Abstract
We have used cDNA arrays to compare the activation of various cellular genes in response to infection with Sendai viruses (SeV) that contain specific mutations. Three groups of cellular genes activated by mutant SeV infection, but not by wild-type SeV, were identified in this way. While some of these genes are well known interferon (IFN)-stimulated genes, others, such as those for interleukin-6 (IL-6) and IL-8, are not directly induced by IFN. The gene for beta IFN (IFN-β), which is critical for initiating an antiviral response, was also specifically activated in mutant SeV infections. The SeV-induced activation of IFN-β was found to depend on IFN regulatory factor 3, and the activation of all three cellular genes was independent of IFN signaling. Mutations that disrupt four distinct elements in the SeV genome (the leader RNA, two regions of the C protein, and the V protein) all lead to enhanced levels of IFN-β mRNA, and at least three of these viral genes also appear to be involved in preventing activation of IL-8. Our results suggest that SeV targets the inflammatory and adaptive immune responses as well as the IFN-induced intracellular antiviral state by using a multifaceted approach.
Alpha/beta interferons (IFN-α/β) are cytokines that act in a pleiotropic manner to limit viral replication and spread (2, 57). In fibroblasts (e.g., the bronchial epithelial target of many paramyxoviruses), the product of the single IFN-β gene is directly induced by viral infection, and IFN-β feeds back onto cells in an autocrine manner to induce multiple IFN-α genes and in a paracrine manner to prime neighboring cells for their possible infection (60). Since most viruses induce IFN-β to some extent, intracellular double-stranded RNA (dsRNA) generated from the viral genome is traditionally assumed to be the common signature of virus replication that sets the IFN system in motion (22, 32). dsRNA is thought to induce the formation of an enhanceosome at the IFN-β promoter that includes IFN regulatory factor 3 (IRF-3) and NF-κB (among other transcription factors) (65). IFNs induce a cellular state that is nonconducive for viral replication by signaling through their cell surface receptor, leading to the phosphorylation of cytoplasmic STAT proteins and their nuclear translocation. IFN-α/β responses are regulated primarily via IFN-stimulated gene (ISG) factor 3, a heterotrimeric transcription factor composed of STAT1, STAT2, and IRF-9 (p48). ISG factor 3 binds to a DNA element (IFN-stimulated response element) in the promoters of ISGs and activates their expression (7).
The extravasation of neutrophils, eosinophils, basophils, and mononuclear cells is the salient feature of the innate response to microorganisms in the lung. Localized and systemic pro- and anti-inflammatory cytokines thus also play an important role in the outcome of viral infection and pathogenicity of this organ (58). The CC chemokine interleukin-8 (IL-8) is secreted from epithelial surfaces in a polar fashion during infection with pathogenic bacteria such as Salmonella enterica serovar Typhimurium and sets up a subepithelial chemotactic gradient directing neutrophils and other immune cells to the site of infection (27). In polarized epithelial monolayers, S. enterica serovar Typhimurium-induced IL-8 expression is controlled via the activation of the mitogen-activated protein kinase cascade and IκBα kinase, followed by NF-κB translocation to the nucleus and production of IL-8 mRNA. IL-8 secretion by primary human monocytes in response to dengue virus infection is also tightly linked to NF-κB activation (3). Sendai virus (SeV) infection of human embryonic kidney 293 cells induces the expression of the CXC chemokine RANTES in an IRF-3- and NF-κB-dependent manner (23, 41). NF-κB, like IRF-3, is found in the cytoplasm of unstimulated cells, retained in a complex with the inhibitory IκB proteins. Upon stimulation with many inducers, including dsRNA and virus infection, IκB is rapidly phosphorylated and degraded, resulting in NF-κB release and translocation to the nucleus (30, 33).
Given the importance of the host innate immune response to virus infection, viruses have, during their coevolution with cells, developed strategies to regulate cytokine synthesis and action. SeV, a model paramyxovirus and respiratory pathogen of mice, is known to use its C protein to evade the host interferon response by at least two mechanisms. (i) C binds STAT1, preventing its activation in response to IFN, and the carboxyl part of the C protein (i.e., residues 24 to 204, or the Y proteins) is sufficient for this activity. A phenylalanine at position 170 of C is also critical for blocking STAT1 activation (18, 59). (ii) C also targets STAT1 for degradation, and the amino-terminal residues of the C proteins (residues 1 to 23, which are absent in the Y proteins) are essential for this activity (reference 17 and references therein).
This paper reports that SeVs carrying specific mutations in the C gene, in contrast to wild-type SeV (SeV-wt), activate IL-8 and IFN-β expression as well as that of several ISGs. Our results suggest that the products of virtually all of the viral accessory genes (C and V proteins and leader RNA) act to prevent the expression of these cellular genes that are central to the overall host antiviral response.
MATERIALS AND METHODS
Cells and viruses.
2C4 cells (39), 2fTGH cells (48), and their derived cell lines U3A (45) and U5A and U5A+IFNAR (43) were obtained from IM Kerr (Imperial Cancer Research Fund, London, United Kingdom) and grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in the presence of the relevant maintenance drug (hygromycin at 250 μg/ml or G418 at 400 μg/ml). The generation of recombinant SeV (rSeV) expressing alternate C and V (and P) proteins is described elsewhere (8, 19, 37, 38). All SeV stocks were grown in the allantoic cavities of 10-day-old embryonated chicken eggs. Virus present in the allantoic fluid was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Coomassie blue staining after virus pelleting. Virus titers were determined by plaquing on LLC-MK2 cells.
Virus infections.
Cells were infected at a multiplicity of infection of 20 in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. After an absorption period of 1 to 2 h, the inoculum was removed and replaced with fresh medium.
Plasmids, transient transfections, and luciferase assay.
IRF-3 (54), IRF-3ΔN (41), and IRF-3-ribozyme (67) were obtained from John Hiscott and Paula Pitha. pDsRed2, expressing red fluorescent protein (RFP), was from Clontech. The reporter plasmid with the firefly luciferase gene under the control of the human IFN-β promoter was described by King and Goodbourn (35) and is referred to here as pβ-IFN-fl-lucter. pTK-rl-lucter, used as a transfection standard, contains the herpes simplex virus thymidine kinase promoter region upstream of the Renilla luciferase gene (Promega). For transfections, 100,000 cells were plated in six-well plates 20 h before transfection with 1 μg of pβ-IFN-fl-lucter, 0.3 μg of pTK-rl-lucter, 1 μg of IRF-3-expressing plasmid, and 6.9 μl of Fugene (Roche) according to the manufacturer's instructions. At 24 h posttransfection, the cells were (or were not) infected with SeV recombinants or treated with 50 μg of poly(I)-poly(C) (Sigma, St. Louis, Mo.) per ml. Twenty hours later, cells were harvested and assayed for firefly and Renilla luciferase activities (dual-luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those of Renilla luciferase.
RNA extraction and quantification.
Total RNA was extracted with Trizol reagent (Invitrogen), and two dilutions were electrophoresed on agarose-HCHO gels. The gels were stained with ethidium bromide, and the intensities of the 18S and 28S rRNA bands were quantified by using the ChemiDoc System (Bio-Rad) and One-D-scan software. All samples were diluted to a final concentration of 1 μg/μl so that their subsequent transcription into DNA, if not quantitative, would be little influenced by this parameter.
