Abstract
During infection with adenovirus, massive changes in the transcription of virus genes are observed, suggesting that the expression of cellular genes may also be modulated. To characterize the levels of cellular RNA species in infected cells, cDNA arrays were screened 24 h after infection of HeLa cells with wild-type adenovirus type 5, strain dl309. Despite complete transduction of the cells, fewer than 20 cellular genes (out of 4,600 analyzed and 1,200 found detectable and expressed above background) were altered more than threefold in their corresponding RNA levels compared to mock-infected cells. In particular, the expression of the myc oncogene was reduced at the mRNA level. This reduction was dependent on the replication of virus DNA and partially dependent on the presence of the adenovirus gene products E1B-55 kDa and E4orf6, but not E4orf3. On the other hand, MYC protein had an increased half-life in infected cells, resulting in roughly constant steady-state protein levels. The adenovirus E1A gene product is necessary and sufficient to stabilize MYC. Overexpressed MYC inhibited adenovirus replication and the proper formation of the virus replication centers. We conclude that adenovirus infection leads to the stabilization of MYC, perhaps as a side effect of E1A activities. On the other hand, myc mRNA levels are negatively regulated during adenovirus infection, and this may avoid the detrimental effect of excessive MYC on adenovirus replication.
Adenovirus expresses a variety of factors that can directly or indirectly affect the expression of viral and cellular genes (33). These include the E1A proteins and E1B-55 kDa, which modulate the activity of growth-regulatory transcription factors, namely, E2F proteins and p53 (48). These activities are further modified by E4 proteins, in particular E4orf3, E4orf6, and E4orf6/7 (37, 38). In addition, E1A is capable of directly interacting with the basal transcription initiation factors TBP and YY1 (13, 33). The most dramatic change in mRNA synthesis, however, occurs when the virus switches from early to late phase. At this time, viral DNA is replicated in distinct centers within the nucleus (33). The adenovirus IVa2 gene product is then highly expressed and binds to intragenic sequences within the adenovirus major late expression unit (25, 40). As a result, the promoter is activated 20- to 30-fold, and eventually, mRNA species derived from the L genes represent a large proportion of all mRNA molecules within the infected cell. Hence, the infectious cycle of adenovirus is largely characterized by massive temporal changes in the transcriptional regulation of virus genes. It has been suggested elsewhere that at least some components of the basal transcription machinery are tethered to the major late promoter to a large extent, thus becoming limiting for other transcription units within the infected cell (11).
Some of the factors expressed by the virus have been studied extensively regarding their impact on cellular transcription, mostly after overexpression of single proteins (33). However, little is known about the activity of cellular genes in the context of a productive adenovirus infection. Although one would intuitively assume that the massive activation of the major late promoter within the replication centers would exhaust a number of cellular transcription factors and thereby widely affect the expression of cellular genes, only a small set of cellular mRNA species was previously analyzed individually in this regard (24, 32). cDNA arrays represent a novel tool to perform a search of differentially expressed genes on a large scale, and this technology appears suitable to identify cellular genes that respond to adenovirus infection.
The most widely analyzed system to study adenovirus infection is represented by HeLa cells, infected with adenovirus type 5, and one of the commonly used strains of this virus is dl309 (18). HeLa cells are derived from a cervical carcinoma, and this tumor species was the first target of an attempt to perform oncolytic therapy with adenovirus (34). As a starting point to reveal the impact of adenovirus infection on cellular gene expression, we used this system and compared cellular gene expression between mock-infected and adenovirus-infected cells after 24 h, using cDNA microarrays. Relatively few genes were found differentially regulated, whereas most genes analyzed largely maintained their expression levels despite the presence of replicating adenovirus.
myc was one of the genes downregulated in infected cells. The product of this gene is a widely studied oncoprotein, overexpressed in numerous tumor species. It is capable of regulating cell proliferation, apoptosis, transcription, and possibly DNA repair (1, 10, 26). In the past, conflicting results had been reported regarding the positive or negative regulation of myc by adenovirus and its E1A gene product (9, 16, 23, 24, 32, 39, 42). However, it is clear that E1A and MYC have overlapping functions and interaction partners. Both proteins can stimulate cell proliferation, and both are inducers of apoptosis (5). Further, both activate the expression of p14/ARF and induce accumulation of p53 (8). Also, both induce the expression of transcriptionally active p73 (47). Finally, E1A as well as MYC was found to interact with a cofactor of transcription, TRRAP (12, 28). However, little is known about the possibility of mutual regulation between adenovirus gene products and MYC.
