analysis of the expression of Zta given by using the monoclonal antibody BZ1 (38) is shown in panel D.
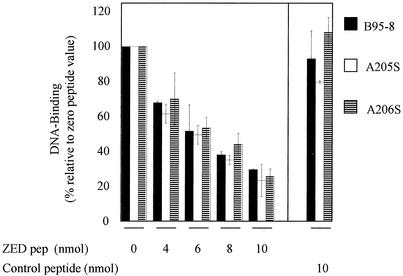
A detailed evaluation of the ability of ZEDpep to interfere with the ability of the full-length Zta proteins B95-8, A205S, and A206S was then undertaken (Fig. 4). The addition of between 4 and 10 nmol of ZEDpep compromised the ability of all three forms of Zta to interact with DNA in a dose-dependent manner, whereas another coiled coil peptide of unrelated sequence, SKIP1 (13), was unable to disrupt Zta complex formation with DNA. Thus, it appears that the zipper of Zta can be specifically targeted by a short synthetic peptide and that the coiled coil region of Zta can be considered a good candidate for future drug design.
FIG. 4.
The synthetic peptide is specific for Zta and can disrupt all three natural variants. The ability of ZEDpep or of an unrelated coiled coil control peptide, SKIP1 (IAALERKNAALEQKAIAALEYKIAALEKK [13]), to disrupt the ability of each of the three Zta full-length proteins to interact with AP1 sites was evaluated with EMSA. The DNA binding relative to the no-peptide control value is indicated. The error bars represent the standard deviation from two independent experiments.
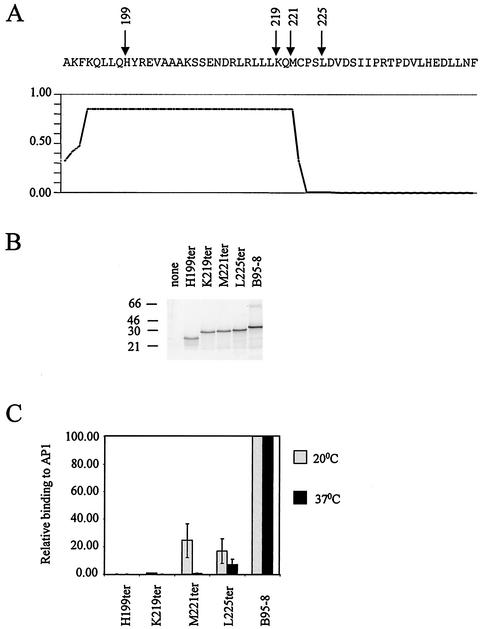
Several studies suggest that the dimerization domain of Zta is limited to amino acids 196 to 227; however, our previous analysis of the dimerization region predicted that the coiled coil is unlikely to extend beyond residue M221 (illustrated in Fig. 5A). We therefore reassessed the extent of the zipper and C-terminal region required to mediate the ZRE binding function of Zta in vitro and for transactivation through ZRE-dependent promoters in vivo. We approached this by making a series of C-terminal deletion mutants. These were expressed in vitro (Fig. 5B), and their ability to interact with two ZREs was evaluated by electrophoretic mobility shift assays (EMSA) (Fig. 5C). Data obtained from one ZRE (AP1) are shown; equivalent results were obtained for a second ZRE (M) (data not shown). Initial results from DNA-binding assays undertaken at 20°C did not fully correlate with the transactivation activity that we observed in vivo, which prompted us to further analyze DNA binding at the physiological temperature of 37°C (Fig. 5C). Truncation at 199H, carboxy terminal to the DNA-binding motif, resulted in negligible DNA binding at either temperature, emphasizing the requirement for the zipper region. Interestingly, truncation at K219 abolished DNA binding at both temperatures, but truncation at M221 retained some DNA binding at 20°C. This suggests that the zipper facilitates DNA binding but that this is insufficient at 37°C. In vivo transactivation assays on the reporter construct containing seven ZREs (Fig. 6A) and the natural BSLF2+BMLF1 promoter (Fig. 6B) revealed that only full-length Zta is capable of transactivating. It can be concluded that, although the coiled coil region is sufficient to direct DNA complex formation at 20°C, sequences C terminal to L225 (the CT region) are required for both DNA complex formation and transactivation at 37°C.