RT and real-time PCR via TaqMan.
Ten microliters of total RNA was mixed with 0.5 μg of random hexamer primer (Promega) and subjected to a reverse transcription (RT) reaction with Superscript enzyme (Gibco), as described by the manufacturer, in a total volume of 50 μl. Two microliters of each cDNA was then combined with 1 μl of internal control (either 20× ribosomal 18S or human glyceraldehyde-3-phosphate dehydrogenase [GAPDH] [Applied Biosystems]), 11 μl of MasterMix (Eurogentec), 20 pmol (each) of forward and reverse primers, and 4.4 pmol of TaqMan probe in a total volume of 22 μl. The following primers and probes (Eurogentec or Microsynth) were used: for the IFN-β gene, 5′-CAGCAATTTTCAGTGTCAGAAGCT-5′ (forward), 5′-TCATCCTGTCCTTGAGGCAGT-3′ (reverse), and 5′-CTGTGGCAATTGAATGGGAGGCTTGA-3′ (probe); for the IL-8 gene, 5′-CGGTGGCTCTCTTGGCAG-3′ (forward), 5′-TTAGCACTCCTTGGCAAAACT-3′ (reverse), and 5′-CTTCCTGATTTCTGCAGCTCTGTGTGAAGGT-3′ (probe); for the IL-6 gene, 5′-TCCAGGAGCCCAGCTATGAA-3′ (forward), 5′-CCCAGGGAGAAGGCAACTG-3′ (reverse), and 5′-TCCTTCTCCACAAGCGCCTTCGG-3′ (probe); for the 6-16 gene, 5′-CCTGCTGCTCTTCACTTGCA-3′ (forward), 5′-AGCCGCTGTCCGAGCTC-3′ (reverse), and 5′-TGGAGGCAGGTAAGAAAAAGTGCTGCTCGG-3′ (probe); for the N gene of SeVZ, 5′-GCAATAACGGTGTCGATCACG-3′ (forward), 5′-GATCCTAGATTCCTCCTACCCCA-3′ (reverse), and 5′-CGAAGATGACGATACCGCAGCAGTAGC-3′ (probe); and for the N gene of SeVM, 5′-CGAAGAGGATGATGCCGC-3′ (forward), 5′-GGGTCATGTATCCTAAATCCTCGT-3′ (reverse), and 5′-CAGCAGCTGGGATGGGAGGAAT-3′ (probe). Real-time PCR was carried out in a 7700 sequence detector (Applied Biosystems, Foster City, Calif.).
Generation of customized cDNA arrays.
Macroarrays were prepared as described previously (55). 5′ IMAGE clones 0.5 to 0.8 kb in length were chosen and obtained from the Human Genome Mapping Project (Hinxton, United Kingdom), plated onto L agar plates and grown overnight at 37°C. Single colonies were picked and propagated overnight in Luria-Bertani medium containing 50 μg of ampicillin per ml. Bacterial lysates were generated by 1:10 dilution in distilled water. From these lysates, inserts were amplified by PCR as described above. After purification (QIAquick PCR purification kit; Qiagen, Crawley, United Kingdom), PCR products were sequenced (ABI Prism; Applied Biosystems). PCR-amplified cDNAs were transferred into 96-well plates and spotted manually onto dry nylon membranes (Hybond N+; Amersham Pharmacia, Little Chalfont, United Kingdom) in triplicates by using 96-pin replicators (Nalge Nunc, Naperville, Ill.; V&P Scientific, San Diego, Calif.). Membranes were air dried, denatured by alkaline treatment, and then neutralized. The membranes were again air dried and UV cross-linked prior to the experiment.
Generation of labeled cDNA, hybridization, washing of membranes and analysis.
Radiolabeled cDNA was generated from 10 μg of total RNA by RT with 400 U of reverse transcriptase (Superscript II; Gibco) in the presence of 30 μCi of [α-33P]dCTP. After RT, residual RNA was hydrolyzed by alkaline treatment at 70°C for 20 min. For removal of unincorporated nucleotides, the cDNA was purified by using G-50 columns (Amersham Pharmacia) according to the instructions of the manufacturer. Before hybridization to the arrays, the labeled cDNA was mixed with 50 μg of COT-DNA (Gibco) and 10 μg of poly(A) DNA (Sigma), denatured at 95°C for 5 min, and hybridized for 1 h to minimize nonspecific binding. The cDNA was then added directly to the membranes, which had been prehybridized in 20 ml of hybridization buffer for at least 30 min. The membranes were hybridized for 16 h at 65°C in hybridization bottles (Amersham Pharmacia) in a rotary hybridization oven. After hybridization, the hybridization buffer was discarded and replaced by 150 ml of washing buffer. The membranes were washed once in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS, twice in 0.2× SSC-0.1% SDS, and once in 0.1× SSC-0.1% SDS for 20 min each at 65°C. The membranes were then exposed to phoshorimage screens for 48 h and scanned with a phosphorimager (Storm; Molecular Dynamics, Little Chalfont, United Kingdom). For analysis, images were analyzed with ImageQuant (Molecular Dynamics). Further data analysis was performed with Excel (Microsoft).
IL-8 assay.
Levels of IL-8 in supernatants and in cell lysates were determined by a sandwich enzyme-linked immunosorbent assay with paired monoclonal antibodies (Pierce) as described by the manufacturer. The IL-8 concentrations were normalized by using total protein levels in the lysate.
RESULTS
In our experiments we have used two strains (or lineages) of SeV whose history is relevant to studies of virulence. The natural host of SeV has not been clearly identified, and this virus is sometimes referred to as murine parainfluenza virus type I because it efficiently infects mice, causes disease, and spreads readily to uninfected animals. However, there is no virologic or serologic evidence of SeV in wild mouse populations (29). There are two known lineages of SeV, Z/H/Fushimi and Ohita M/Hamanatsu (12, 31, 53, 62). The nucleotide sequences within each lineage are 99% identical, and they are 89% identical between lineages. Z/H/Fushimi comes from viruses isolated in the early 1950s after an epidemic of pneumonitis of newborn infants in Sendai, Japan (29, 56). These SeVs have been passaged extensively in eggs in various labs since the 1950s, and they are moderately virulent for mice (50% lethal dose [LD50] = 103 to 104 PFU). All of the SeVZ strains used in this study (including the wt) are recombinants.
Ohita M (SeVM) and Hamanatsu, in contrast, are highly virulent (LD50, <102), low-egg-passage (nonrecombinant) viruses isolated from two completely separate, very severe epidemics of animal houses in Japan. This lineage is presumably closer to the virus in its natural (unknown) host, and it is known that SeV passage in eggs attenuates its virulence in mice. SeVM grew poorly in cell culture, and a clear-plaque variant emerged that was avirulent (LD50, >105) and contained only two amino acid substitutions, CF170S and LE2050A (31). When placed in the rSeVZ background, the CF170S mutation was found to account for most or all of the loss of virulence (15). This virus is referred to here as SeVM-CF170S. SeVM-CF170S appeared to initiate the infection of mice normally, but the infection was limited to the first day. This was the first evidence that the C gene, like the SeV V gene (34), was involved in countering host innate defenses. Although SeVZ-wt is attenuated relative to SeVM-wt in laboratory mice, rSeVZ-wt still replicates as efficiently in the monkey and chimpanzee models of human respiratory disease as human parainfluenza virus type 1 (56), the virus which is most closely related to SeV and which is endemic in children.