We found that adenovirus infection influences the expression of myc in two opposite ways, i.e., reduction of its mRNA and stabilization at the protein level. When MYC was allowed to accumulate in excess, it inhibited the replication of adenovirus DNA. Hence, adenovirus and MYC interact at multiple levels, and these interactions may affect the efficiency of virus replication.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells (American Type Culture Collection) were maintained in Dulbecco's modified eagle medium (Life Technologies) with 10% fetal bovine serum. Transfections were done using Lipofectamine 2000 (Life Technologies). Adenovirus and adenovirus-derived vectors were propagated and the titers were determined as described previously (20, 43).
Array hybridization and evaluation.
The expression of cellular genes during adenovirus infection was analyzed with microarrays. Experimental procedures are described as follows, according to the “minimum information about a microarray experiment (MIAME)” standards (4).
(i) Experimental design.
HeLa cells (1.4 × 107) were infected with adenovirus type 5, strain dl309, at a multiplicity of infection of 10. Detection of the adenovirus E2A protein by immunofluorescence (43) in a separate experiment revealed that virtually 100% of the cells were infected under these conditions (data not shown). We analyzed the same number of mock-infected cells in parallel. After 24 h, the cells were harvested and total RNA was prepared, yielding two samples. In a second experiment, another set of two samples was prepared according to the same protocol but independently of the first experiment. Each sample of RNA was labeled by reverse transcription and incorporation of two different fluorescent dyes. Four cDNA microarrays were hybridized on two different days. Each day, a dye-swap experiment was done. Since each cDNA species was spotted twice on each array, eight hybridizations were carried out for every cDNA clone, each with differently labeled probes derived from infected and noninfected samples.
(ii) Array design.
Glass slides (Corning Inc.; GAPS amino silane-coated slides, catalog no. 2549) were spotted with PCR-amplified cDNA clones. The templates used were obtained from Invitrogen/Research Genetics, and their identity corresponds to the GF200 series available from this company, with some modifications. The exact list of genes examined and the results obtained for each of them will be made available at http://www.med.uni-marburg.de/wwwmzh/viro/dobbelst/agdobb.htm. Each PCR product was spotted twice, in different regions of one array.
(iii) Samples.
Total RNA was prepared from mock-infected or adenovirus-infected HeLa cells with Trizol reagent (Invitrogen). Fifty micrograms of RNA was annealed to 4 μg of an oligo(dT) 15-mer in 37 μl of water by being heated to 65°C and cooled to room temperature. Reverse transcription was carried out using Superscript II (Invitrogen) reverse transcriptase in a 62-μl volume of the manufacturer's buffer, in the presence of dATP, dGTP, and dTTP (161 μM each), as well as 16 μM nonconjugated dCTP and 16 μM dCTP that was coupled to the dye Cy3 or Cy5 (Amersham Pharmacia Biotech; PA 53021 and PA 55021, respectively). After incubation for 10 min at 25°C and 120 min at 37°C, the obtained polynucleotides were purified by being annealed to a silica matrix, with a PCR purification kit (Qiagen). Each sample was mixed with 10 μg of human cot1 DNA (Invitrogen) and ethanol precipitated. The precipitate was dissolved in 10 mM Tris-Cl (pH 8.0), boiled for 3 min, chilled rapidly on ice, further denatured in 200 mM NaOH for 10 mM at 37°C, and ethanol precipitated. Before hybridization, the precipitate was dissolved in 24 μl containing 4.2× SSC (1× SSC corresponds to 150 mM sodium chloride and 15 mM sodium citrate, pH 7) and 1.7% sodium dodecyl sulfate (SDS), as well as polydeoxyadenosine (octamers and 20-mers, 0.21 mg/ml each). Each of the two pairs of RNA samples was reverse transcribed, once incorporating Cy3 and once incorporating Cy5 dye.
(iv) Hybridizations.