FIG. 5.
The relevance of the C-terminal region of Zta for DNA binding was investigated. A series of truncations of the Zta coding sequence were generated (based on the B95-8 sequence) by using site-directed mutagenesis. The positions where the ter codons were introduced are indicated on the sequence (residues 191 to 245) in panel A. The prediction of the extent of coiled coil formation through this region is indicated graphically below. The x axis is aligned with the amino acid sequence above, and the y axis represents the predictive value of forming a coiled coil (17). The ter mutant series, together with B95-8 sequence, were transcribed and translated in vitro as described in the Fig. 1 legend. The resulting proteins were fractionated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis and were analyzed by phosphorimaging (Storm) (B). The migration of molecular weight markers is indicated on the left in kilodaltons. Following normalization for protein concentration (taking into account the number of methionine residues in each protein), their ability to specifically interact with a ZRE (AP1) was assessed by using EMSA at both 20 and 37°C. Following phosphorimaging, the amount of DNA-binding activity relative to B95-8 was determined (C). Error bars represent the standard deviations derived from duplicate experiments.
FIG. 6.
HeLa cells were transfected with a total of 2 μg of DNA containing the reporter vectors and eukaryotic expression vectors for the Zta sequences. DG75 cells were transfected by using electroporation with a total of 20 μg of DNA containing the reporter vectors, SCAT (5.0 μg) (27) and HSVTK luciferase (5.0 μg) (Promega) (A), and either a Zta expression vector containing the Zta sequences indicated in Fig. 6A (10.0 μg) or the respective “empty” vector, pBabe (22) (10.0 μg), as a control. Forty-eight hours later, the chloramphenicol acetyltransferase and luciferase activities were determined (Promega). The relative activation (n-fold) after correction for different transfection efficiencies is shown above the chloramphenicol acetyltransferase activity data in panels B and C. Western blot analysis of the expression of Zta given by using the polyclonal antibody EE is shown in panel C.
Small differences in the stability of the coiled coil structure of the natural variants of Zta that suggested that they might differ in their ability to direct stable dimer formation and thus DNA-binding-dependent functions were previously identified (12). In addition, in a recent study a 48% decrease in dimer formation was observed for A206S compared to the result for B95-8 (18). Here we assessed the potential impact of these coiled coil variants within the context of full-length Zta protein. No significant differences were found in their ability to interact with two ZREs or to direct transcription through two further ZREs. This suggests that the small differences in stability of these coiled coil structures observed by Hicks et al. and by Martel-Renoir et al. (12, 18) do not reflect differences in the function of the coiled coil when it is within the context of full-length protein. Interestingly, both of the previous studies measured dimerization in the absence of DNA and so neither would reflect any potential effects of increased stability of the coiled coil due to interaction of the adjacent basic region with DNA.
Based on our previous structural analysis of the coiled coil region of Zta, we designed a short synthetic peptide that was predicted to dimerize with Zta. This successfully disrupted the ability of the full-length Zta protein to form DNA complexes, which emphasizes the relevance of the coiled coil region for the function of Zta. In addition, it establishes the principle that the coiled coil region of Zta is a suitable target for drug design. The 50% inhibitory concentration for the Zta peptide was equivalent for all three of the naturally occurring coiled coil sequence variants of Zta, which suggests that this approach may be effective for all isolates of EBV; however, since they are all in the high-micromolar range, the synthetic peptide is unlikely to be a drug candidate itself.