We have used a cDNA array designed to study the human cell response to IFN-α/β (55) to monitor the effects of various SeV infections on host mRNA levels. Around 150 genes of interest were selected from the UniGene database. These genes comprise known ISGs and genes of intrinsic interest which might or might not be induced by IFNs in different cell systems. They include genes involved in cell proliferation, immune responses, and the responses to a variety of cytokines (see Table 1 of reference 55). We compared matched sets of SeV carrying two different mutations in the C gene (SeVM-CF170S and SeVZ-CΔ10-15), whose products interact with STAT1 in different ways (to interfere with IFN signaling and to induce STAT1 instability, respectively) (Fig. 1D). We also used matched sets of SeV carrying mutations in the viral replication promoters (SeVZ-GP1-42 and -GP31-42) that prevent apoptosis and lead to persistent infections (the numbers refer to the nucleotides of the genomic promoter that have been replaced with the equivalent sequences of the antigenomic promoter) (16, 20). Promoter mutations are thought to act via mutant leader RNAs that are abundantly transcribed from the genomic replication promoter and which bind to cellular RNA-binding proteins that regulate mRNA fate (28).
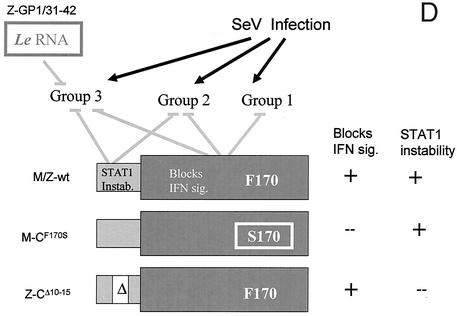
FIG. 1.
Comparison of host mRNA levels in 2fTGH cells infected with various SeVs. (A to C) Parallel cultures of 2fTGH cells were infected with 20 PFU of the various SeVs per cell. Total cytoplasmic RNA was prepared with Trizol at 24 hpi, and equal amounts (10 μg) were used as a template for oligo(dT)-primed [33P]cDNA synthesis. The [33P]cDNA was annealed to triplicate DNAs arrayed on nylon membranes, and the [33P]cDNA bound was quantitated in a PhosphorImager. The graphs show the fold increase in each mRNA relative to the mock control. (D) Schematic representation of the viral mutations and their effects on host gene activation. The C proteins are shown as two telescoping boxes representing the longer (C′ and C) and shorter (Y1 and Y2) C proteins, whose activities during infection, and the mutations investigated, are indicated. The promoter mutation GP42 is thought to exert its effect via mutant leader (Le) RNA. The presumed requirement for the various wt genetic elements to prevent host gene activation is shown. The names of the mutant SeVs used are also indicated. Instab., instability; sig., signaling.
Upon infection with SeVM-wt or SeVZ-wt, many of these mRNA levels remain unchanged (Fig. 1A to C). This lack of response is presumably due in part to active SeV countermeasures that neutralize the cell's antiviral response (24). Upon infection with the mutant viruses, the mRNA levels of 15 of the 150 genes examined were elevated, and three patterns of gene activation were seen (all values are triplicates, and a twofold difference is very significant [55]). One series of genes (group 1, nine genes) (Fig. 1A) is activated by SeV-CF170S infection alone; these mRNA levels are unchanged in SeV-CΔ10-15 and SeV-GP1/31-42 infections. The CF170S substitution inactivates the ability of all four C proteins (C′, C, Y1, and Y2) to stably bind STAT1 and to interdict IFN signaling (18, 59). According to this view, any of the four C proteins may function to prevent these mRNA levels from increasing during SeV infection (Fig. 1D) (14). The IL-6 gene is the sole representative of group 2; it is activated by SeV-CΔ10-15 as well as SeV-CF170S, but not by the promoter mutants or the wt viruses (Fig. 1B). According to this view, a second function of the C gene, specific to the NH2-terminal 23 amino acids present only in the longer C proteins, is also required for SeV to prevent IL-6 activation. The third group, consisting of five genes, is activated by SeV-CΔ10-15 and SeV-GP1-42 infections as well as SeV-CF170S, but not by SeV-GP31-42 or SeV-wt infections (Fig. 1C). Apparently, a third function provided specifically by the first 30 nucleotides (nt) of the genomic promoter (or leader RNA) is also required to prevent activation of genes such as that of IL-8 (Fig. 1C). This third function is not the ability of mutant leader RNA to bind TIAR, a host RNA-binding protein important for virus-induced apoptosis, as this occurs with SeV-GP31-42 as well (28). In summary, comparative analysis of host gene activation with SeV with specific mutations has identified three groups of cellular genes that respond differently to SeV infection.
Real-time RT-PCR estimations of mRNA levels.
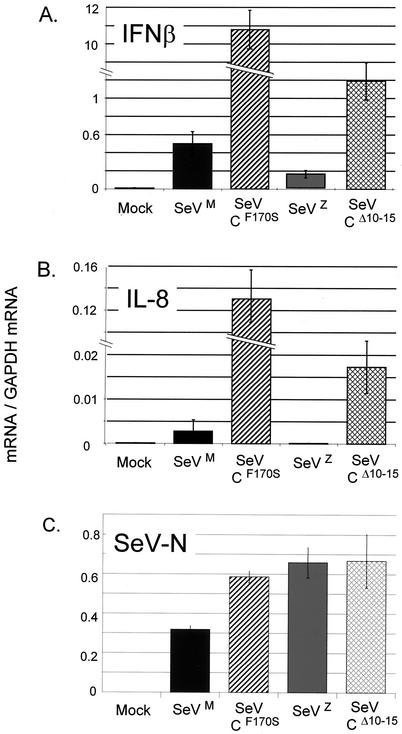
IFN signaling through the JAK/STAT pathway activates many ISGs (such as 6-16, PKR, etc.) that collectively contribute to the cellular antiviral response. SeVs that interdict IFN signaling would therefore also prevent the activation of these ISGs. While many of the genes activated by the mutant SeVs are well known ISGs, IL-6 and IL-8 are known to be non-ISGs; treatment of 2fTGH cells with 1,000 IU of IFN-α does not increase IL-6 or IL-8 mRNA levels over those of the untreated control (55). We therefore continued our study of selected host genes activated by SeV by real-time RT-PCR (TaqMan), a method that is more quantitative than DNA arrays. We first examined IL-8 (a chemokine) and IFN-β, an early host response protein whose gene was absent in the DNA array. 2C4 cells (a 2fTGH-derived cell line) were infected with 20 PFU of the various SeVs per cell, and the levels of various host mRNAs were determined, including that of GADPH as an internal control (see Materials and Methods). All four SeVs (SeVM-wt, CF170S, SeVZ-wt, and CΔ10-15) grow relatively well in 2C4 cells, as indicated by their accumulated N mRNAs (Fig. 2 and 3) or N proteins (data not shown); however, the SeVM-wt infections accumulated ca. 2-fold less N mRNA than the three other SeV infections (Fig. 2C). The relative levels of virus replication are presumably important in stimulating the host response, and SeVM-wt infections might therefore be expected to activate IL-8 and IFN-β less strongly than the other SeVs for this reason alone. We found that IL-8 and IFN-β mRNA levels were elevated >20-fold in SeVM-CF170S versus SeVM-C-wt infections. A strong difference was also found between SeVZ-CΔ10-15 and SeVZ-C-wt infections, where N mRNAs had accumulated identically (Fig. 2). Moreover, IL-8 and IFN-β mRNA levels were also elevated in other SeVM-CF170S versus SeVM-C-wt infections of 2C4 cells, where N mRNAs had accumulated identically (Fig. 3). Thus, specific mutations in two different regions of the SeV C proteins lead to increased activation of IL-8 and IFN-β. In all cases, IL-8 and IFN-β mRNA levels were more strongly increased by SeVM-CF170S than by SeVZ-CΔ10-15.