Arrays were prehybridized for 20 min at 56°C in a solution containing 1% bovine serum albumin, 3× SSC, and 0.1% SDS. After being washed with water, the array was dried by brief centrifugation. The hybridization sample (specified above) was boiled for 2 min and immediately incubated with the array for approximately 16 h at 56°C in a humid chamber. Four washing steps were carried out, each for 5 min at room temperature: 1× SSC-0.1% SDS, 0.1× SSC-0.1% SDS, 0.1× SSC, and H2O. The array was dried by brief centrifugation.
(v) Measurements.
The arrays were scanned using a laser source and fluorescence detection device (Genitic Micro Systems, now Affymetrix; array scanner, model 418). Quantification matrices were obtained by extracting the spot intensities from scanned images with ImaGene 3.0 software (BioDiscovery Inc., Marina Del Rey, Calif.). Eight spots corresponding to each cDNA clone were analyzed, for comparison of infected and noninfected cells. Since the labeling reaction was carried out using an oligo(dT) primer, mRNA species that do not contain a poly(A) tail (i.e., histone mRNAs) were not further evaluated.
(vi) Evaluation and normalization.
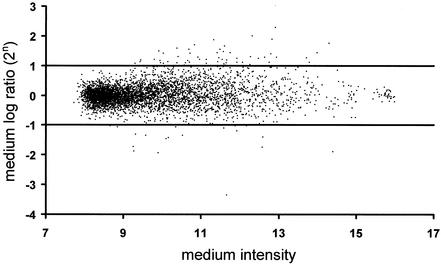
For each spot, median signals and background intensities for both channels were obtained. To account for spot differences, the background corrected ratios of the two channels were calculated. Following the annotation of reference 46, we used the log ratio M = log2R/G and the mean log intensity A = log2[(RG)0.5], where R and G denote the measured fluorescence intensities after background subtraction for the Cy5 and Cy3 dyes, respectively. To balance the fluorescence intensities for the two dyes, as well as to allow the comparison of expression levels across experiments, the raw data were standardized. We used an intensity-dependent standardization as described in reference 46 to correct for inherent and random bias on each chip (the Lowess scatter-plot smoother). As each gene was spotted twice on the chip, and four arrays were analyzed, mean log ratios M for each gene were calculated. To find differently expressed genes, the genes were sorted by the d statistic introduced by Tusher et al., and the computed exchangeability factor s0 for this experiment was 1.26 (41). All chips had a signal-to-background ratio above 3 (a ratio below 2 indicates poor-quality chips) and an average local background intensity below 500 (arbitrary units, 9 on the log scale in Fig. 1). Less than 1.6% of the spots had poor within-chip reproducibility (more-than-fourfold differences between spot replicates or negative values).
FIG. 1.
Expression levels of 4,600 genes in response to adenovirus infection. Arrays were hybridized with cDNA pools derived from adenovirus-infected and mock-infected HeLa cells. Cells were infected at a multiplicity of infection of 10, for 24 h, followed by RNA preparation and array hybridization. The mean log2 ratio M of signal intensities (with virus versus without virus, eight spots for each gene on four different microarrays) is plotted against the average signal intensity A (logarithmic scale, arbitrary units) for each gene under examination.
Reverse transcription-PCR (RT-PCR).