The contribution of residues within the CT region (amino acids 222 to 245) of Zta to its function has been addressed previously, but the various studies reached different conclusions. Deletion of the carboxy-terminal 18 amino acids did not prevent either DNA binding in vitro or transactivation of a ZRE-dependent promoter construct in vivo (23). In contrast to this, in a recently published study of a hybrid protein containing part of Zta, a strong drop in transactivator function was observed when the carboxy-terminal 5 amino acids were deleted (pZ95dletaC5) (18). Our demonstration of temperature dependence by some of the C-terminal mutants of Zta (described in Fig. 6) may explain why the contribution from the CT region had been overlooked previously. DNA complex formation is normally assayed at temperatures between 4 and 20°C, where M221ter and L225ter retain some ability to form DNA complexes. The data presented here clearly demonstrate that, under the stringent assay conditions (DNA-binding assays at 37°C), the CT region is required for the DNA-binding-dependent functions of Zta. Together this demonstrates a clear role for the CT region for dependent functions of Zta both in vitro and in vivo.
It is important that some functions of Zta are independent of complex formation with ZREs (reviewed in reference 30) and that the contribution of the CT region to those functions remains unknown. The CT region has no homology to other members of the bZIP family or other proteins in available species-wide databases (as of January 2003), so further investigation is required to ask whether the CT region may act to stabilize the coiled coil of Zta or to enhance DNA-binding function through a different mechanism.
Acknowledgments
M.R.H. and S.S.A. contributed equally to this work.
We thank Gillian McEnroe, Kerensa Jones, Kevin Clarke, Cahora Medina Palazon, Elizabeth Woods, and Victoria Frost for molecular biology assistance; Paul Farrell, Hartmut Land, Martin Rowe, and Michael Carey for reagents; and Dek Woolfson for advice.
This work was supported by grants from the Medical Research Council and The Wellcome Trust.
REFERENCES
- 1.Carey, M., J. Kolman, D. A. Katz, L. Gradoville, L. Barberis, and G. Miller. 1992. Transcriptional synergy by the Epstein-Barr virus transactivator ZEBRA. J. Virol 66:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cayrol, C., and E. Flemington. 1996. G(0)/G(1), growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J. Biol. Chem. 271:31799-31802. [DOI] [PubMed] [Google Scholar]
- 3.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G(0)/G(1) cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled-coil of HIV type 1 GP41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y.-N., D. L.-Y. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZip family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford, D. H. 2001. Biology and disease associations of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B 356:461-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrell, P., D. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flemington, E., and S. Speck. 1990. Evidence of coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc. Natl. Acad. Sci. USA 87:9459-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flemington, E., and S. H. Speck. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks, M. R., S. Balesaria, C. Medina-Palazon, M. J. Pandya, D. N. Woolfson, and A. J. Sinclair. 2001. Biophysical analysis of natural variants of the multimerization region of Epstein-Barr virus lytic-switch protein BZLF1. J. Virol. 75:5381-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicks, M. R., J. Walshaw, and D. N. Woolfson. 2002. Investigating the tolerance of coiled-coil peptides to nonheptad sequence inserts. J. Struct. Biol. 137:73-81. [DOI] [PubMed] [Google Scholar]
- 14.Kouzarides, T., G. Packham, A. Cook, and P. J. Farrell. 1991. The BZLF1 protein of EBV has a coiled coil dimerization domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene 6:195-204. [PubMed] [Google Scholar]