FIG. 2.
Effects of SeV C gene mutations on IFN-β and IL-8 mRNA levels during infection. Parallel cultures of 2C4 cells were infected (or not) in triplicate for 24 h with 20 PFU of the various SeVs per cell. Total cytoplasmic RNA was prepared from each culture, and the same amount of RNA (ca. 1 μg) was transcribed into cDNA with random hexadeoxynucleotides and murine leukemia virus reverse transcriptase. The relative amounts of IFN-β and IL-8 gene sequences, relative to that of GADPH as an internal control, were determined by real-time PCR (see Materials and Methods). The average levels of the mRNAs and their deviations in the triplicate infections are shown. The relative SeV N mRNA levels of the SeVZ-C-wt and -CΔ10-15 infections were also determined (SeVZ and SeVM are 10% different in sequence, and their detection requires different primers and probes).
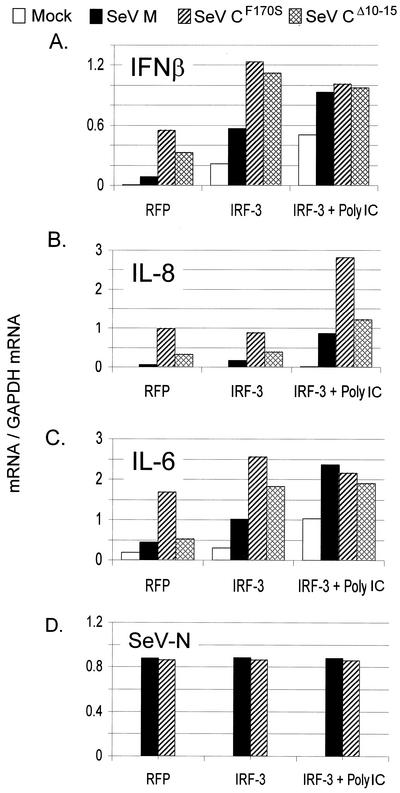
FIG. 3.
Effects of IRF-3 and dsRNA treatment on SeV-induced host gene activation. Parallel cultures of 2C4 cells were transfected with pRFP or pIRF-3. The cultures were then infected (or not) with 20 PFU of the various SeVs per cell at 20 h posttransfection, and some of the cultures were also treated with 50 μg of poly(I)-poly(C) per ml. The cells were harvested at 24 hpi. The relative amounts of IFN-β, IL-6, and IL-8 gene sequences present were determined as described for Fig. 2.
We also examined the effects of various SeV infections of cells transfected with plasmids expressing IRF-3 (or RFP as a neutral control), and in some cases the cells were also treated with 50 μg of poly(I)-poly(C) (dsRNA) per ml. Elevated IRF-3 levels should enhance the antiviral response of the cells to SeV infection, and the combined treatment is thought to approximate a virus infection in itself and should potentiate the antiviral response. This appears to be so, as the levels of IL-6, IL-8, and IFN-β mRNAs induced by SeV infection in general increased with increasing proresponse treatment (i.e., RFP, IRF-3, and IRF-3 plus dsRNA) (Fig. 3). IRF-3 overexpression (Fig. 3A to C, IRF-3 versus RFP) enhanced IFN-β and IL-6 activation by SeV but did not enhance that of IL-8. The additional dsRNA treatment had little effect on further enhancing IFN-β and IL-6 mRNA levels but strongly enhanced IL-8 mRNA levels. These differences in the enhancement of IL-6 and -8 and IFN-β activation upon treatment with IRF-3 with or without dsRNA presumably reflect different activation pathways in response to SeV infection. It is possible that IL-8 activation requires both IRF-3 and dsRNA, whereas IRF-3 is sufficient for IL-6 and IFN-β activation.
SeV activation of IL-6, IL-8, and IFN-β is independent of IFN signaling.
2fTGH human fibrosarcoma cells were chosen for these experiments because sublines defective in specific components of the IFN signaling system have been generated from these cells by X irradiation (48). U5A cells, for example, are defective in the IFN-α/β receptor 2 chain, which is essential for IFN-α/β signaling, and these cells have been restored to IFN sensitivity by complementation with the IFNAR2 gene (U5A+IFNAR2 cells) (43). Even though IL-6 and -8 are not activated upon simple IFN treatment of uninfected cells, IFN secreted during SeV infection may act differently, as additional signaling pathways are being induced by the virus infection. Moreover, CF170S (in contrast to CΔ10-15) does not prevent IFN signaling, and it is important to know whether this phenotype is responsible for the activation of IL-6 and -8. We therefore examined the various SeV infections of U5A as well U5A+IFNAR2 cells to determine whether activation of IL-6 and IFN-β by SeV-CF170S and SeV-CΔ10-15 required IFN signaling. We also examined the activation of the 6-16 gene, a known ISG, as a positive control (10). As shown in Fig. 4, both SeV-CF170S and SeV-CΔ10-15 activated IL-6, IL-8, and IFN-β in U5A cells relative to SeV-wt infection. The IFNAR2-complemented cell line yielded similar results, except that the activation of these genes was paradoxically reduced in U5A+IFNAR2 cells relative to U5A cells. In contrast to the case for IL-6, IL-8, and IFN-β, little or no activation of 6-16 occurred in SeV-C mutant-infected U5A cells, whereas a modest activation was evident in U5A+IFNAR2 cells. Moreover, 6-16 was the only mRNA whose levels in U5A+IFNAR2 cells exceeded those in U5A cells. The activation of IL-6, IL-8, and IFN-β during SeV infection, in contrast to that of 6-16, is thus largely independent of IFN signaling.
FIG. 4.
SeV infection of cells defective for the IFN receptor and their complemented pseudo-wt derivatives. (A to D) Parallel cultures of U5A or U5A+IFNAR2 cells were infected (or not) in duplicate with 20 PFU of the various SeVs per cell for 24 h. The relative amounts of IFN-β, IL-6, IL-8, and 6-16 mRNAs present were determined as described for Fig. 2. (E and F) Equal samples of the culture supernatants (F) or cytoplasmic extracts of the cultures (E) were analyzed for IL-8 protein levels by enzyme-linked immunosorbent assay.
STAT1-defective U3A cells.