Total RNA was isolated from HeLa cells (Trizol reagent; Life Technologies), and mRNA levels were determined by reverse transcription and semiquantitative PCR, essentially as described previously (7). When the primers did not span an intron sequence, i.e., in the cases of TOB-1, CD24, SPUVE, and NPTX1, any residual genomic DNA was removed by DNase I treatment. Reverse transcription was performed with Superscript II polymerase (Life Technologies), and PCR amplification was performed with Expand HiFi DNA polymerase (Roche). The PCR consisted of a 3-min denaturation step at 96°C, followed by the indicated numbers of cycles at 96°C for 30 s, 57°C for 30 s, and 70°C for 50 s. Specific oligonucleotides were employed for reverse transcription, as well as for PCR, and their sequences are specified as follows, in each case the gene name being followed by the reverse transcription primer and the two PCR primers: SPUVE, GCT TCT GGG TTC CTT TCA CAT AGG, GGA AAC CCA CTT GGC CTG CAT ACC, and TCC ATC GTG TAT GCA GTG GGC AGC; LOXL-1, CTT GAG GAT GTA GTT CCC AGG CTG, GTC TCC CTG ACT TGG TCC CAG ACC, and CCG CAT TGT AGG TGT CAT AGC AGC; CD24, ACG TGG AGG AAT TAC AGT AAC ACC, TGC TGG CAC TGC TCC TAC CCA CGC, and AAT CTC CAT TCC ACA ATC CCA TCC; myc, GCC ACC GCC GTC GTT GTC TC, AGC CAG CGG TCC GCA ACC CTT GCC, and AGC TCG AAT TTC TTC CAG ATA TCC; NPTX1, CTG TAC TGC TCG AGG TTC TCC AGG, GCC GAC GCG CTT CAT CTG CAC TTC, and GTT TTG AGC GAT TGC AAA GTT TGC; TOB-1, TAC AGC AGC AGA GTG ACC AAA AGG, TAT GCA GCT TGA AAT CCA AGT AGC, and TGG AGA GCT GGC CAC TGA TGA GGC; HSOBRGRP, GCA CTG CAT AAG GTG GCA GCT GCC, GGC GGG CGT TAA AGC TCT CGT GGC, and TAA AGT GCT ACC ACT GCT CCC AGC; HPRT1, CTT CGT GGG GTC CTT TTC ACC AGC, ACC TTG ACC ATC TTT GGA TTA TAC, and ACC TTG ACC ATC TTT GGA TTA TAC; E1A, GGT GAT GTC GGG CGT CTC AGG, GCG GTT TCG CAG ATT TTT CCC G, and GCA GGC GCC ATT TTA GGA CG; E2A, GCT GAA ACC CAC CAT TTG TAG CGC, GAT CTT CGC TTT TGT GAT ACA GGC, and GAC GCA ATG GCC AAA TCC GCC GTC.
Assessment of adenovirus DNA replication.
Infected cells were harvested, followed by preparation of genomic DNA (Qiagen) and semiquantitative PCR. The primers and PCR conditions were the same as those used to quantify the E1A mRNA levels (see above).
Immunoblotting.
Proteins were separated on SDS-polyacrylamide gels and transferred to nitrocellulose, followed by incubation with antibodies in phosphate-buffered saline (PBS) containing 5% milk powder and 0.1% Tween 20. Peroxidase-coupled secondary antibodies (whole immunoglobulin G; Jackson) were then detected by chemiluminescence (Pierce). Monoclonal mouse antibodies were against adenovirus E1A (Ab-1; Calbiochem), adenovirus E2A (clone B6-6; obtained from J. Flint), MYC (clone 9E10; Santa Cruz Biotechnology), and actin (clone C-2; Santa Cruz Biotechnology).
Immunofluorescence.
Cells were seeded in chamber slides (Nunc) suitable for microscopy. They were infected as described above, followed by fixation with paraformaldehyde (4% in PBS; 15 min). They were permeabilized with Triton X-100 (0.2% in PBS; 25 min) and incubated with a monoclonal antibody to adenovirus E2A-72 kDa (clone B6-6). The primary mouse antibody was visualized with a secondary antibody coupled to the dye Alexa 594 (Molecular Probes). Before being mounted (Fluoprep; bioMérieux), the cell nuclei were briefly stained with 4′,6′-diamidino-2-phenylindole (DAPI).
RESULTS
A limited number of cellular mRNA species are differentially expressed in the presence of replicating adenovirus.
To identify cellular genes that are differentially expressed during the late phase of adenovirus infection, HeLa cells were infected at a multiplicity of infection of 10 for 24 h. Immunofluorescence staining of the viral E2A-72-kDa protein confirmed that virtually all cells were infected under these conditions (data not shown). In parallel, HeLa cells were mock infected. RNA was prepared and processed for cDNA synthesis and hybridization of microarrays. Thereby, the expression of cellular genes upon virus infection was compared to that for mock-infected cells. The results of array hybridization are depicted in Fig. 1. Out of 4,600 genes, approximately 1,200 were detectable with reproducibility and above background (average intensity > 9). However, only a small subset of these genes was found to be differentially regulated. Seventy-five cDNA clones corresponded to a more-than-twofold differential expression, and only 14 genes had expression levels that differed more than threefold. The named genes that did respond to infection most strongly, yielding a d value (score) (41) of more than 1, are listed in Table 1. Most of them were downregulated upon infection with adenovirus, such as the oncogene myc (alias c-myc) and TOB-1 (alias APRO6), a gene with an antiproliferative product of the Btg family (27). Only the LOXL-1 mRNA was detected at increased levels. This gene encodes a product with homology to lysyl oxidase (Lox), an enzyme catalyzing cross links in elastin and collagens (35).