- 15.Krylov, D., K. Kasai, D. R. Echlin, E. J. Taparowsky, H. Arnheiter, and C. Vinson. 1997. A general method to design dominant negatives to B-HLHZip proteins that abolish DNA binding. Proc. Natl. Acad. Sci. USA 94:12274-12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 18.Martel-Renoir, D., M. Wesner, and I. Joab. 2000. Dimerization of the Epstein-Barr virus ZEBRA protein in the yeast two-hybrid system. Comparison of a ZEBRA variant with the B95-8 form. Biochimie 82:139-145. [DOI] [PubMed] [Google Scholar]
- 19.Mauser, A., E. Holley-Guthrie, D. Simpson, W. Kaufmann, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 induces both a G2 and a mitotic block. J. Virol. 76:10030-10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mauser, A., E. Holley-Guthrie, A. Zanation, W. Yarborough, W. Kaufmann, A. Klingelhutz, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 induces expression of E2F-1 and other proteins involved in cell cycle progression in primary keratinocytes and gastric carcinoma cells. J. Virol. 76:12543-12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, G. 1989. The switch between EBV latency and replication. Yale J. Biol. Med. 62:205-213. [PMC free article] [PubMed] [Google Scholar]
- 22.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Packham, G., A. Economou, C. M. Rooney, D. T. Rowe, and P. J. Farrell. 1990. Structure and function of the Epstein-Barr virus BZLF1 protein. J. Virol. 64:2110-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 25.Rodriguez, A., E. J. Jung, and E. K. Flemington. 2001. Cell cycle analysis of Epstein-Barr virus-infected cells following treatment with lytic cycle-inducing agents. J. Virol. 75:4482-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez, A., E. J. Jung, Q. Yin, C. Cayrol, and E. K. Flemington. 2001. Role of c-myc regulation in Zta-mediated induction of the cyclin-dependent kinase inhibitors p21 and p27 and cell growth arrest. Virology 284:159-169. [DOI] [PubMed] [Google Scholar]
- 27.Rooney, C. M., D. T. Rowe, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarzmann, F., M. Jager, N. Prang, and H. Wolf. 1998. The control of lytic replication of Epstein-Barr virus in B lymphocytes. Int. J. Mol. Med. 1:137-142. [DOI] [PubMed] [Google Scholar]
- 29.Sharma, V. A., J. Logan, D. S. King, R. White, and T. Alber. 1998. Sequence-based design of a peptide probe for the APC tumor suppressor protein. Curr. Biol. 8:823-830. [DOI] [PubMed] [Google Scholar]
- 30.Sinclair, A. J. bZIP proteins of human gamma herpesviruses. J. Gen. Virol., in press. [DOI] [PubMed]
- 31.Sinclair, A. J., and P. J. Farrell. 1992. Epstein-Barr virus transcription factors. Cell Growth Differ. 3:557-563. [PubMed] [Google Scholar]
- 32.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 33.Stone, S. R., M. J. Hughes, and J.-P. Jost. 1991. Qualitative and quantitative studies of protein-DNA interactions by gel mobility shift assay, p. 1-10. In J.-P. Host and H. P. Saluz (ed.), A laboratory guide to in vitro studies of protein-DNA interactions, vol. 5. Birkhauser Verlag, Basel, Switzerland.
- 34.Taylor, N., E. Flemington, J. L. Kolman, R. P. Baumann, S. H. Speck, and G. Miller. 1991. ZEBRA and a Fos-GCN4 chimeric protein differ in their DNA-binding specificities for sites in the Epstein-Barr virus BZLF1 promoter. J. Virol. 65:4033-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urier, G., B. M., P. Chambard, and A. Sergeant. 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, F. Y., H. Chen, S. E. Wang, C. M. J. apRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein α interacts with ZTA and mediates ZTA-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao, S., M. Brickner, E. I. Pares-Matos, and J. Chmielewski. 1998. Uncoiling c-Jun coiled coils: inhibitory effects of truncated Fos peptides on Jun dimerization and DNA binding in vitro. Biopolymers 47:277-283. [DOI] [PubMed] [Google Scholar]
- 38.Young, L. S., R. Lau, M. Rowe, G. Niedobitek, G. Packham, F. Shanahan, D. T. Rowe, D. Greenspan, J. S. Greenspan, A. B. Rickinson, et al. 1991. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J. Virol. 65:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]