The SeV C proteins interact with STAT1 in two ways (Fig. 1D). C and STAT1 form a stable complex in vitro and during SeV infection, and this complex is associated with a loss of IFN signaling. These events are blocked by the CF170S mutation but not by CΔ10-15. The shorter Y proteins are also active in this respect. The longer C proteins alone also induce STAT1 instability, and in contrast to their effects on IFN signaling, this effect does not require F170 (Fig. 1D). To examine whether SeV-C mutant-induced activation of IL-6, IL-8, and IFN-β requires STAT1, U3A cells, which are known to be defective for STAT1, were examined (45). However, we were unable to examine the companion U3A+STAT1 cells, as these cells were found to have lost STAT1 expression. Moreover, attempts to recomplement U3A cells with STAT1 failed (data not shown).
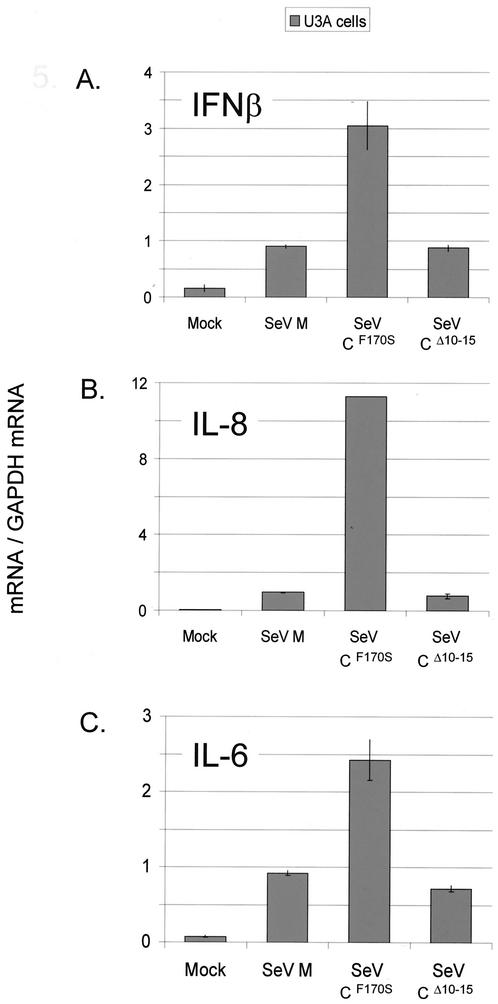
The results of the U3A cell infections are shown in Fig. 5. As before, IL-6 and -8 and IFN-β mRNA levels were all clearly increased in SeV-CF170S infections relative to SeV-wt infections. However, in contrast to the case for U5A and 2fTGH cells, SeV-CΔ10-15 infections did not contain enhanced mRNA levels relative to SeV-wt infections. Given that STAT1 is probably not the only gene that has been lost in U3A cells that have been X irradiated (which randomly destroys chromosomal DNA), we can conclude only that enhanced mRNA levels due to SeV-CΔ10-15 infection require STAT1 and/or another, unknown gene, whereas activation due to SeV-CF170S requires neither STAT1 nor any other gene destroyed in U3A cells.
FIG. 5.
SeV infection of cells defective for STAT1. Parallel cultures of U3A cells were infected (or not) in duplicate with 20 PFU of the various SeVs per cell for 24 h. The relative amounts of IFN-β, IL-6, and IL-8 mRNAs present were determined as described for Fig. 2.
IL-8 gene activation and IL-8 secretion.
IL-8 is not known to act intracellularly. We therefore examined whether the SeV-induced IL-8 gene activation in U5A/U5A+IFNAR2 cells also led to increased IL-8 protein synthesis and secretion. When IL-8 protein levels in cytoplasmic extracts of the various SeV-infected cells were examined, they were found to roughly mirror the mRNA levels (Fig. 4E). However, when the culture supernatants were examined, a somewhat different result was found (Fig. 4F). Whereas the increased IL-8 mRNA level of SeV-CΔ10-15-infected U5A cells was accompanied by strongly increased IL-8 secretion, that of U5A+IFNAR2 cells led to only a modest increase in IL-8 secretion. Moreover, the increased IL-8 mRNA levels in either SeV-CF170S-infected cell line did not lead to clearly increased IL-8 secretion.
IL-8 expression can be controlled at both the transcriptional and posttranscriptional levels. In polarized epithelial monolayers, S. enterica serovar Typhimurium-induced IL-8 secretion requires not only the activation of NF-κB and production of IL-8 mRNA but also the activation of the small, Rho family GTPases Cdc42 and Rac1, which regulate endocytic protein traffic from the Golgi network to the basolateral surface of the cell. In the absence of Cdc42 or Rac1 function, IL-8 mRNA levels increase in response to bacterial infection but IL-8 is not secreted (3), similar to the case for our SeV-CF170S-infected U5A cells. The requirement for Cdc42 and Rac1 activation, moreover, is cell type dependent (9). The CF170S and CΔ10-15 mutations thus appear to affect IL-8 secretion differently.
SeV prevents IFN-β gene activation in several ways.
The manner in which IFN-β transcription is induced by virus infection is well studied, and the activation of IRF-3 is central to this process. IRF-3 is expressed constitutively and is found in the cytoplasm in an inactive, unphosphorylated state. Upon virus infection or dsRNA treatment of cells, IRF-3 is phosphorylated by an unknown kinase and translocates to the nucleus, where, together with other transcription factors such as NF-κB (which is itself also directly activated by virus infection or dsRNA), it activates IFN-β transcription (41, 65, 68). Phosphorylation of IRF-3 after viral infection is the first step in the activation of a gene program that includes a positive feedback loop of IFN-α/β and IRF family members (60). The results described above suggest that the SeV C gene encodes functions that prevent virus-induced IFN-β transcription (directly or indirectly). During the course of this work, it was reported that the SeV V protein, as well as the V proteins of the rubulaviruses SV5 and hPIV2, also prevented IFN-β transcription (26, 50, 64).
Given that SeV appears to use two viral genes (C and V) to neutralize IFN-β expression, we have examined a broader panel of mutant rSeV infections for their relative activation of the IFN-β promoter compared to that of dsRNA treatment. Besides SeV-CF170S and CΔ10-15, we examined two promoter mutants, SeV-AGP55, in which the first 55 nt of the antigenomic promoter is replaced with the equivalent leader sequences of the genomic promoter (38). SeV-AGP55 transcribes leader RNA from both promoters (and no trailer RNA). The converse SeV-GP48 has the first 48 nt of the genomic promoter replaced with the equivalent trailer sequences, and SeV-GP48 transcribes basically trailer RNA from both promoters (and no leader RNA; GP48 and GP1-42 are identical in this respect [data not shown]) (19, 20). Finally, we examined SeV-V−/W++, which contains a stop codon at the beginning of the V open reading frame (ORF), such that edited V mRNAs are translated into W-like proteins, and specifically no V protein is expressed (8).