TABLE 1.
Cellular genes differentially expressed upon adenovirus infectiona
| Gene | Alternate name | Database accession no.
|
Score | Log2 fold change | Fold activation (↑) or repression (↓) | |
|---|---|---|---|---|---|---|
| Expressed sequence tag | Full length | |||||
| SPUVE | Serine protease, umbilical endothelium | R76394 | NM_007173 | 1.812 | −2.276 | 4.8 ↓ |
| LOXL-1 | Lysyl oxidase-like 1 | AA405804 | NM_017526 | 1.385 | +1.939 | 3.8 ↑ |
| CD24 | CD24 antigen (small cell lung carcinoma cluster 4 antigen) | H59915 | BC007674 | 1.303 | −2.007 | 4.0 ↓ |
| myc | MYC | AA514409 | XM_037660 | 1.216 | −1.887 | 3.7 ↓ |
| NPTX1 | Neuronal pentraxin I | H22481 | NM_002522 | 1.194 | −1.866 | 3.6 ↓ |
| TOB-1 | Transducer of ERBB2 1 | AA490213 | XM_038452 | 1.141 | −1.536 | 2.9 ↓ |
| HSOBRGRP | Homo sapiens mRNA for leptin receptor gene-related protein | H51066 | NM_017526 | 1.085 | −1.432 | 2.7 ↓ |
| myc | v-myc avian myelocytomatosis viral oncogene homolog | W87741 | XM_037660 | 1.023 | −1.465 | 2.7 ↓ |
HeLa cells were infected with adenovirus or mock infected as described in the legend to Fig. 1, followed by RNA preparation and array hybridization. Differentially expressed genes were sorted by the d statistic introduced by Tusher et al. (41), and named genes with a d value (score) above 1 are listed in the table. Note that two different cDNA clones derived from myc independently yielded a high score.
Differential gene expression is mostly conserved in two different cell lines.
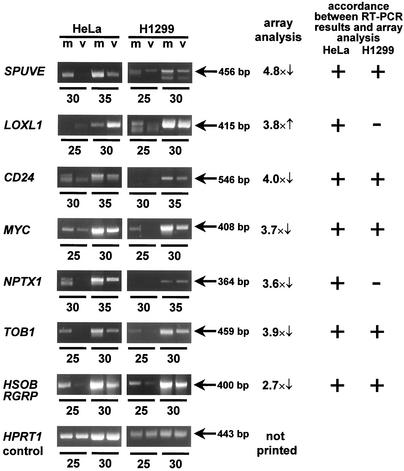
To verify the differential expression of the genes listed in Table 1, their mRNA levels were determined by reverse transcription, followed by semiquantitative RT-PCR. The differential expression of all genes tested in this way was confirmed in HeLa cells (Fig. 2), validating the results obtained by cDNA array analysis. To find out whether adenovirus can affect the same genes in a different cell line, H1299 cells (human lung adenocarcinoma) were infected and RNA was prepared as described above for HeLa cells, followed by RT-PCR analysis. H1299 cells did not show the upregulation of LOXL-1 and the suppression of NPTX1 seen in HeLa cells. However, the genes SPUVE, CD24, myc, TOB-1, and HSOBRGRP were each downregulated in HeLa cells, as well as in H1299 cells (Fig. 2), indicating that the pattern of regulation of cellular genes by adenovirus infection is not entirely restricted to one particular cell type.
FIG. 2.