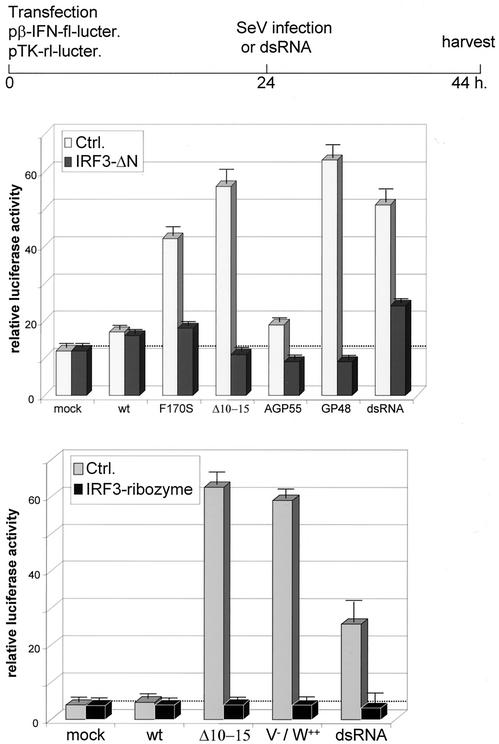
2fTGH cells were transfected with a reporter plasmid in which luciferase is controlled by the IFN-β promoter (pIFNβ-luciferase). To determine whether virus-induced IFN-β transcription required IRF-3 activation, the cells were cotransfected with either a dominant-negative mutant of IRF-3 (IRF-3ΔN), an anti-IRF-3 ribozyme, or an empty control plasmid (see Materials and Methods). The transfected cultures were then infected (or not) with the various SeVs (or treated with dsRNA) 24 h later and were harvested at 20 h postinfection (hpi). As shown in Fig. 6, with the notable exception of SeV-AGP55, all of the mutant SeV infections induced the reporter more strongly than SeV-wt and as well as dsRNA treatment. In all cases, the coexpression of IRF-3ΔN or an anti-IRF-3 ribozyme prevented the SeV-induced expression of the reporter. Thus, mutations in two regions of the C protein that carry out different functions, as well as the loss of leader RNA expression or the expression of the V protein, all lead to IFN-β promoter activation in an IRF-3-dependent manner. Overexpression of the W protein cannot compensate for the lack of V protein, so the highly conserved Cys-rich carboxyl domain of V is specifically required. Only the loss of trailer RNA expression (SeV-AGP55) did not result in IFN-β activation.
FIG. 6.
Effect of SeV infection on IFN-β promoter reporter gene expression. Parallel cultures of 2fTGH cells were transfected with a mixture of pINFβ-fl-lucter and pTK-rl-lucter along with pIRF-3-ΔN, pAnti-IRF-3-ribozyme, or an empty plasmid (Ctrl), as indicated in Materials and Methods. Duplicate cultures were infected (or not) with 20 PFU of the various SeVs per cell or treated with dsRNA and harvested at 20 hpi. The relative activities of the fl- and rl-luciferases were determined. Error bars indicate standard deviations.
Leader and trailer RNAs, the promoter-proximal products of viral RNA synthesis, are AU rich and are thought to bind to cellular RNA-binding proteins that bind AU-rich elements (28). In contrast to leader RNA, trailer RNA is expressed relatively late in infection (upon antigenome accumulation) and would not be expected to counteract immediate-early response genes. However, many of the other viral products that do not have a dedicated essential role in the replication machinery per se (C and V proteins and leader RNA) appear to be pressed into service to coordinately prevent IFN-β activation.
DISCUSSION
We have used cDNA arrays to compare the activation of various cellular genes in response to infection with SeVs that contain mutations in their C proteins or leader regions. Three groups of cellular genes were identified in this way (Fig. 1). Of the mutant SeV examined by DNA arrays, only the leader(31-42) mutation failed to activate any of the cellular genes relative to SeV-wt, and consistent with this failure, SeV-GP31-42 was the only one of the four mutant SeVs not to have lost virulence in mice (M. Itoh, unpublished data). The CF170S mutation is associated with a ca. 5-log-unit increase in LD50 (31), and this mutation appears to be the most important in cell culture infections as well as in mice (15). This mutation activates IL-6, IL-8, and IFN-β more strongly than CΔ10-15, and only this mutation activates all three groups of cellular genes that respond to SeV infection.
The IFN-β gene is both a primary response gene and an ISG, and it plays a central role in initiating the IFN-induced antiviral response. This is the first report that the SeV C proteins and leader RNA counteract the expression of this key primary response gene, and it confirms that the report of Poole et al. (50) that the product of our hemagglutinin-tagged V gene is active in this respect. As expected, activation of IFN-β required activation of IRF-3 (Fig. 6) and was independent of IFN signaling (Fig. 4), but the details of how this occurs remain to be elucidated. We have also provided evidence that the SeV C proteins and leader RNA counteract the expression of the chemokine IL-8. Infections by other viruses, e.g., respiratory syncytial virus (RSV) (44), dengue virus (3), hepatitis C virus (49), and human immunodeficiency virus type 1 (46, 52), are known to induce IL-8 secretion, as do infections by bacteria and parasites, including Mycobacterium tuberculosis (66). In a related vein, cytomegalovirus, a large DNA virus, encodes a chemokine receptor that may facilitate virus replication (13), and human herpesvirus 8/Kaposi's sarcoma virus carries four ORFs whose products are related to chemokines (42). Viral modulation of chemokine expression presumably represents one aspect of the continuous battle between viral parasites and antiviral, inflammatory, and immune responses of the host. SeV infection has been reported to induce the CXC chemokine RANTES via the activation of IRF-3 and NF-κB (23, 40). IL-8 may not have been noticed in these earlier studies, because SeV-wt induces very little IL-8 (Fig. 2). Our results suggest that SeV targets the inflammatory and adaptive immune responses (IL-6 and IL-8) as well as the IFN-induced intracellular antiviral state (IFN-β and STAT1). As IFN-β and IL-8 transcription both depend on NF-κB activation, SeV may target this key transcription factor as well.
The leader(1-42), CΔ10-15, CF170S, and V−/W++ mutations appear to disrupt four distinct elements in the SeV program to counteract the cellular antiviral response. The facts that they all lead to enhanced levels of IFN-β mRNA and that at least three of them increase IL-8 mRNA levels suggest that SeV employs a multifaceted approach to inhibit viral clearance by inflammatory cells as well as to prevent the IFN-induced antiviral state, sometimes using the same viral macromolecules due to its limited coding capacity. The best-studied example of paramyxovirus-induced activation of IL-8 is that of RSV (21, 69). The IL-8 promoter in A549 cells can be induced by RSV infection in at least three distinct pathways: via tumor necrosis factor alpha (which requires only an intact NF-κB binding site), directly by intracellular RSV replication (which also requires other transcription factor binding sites) (5), and via the interaction of the viral F protein with Toll-like receptor 4 (in which IRF-3 plays an important role) (36). Measles virus H protein interaction with Toll-like receptor 2 also activates IL-8 (1). If all three parallel cellular pathways for IL-8 expression operate during SeV infection of 2fTGH cells, several different SeV products will be required to effectively prevent IL-8 activation (Fig. 1D).