Independent quantification of selected mRNA species by RT-PCR. The mRNA levels of the indicated genes from mock-infected (m) and adenovirus-infected (v) HeLa and H1299 cells were determined in parallel by reverse transcription and semiquantitative PCR. Where possible, PCR primers were designed to span at least one intron, and in these cases, the expression levels determined reflect mRNA. In other cases, i.e., TOB-1, CD24, SPUVE, and NPTX1, PCR products correspond to a sequence within one intron, therefore reflecting pre-mRNA and mRNA from the respective genes. The reaction was allowed to proceed for the indicated numbers of PCR cycles, followed by agarose gel electrophoresis and staining with ethidium bromide. The factor of differential expression, as determined by array hybridization, is indicated for comparison, with arrows indicating up- or downregulation upon virus infection. Agreement between the result of array analysis and that of RT-PCR is indicated by “+.”
Adenovirus mutations and the inhibition of virus DNA replication affect cellular gene expression.
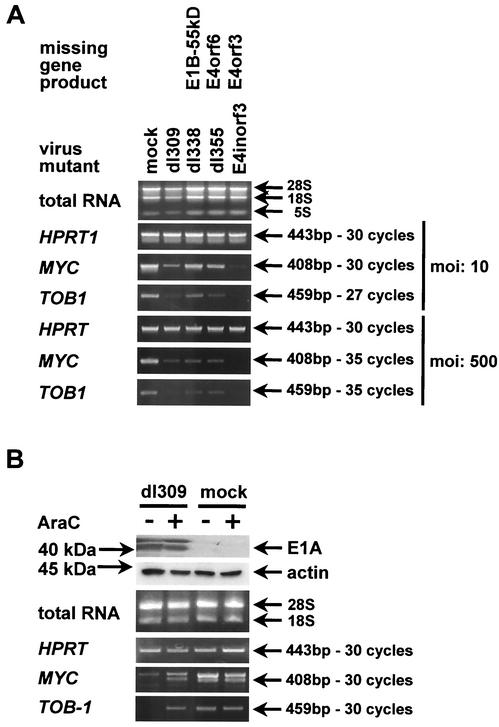
We asked whether the alteration of cellular gene expression levels by adenovirus infection was dependent on particular adenovirus gene products. To test this, HeLa cells were infected with adenovirus dl309 (18) and the mutants dl338 (lacking E1B-55 kDa [31]), dl355 (lacking E4orf6 [30]), and E4inorf3 (lacking E4orf3 [17]). E1B-55 kDa and E4orf6 deletion mutants fail to induce the shutdown of cellular protein synthesis (2, 15, 22, 31), and this in turn might affect cellular transcription. A mutant lacking E4orf3 was included because E4orf3 was shown earlier to modulate the expression of metallothionein (44), arguing that it might be involved in the regulation of other cellular genes as well. Twenty-four hours after infection, the relative expression levels of myc and TOB-1 were determined by RT-PCR. All adenovirus mutants suppressed the expression of these genes at a high multiplicity of infection (Fig. 3A), indicating that neither E1B-55 kDa nor E4orf6 nor E4orf3 is strictly required for downregulation of myc and TOB-1. At lower multiplicities of infection, mutants lacking E1B-55 kDa or E4orf6 reduced the expression of myc and TOB-1 only to a lesser extent than did wild-type dl309, suggesting that host cell shutdown may enhance the suppression of myc and TOB-1 but is not required for it. The lack of E4orf3 appeared to increase the inhibitory effect of adenovirus on the expression of myc and TOB-1 for reasons that we do not understand at present. Possibilities include direct effects of E4orf3 on transcription, e.g., by relocalizing transcription factors within the nucleus (44), or a dysregulated expression of those viral genes that directly contribute to the downregulation of myc and TOB-1.
FIG. 3.
Influence of adenovirus mutants and DNA replication on cellular gene expression. (A) Impact of adenovirus mutants on the levels of selected cellular mRNA species. HeLa cells were infected with the indicated mutants (lacking E1 or E4 gene products as marked) at the indicated multiplicity of infection, for 24 h, followed by RNA preparation (the positions of 28S, 18S, and 5S rRNAs are indicated) and semiquantitative RT-PCR amplification of the indicated mRNA species, as described in the legend to Fig. 2. (B) Dependence of selected cellular mRNA levels on adenovirus DNA replication. HeLa cells were infected with adenovirus dl309 for 24 h (multiplicity of infection = 500), and ara-C was added 3 h postinfection to a final concentration of 20 μg/ml where indicated. This was followed by RNA preparation and semiquantitative RT-PCR amplification of the indicated RNA species, as described in the legend to Fig. 2. In parallel, the levels of E1A protein were determined by immunoblot analysis.
Next, we analyzed whether the replication of adenovirus DNA and the consecutive early-to-late switch of gene expression are required for the modulation of myc and TOB-1 expression. HeLa cells were infected in duplicate with adenovirus dl309 or mock infected. In each case, an inhibitor of viral DNA polymerase, cytosine arabinoside (ara-C), was added to the culture medium in one of the wells 3 h after infection. After 24 h, the cells were harvested, and cellular RNA was analyzed for expression of myc and TOB-1 by RT-PCR (Fig. 3B). While ara-C did not detectably influence gene expression in mock-infected cells, it completely restored the levels of myc and TOB-1 in adenovirus-infected cells. Western blot analysis revealed that the expression of viral E1A proteins was not impaired by ara-C. We conclude that the early gene products of adenovirus, including E1A, are not sufficient to reduce the levels of myc and TOB-1. While it is formally possible that E1A alone would have suppressed myc mRNA whereas some other adenovirus gene product counteracted this effect in the absence of replication, we strongly favor the interpretation that E1A alone is not capable of repressing myc mRNA expression.
MYC is stabilized during adenovirus infection.
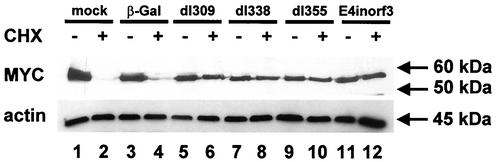
Since MYC is known as a regulator of cell growth and transcription, sharing a number of properties with the adenovirus E1A oncoprotein, we decided to explore the interplay of adenovirus and MYC in more detail. We asked whether the negative regulation of myc mRNA by adenovirus infection was accompanied by a corresponding change in the MYC protein levels. To test this, MYC was detected by immunoblot analysis in mock-infected and adenovirus-infected cells. Surprisingly, it was found that the amount of MYC protein was only marginally affected by adenovirus infection (Fig. 4, compare lanes 1 and 5). This discrepancy between protein and mRNA levels suggested that a change in the biological half-life of MYC protein might compensate for the reduction of mRNA. To address this, the cells were treated with an inhibitor of protein synthesis, cycloheximide. After 4 h of treatment, the levels of MYC were strongly reduced in mock-infected cells, apparently due to intracellular degradation. In contrast, the levels of MYC remained virtually unchanged when the cells had been infected with adenovirus. This effect was independent of adenovirus E1B-55 kDa, E4orf6, or E4orf3, since virus mutants lacking these genes were equally capable of stabilizing MYC. In contrast, a first-generation adenovirus vector, lacking the E1 genes and unable to replicate, did not detectably affect the degradation of MYC compared to that for mock-infected cells (Fig. 4).
FIG. 4.
Impact of adenovirus infection on the half-life of MYC. HeLa cells were transduced with the indicated adenovirus mutants, at a multiplicity of infection of 20. Twenty hours later, the cells were treated with cycloheximide (CHX; 50 μg/ml) or mock treated as indicated. Four hours later, the cells were harvested, followed by immunoblot detection of MYC. β-Gal, beta-galactosidase.
Adenovirus E1A protein is sufficient to stabilize MYC.
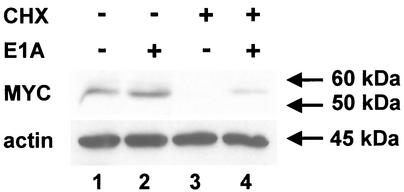
Since a virus lacking the E1 region did not stabilize MYC, whereas a wild-type virus was able to do so, it was conceivable that an E1 gene product might increase MYC stability. Further, since deletion of E1B-55 kDa did not affect the stability of MYC in virus-infected cells (compare lanes 7 and 8, Fig. 4, virus dl338), we raised the hypothesis that an E1A gene product might stabilize MYC. To test this, we transiently transfected HeLa cells with an expression plasmid for the E1A 13S protein of adenovirus type 5. It was found that overexpression of E1A resulted in a considerable increase in the amount of MYC that was retained after abolishing protein synthesis with cycloheximide. The block in MYC degradation was not as complete as that after virus infection, perhaps due to incomplete transfection of the cells. Possibly for the same reason, we detected little if any increase of MYC levels when E1A was expressed but cycloheximide was omitted (Fig. 5, compare lanes 1 and 2). Nonetheless, the stabilizing effect of E1A was clearly observed (Fig. 5, compare lanes 3 and 4) and was highly reproducible in independent experiments (data not shown). We conclude that an adenovirus E1A protein alone can increase the stability of MYC.