Rubulaviruses do not express C proteins, but their V proteins have recently been found to prevent IFN-β expression by preventing the activation of IRF-3 and NF-κB, as well as inducing the degradation of STAT1 or STAT2 (26, 50). The Rubulavirus V proteins thus also counteract more than one arm of the innate antiviral response. The versatility of these viral gene products continues to surprise us. The SeV C proteins have been more intensively studied than SeV V protein or leader RNA. Like the influenza A virus NS1 protein (6, 63) and hepatitis C virus NS5A protein (49), the SeV C proteins are pleiotropic polypeptides that have multiple activities during infection, presumably due to their interaction with various viral and cellular proteins. Their multiple functions, deciphered in large part via C gene mutations, include (i) stimulation of viral RNA synthesis early in infection (SeV-C′/C− infections exhibit a 10-h delay in the accumulation of viral products) (37); (ii) inhibition of viral RNA synthesis in a promoter-specific manner late in infection, by interacting with the P4-L vRdRP (this selective inhibition may promote the switch from mRNA synthesis to genome replication and increase the fidelity of vRdRP promoter recognition) (4, 61); (iii) a role in virion assembly, possibly by interaction with the matrix (M) protein (SeV-4C− particles are poorly infectious and amorphic) (25); (iv) interaction with STAT1 in two separate ways, to inhibit IFN signaling and to induce STAT1 instability (17); and (v) inhibition of the IRF-3-dependent activation of IFN-β and the activation of IL-8 expression in an IFN signaling-independent manner (this work).
How C interacts specifically with all of its viral and cellular partners remains an enigma and is reminiscent of acidic activation domains of transcription factors that interact with multiple partners. Acidic activation domains are “natively disordered” (11, 51), and this property apparently allows them to bind different surfaces with high specificity (multiple induced fits) and limited stability. The NH2-terminal portion of the measles virus P protein that contains the overlapping C protein ORF is, in fact, a recent example of such natively disordered proteins, in accordance with the prediction of algorithms that detect unstructured regions (47). By using the same algorithms (PONDR), the SeV C protein is strongly predicted to be natively disordered, and this property is shared with the common NH2-terminal portions of rubulavirus V, I, and P proteins (17, 26). The CΔ10-15 deletion, moreover, is in a region of C with the highest prediction of disorder. It will be of interest to examine whether purified SeV C proteins are indeed natively disordered.
ADDENDUM IN PROOF
Fujii et al. (Y. Fujii, T. Sakaguchi, K. Kiyotani, C. Huang, N. Fukuhara, Y. Egi, and T. Yoshida, J. Virol. 76:8540-8547, 2002) have shown that mutations in the leader region specifically attenuate virus virulence in mice.
Acknowledgments
We thank Ian Kerr (London, United Kingdom) for useful discussions and for numerous cell lines, Paula Pitha (Baltimore, Md.) and John Hiscott (Montreal, Canada) for the IRF-3 reagents, and the Jerome Pugin lab (University of Geneva) for help with the IL-8 protein assays.
REFERENCES
- 1.Bieback, K., E. Lien, I. M. Klagge, E. Avota, J. Schneider-Schaulies, W. P. Duprex, H. Wagner, C. J. Kirschning, V. ter Meulen, and S. Schneider-Schaulies. 2002. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 76:8729-8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biron, C. A., and G. C. Sen. 2001. Interferons and other cytokines, p. 321-349. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams, and Wilkins, Philadelphia, Pa.
- 3.Bosch, I., K. Xhaja, L. Estevez, G. Raines, H. Melichar, R. V. Warke, M. V. Fournier, F. A. Ennis, and A. L. Rothman. 2002. Increased production of interleukin-8 in primary human monocytes and in human epithelial and endothelial cell lines after dengue virus challenge. J. Virol. 76:5588-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadd, T., D. Garcin, C. Tapparel, M. Itoh, M. Homma, L. Roux, J. Curran, and D. Kolakofsky. 1996. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J. Virol. 70:5067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casola, A., R. P. Garofalo, M. Jamaluddin, S. Vlahopoulos, and A. R. Brasier. 2000. Requirement of a novel upstream response element in respiratory syncytial virus-induced IL-8 gene expression. J. Immunol. 164:5944-5951. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Z., and R. M. Krug. 2000. Selective nuclear export of viral mRNAs in influenza-virus-infected cells. Trends Microbiol. 8:376-383. [DOI] [PubMed] [Google Scholar]
- 7.Darnell, J. E. J. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 8.Delenda, C., S. Hausmann, D. Garcin, and D. Kolakofsky. 1997. Normal cellular replication of Sendai virus without the trans-frame, nonstructural V protein. Virology 228:55-62. [DOI] [PubMed] [Google Scholar]
- 9.Denk, A., M. Goebeler, S. Schmid, I. Berberich, O. Ritz, D. Lindemann, S. Ludwig, and T. Wirth. 2001. Activation of NF-kappa B via the Ikappa B kinase complex is both essential and sufficient for proinflammatory gene expression in primary endothelial cells. J. Biol. Chem. 276:28451-28458. [DOI] [PubMed] [Google Scholar]
- 10.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldmann, H., and H. D. Klenk. 1996. Marburg and Ebola viruses. Adv. Virus Res. 47:1-52. [DOI] [PubMed] [Google Scholar]
- 12.Fujii, Y., K. Kiyotani, T. Yoshida, and T. Sakaguchi. 2001. Conserved and non-conserved regions in the Sendai virus genome: evolution of a gene possessing overlapping reading frames. Virus Genes 22:47-52. [DOI] [PubMed] [Google Scholar]
- 13.Gao, J. L., and P. M. Murphy. 1994. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J. Biol. Chem. 269:28539-28542. [PubMed] [Google Scholar]
- 14.Garcin, D., J. Curran, M. Itoh, and D. Kolakofsky. 2001. Longer and shorter forms of Sendai virus C proteins play different roles in modulating the cellular antiviral response. J. Virol. 75:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin, D., M. Itoh, and D. Kolakofsky. 1997. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology 238:424-431. [DOI] [PubMed] [Google Scholar]
- 16.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcin, D., J. B. Marq, S. Goodbourn, and D. Kolakofsky. 2003. The amino-terminal extensions of the longer Sendai virus C proteins modulate pY701-Stat1 and bulk Stat1 levels independently of interferon signaling. J. Virol. 77:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcin, D., J. B. Marq, L. Strahle, P. Le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256-265. [DOI] [PubMed] [Google Scholar]
- 19.Garcin, D., T. Pelet, P. Calain, L. Roux, J. Curran, and D. Kolakofsky. 1995. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 14:6087-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcin, D., G. Taylor, K. Tanebayashi, R. Compans, and D. Kolakofsky. 1998. The short Sendai virus leader region controls induction of programmed cell death. Virology 243:340-353. [DOI] [PubMed] [Google Scholar]
- 21.Garofalo, R., M. Sabry, M. Jamaluddin, R. K. Yu, A. Casola, P. L. Ogra, and A. R. Brasier. 1996. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J. Virol. 70:8773-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 23.Genin, P., M. Algarte, P. Roof, R. Lin, and J. Hiscott. 2000. Regulation of RANTES chemokine gene expression requires cooperativity between NF-kappa B and IFN-regulatory factor transcription factors. J. Immunol. 164:5352-5361. [DOI] [PubMed] [Google Scholar]
- 24.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 25.Hasan, M. K., A. Kato, M. Muranaka, R. Yamaguchi, Y. Sakai, I. Hatano, M. Tashiro, and Y. Nagai. 2000. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74:5619-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 27.Hobert, M. E., K. A. Sands, R. J. Mrsny, and J. L. Madara. 2002. Cdc42 and Rac1 regulate late events in Salmonella typhimurium-induced interleukin-8 secretion from polarized epithelial cells. J. Biol. Chem. 277:51025-51032. [DOI] [PubMed] [Google Scholar]
- 28.Iseni, F., D. Garcin, M. Nishio, N. Kedersha, P. Anderson, and D. Kolakofsky. 2002. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 21:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishida, N., and M. Homma. 1978. Sendai virus. Adv. Virus Res. 23:349-383. [DOI] [PubMed]
- 30.Israel, A. 2000. The IKK complex: an integrator of all signals that activate NF-kappaB? Trends Cell Biol. 10:129-133. [DOI] [PubMed] [Google Scholar]
- 31.Itoh, M., Y. Isegawa, H. Hotta, and M. Homma. 1997. Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC-MK2 cells. J. Gen Virol. 78:3207-3215. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 33.Karin, M., and Y. Ben Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 34.Kato, A., K. Kiyotani, K. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 36.Kurt-Jones, E. A., L. Popova, L. Kwinn, L. M. Haynes, L. P. Jones, R. A. Tripp, E. E. Walsh, M. W. Freeman, D. T. Golenbock, L. J. Anderson, and R. W. Finberg. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398-401. [DOI] [PubMed] [Google Scholar]
- 37.Latorre, P., T. Cadd, M. Itoh, J. Curran, and D. Kolakofsky. 1998. The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J. Virol. 72:5984-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Mercier, P., D. Garcin, S. Hausmann, and D. Kolakofsky. 2002. Ambisense Sendai viruses are inherently unstable but are useful to study viral RNA synthesis. J. Virol. 76:5492-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lillemeier, B. F., M. Koster, and I. M. Kerr. 2001. STAT1 from the cell membrane to the DNA. EMBO J. 20:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, R., C. Heylbroeck, P. Genin, P. M. Pitha, and J. Hiscott. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19:959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, C., Y. Okruzhnov, H. Li, and J. Nicholas. 2001. Human herpesvirus 8 (HHV-8)-encoded cytokines induce expression of and autocrine signaling by vascular endothelial growth factor (VEGF) in HHV-8-infected primary-effusion lymphoma cell lines and mediate VEGF-independent antiapoptotic effects. J. Virol. 75:10933-10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutfalla, G., S. J. Holland, E. Cinato, D. Monneron, J. Reboul, N. C. Rogers, J. M. Smith, G. R. Stark, K. Gardiner, K. E. Mogensen, et al. 1995. Mutant U5A cells are complemented by an interferon-alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO J. 14:5100-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastronarde, J. G., B. He, M. M. Monick, N. Mukaida, K. Matsushima, and G. W. Hunninghake. 1996. Induction of interleukin (IL)-8 gene expression by respiratory syncytial virus involves activation of nuclear factor (NF)-kappa B and NF-IL-6. J. Infect. Dis. 174:262-267. [DOI] [PubMed] [Google Scholar]
- 45.Muller, M., C. Laxton, J. Briscoe, C. Schindler, T. Improta, J. E. Darnell, Jr., G. R. Stark, and I. M. Kerr. 1993. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 12:4221-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ott, M., J. L. Lovett, L. Mueller, and E. Verdin. 1998. Superinduction of IL-8 in T cells by HIV-1 Tat protein is mediated through NF-kappaB factors. J. Immunol. 160:2872-2880. [PubMed] [Google Scholar]
- 47.Palangat, M., and R. Landick. 2001. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J. Mol. Biol. 311:265-282. [DOI] [PubMed] [Google Scholar]
- 48.Pellegrini, S., J. John, M. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polyak, S. J., K. S. A. Khabar, D. M. Paschal, H. J. Ezelle, G. Duverlie, G. N. Barber, D. E. Levy, N. Mukaida, and D. R. Gretch. 2001. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J. Virol. 75:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 51.Rochat, S., H. Komada, and D. Kolakofsky. 1992. Loss of V protein expression in human parainfluenza virus type 1 is not a recent event. Virus Res. 24:137-144. [DOI] [PubMed] [Google Scholar]
- 52.Roux, P., C. Alfieri, M. Hrimech, E. A. Cohen, and J. E. Tanner. 2000. Activation of transcription factors NF-κB and NF-IL-6 by human immunodeficiency virus type 1 protein R (Vpr) induces interleukin-8 expression. J. Virol. 74:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakaguchi, T., K. Kiyotani, M. Sakaki, Y. Fujii, and T. Yoshida. 1994. A field isolate of Sendai virus: its high virulence to mice and genetic divergence form prototype strains. Arch. Virol. 135:159-164. [DOI] [PubMed] [Google Scholar]
- 54.Schafer, S. L., R. Lin, P. A. Moore, J. Hiscott, and P. M. Pitha. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J. Biol. Chem. 273:2714-2720. [DOI] [PubMed] [Google Scholar]
- 55.Schlaak, J. F., C. M. Hilkens, A. P. Costa-Pereira, B. Strobl, F. Aberger, A. M. Frischauf, and I. M. Kerr. 2002. Cell-type and donor-specific transcriptional responses to interferon-alpha. Use of customized gene arrays. J. Biol. Chem. 277:49428-49437. [DOI] [PubMed] [Google Scholar]
- 56.Skiadopoulos, M. H., S. R. Surman, J. M. Riggs, W. R. Elkins, M. St Claire, M. Nishio, D. Garcin, D. Kolakofsky, P. L. Collins, and B. R. Murphy. 2002. Sendai virus, a murine parainfluenza virus type 1, replicates to a level similar to human PIV1 in the upper and lower respiratory tract of African green monkeys and chimpanzees. Virology 297:153-160. [DOI] [PubMed] [Google Scholar]
- 57.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 58.Strieter, R. M., J. A. Belperio, and M. P. Keane. 2002. Cytokines in innate host defense in the lung. J. Clin. Invest. 109:699-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6:545-557. [DOI] [PubMed] [Google Scholar]
- 60.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 61.Tapparel, C., S. Hausmann, T. Pelet, J. Curran, D. Kolakofsky, and L. Roux. 1997. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J. Virol. 71:9588-9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, X.-L., M. Itoh, H. Hotta, and M. Homma. 1994. A protease activation mutant, MVCES1, as a safe and potent live vaccine derived from currently prevailing Sendai virus. J. Virol. 68:3369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wansley, E. K., and G. D. Parks. 2002. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 76:10109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 66.Yamada, Y., A. Nakamura, M. Hosoda, T. Kato, T. Asano, K. Tonegawa, and M. Itoh. 2001. Cytokines in pleural liquid for diagnosis of tuberculous pleurisy. Respir. Med. 95:577-581. [DOI] [PubMed] [Google Scholar]
- 67.Yeow, W. S., W. C. Au, W. J. Lowther, and P. M. Pitha. 2001. Downregulation of IRF-3 levels by ribozyme modulates the profile of IFNA subtypes expressed in infected human cells. J. Virol. 75:3021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, Y., B. A. Luxon, A. Casola, R. P. Garofalo, M. Jamaluddin, and A. R. Brasier. 2001. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J. Virol. 75:9044-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]