FIG. 5.
Influence of adenovirus E1A protein on the stability of MYC. HeLa cells were transfected with an expression plasmid for E1A or the corresponding empty vector plasmid (pCDNA3). Twenty hours later, the cells were treated with cycloheximide (CHX; 50 μg/ml) or mock treated as indicated. Four hours later, the cells were harvested, followed by immunoblot detection of MYC and actin.
Overexpression of MYC reduces the replication of adenovirus.
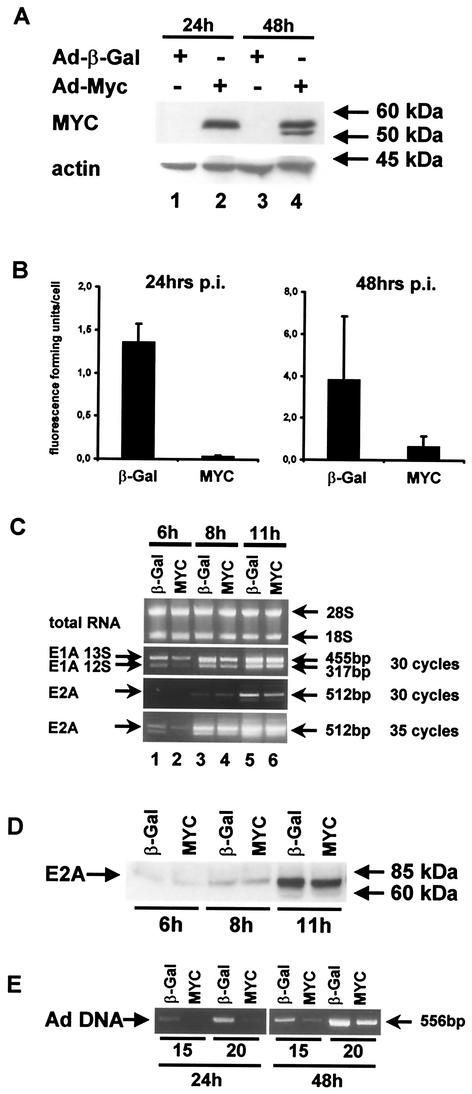
It appears that, during evolution of adenovirus, mechanisms were selected that regulate the transcription of the myc gene as well as the stability of MYC protein (albeit with opposing effects). This raises the possibility that MYC might influence virus replication. To test the ability of adenovirus to replicate in the presence of overexpressed MYC, we infected HeLa cells with wild-type adenovirus dl309, in combination with first-generation expression vectors for beta-galactosidase (control) or MYC. MYC expression was verified by immunoblot analysis (Fig. 6A). Virus yield was determined subsequently. It was found that MYC overexpression led to a more-than-20-fold reduction of virus yield after 24 h and still reduced the amount of harvested virus by a factor of 5 after 48 h (Fig. 6B). To define the stage of virus replication at which MYC displays a negative influence, we prepared the RNA of infected cells at early time points postinfection, followed by quantification of virus mRNA by semiquantitative RT-PCR. It was found that MYC only marginally affected the expression of E1A or E2A early after infection (Fig. 6C), nor did it alter the E2A protein levels (Fig. 6D). In contrast, the replication of virus DNA was strongly impaired by MYC (Fig. 6E). We conclude that MYC inhibits adenovirus replication at a stage after early gene expression but during the replication of virus DNA.
FIG. 6.
Effect of MYC overexpression on adenovirus replication. For all experiments, HeLa cells were infected with a mixture of wild-type adenovirus dl309 (multiplicity of infection = 1) and a first-generation adenovirus vector to express either beta-galactosidase (20) or MYC (29) (each at a multiplicity of infection of 20). (A) Expression of MYC was verified by immunoblot analysis at the time points indicated. Note that endogenous MYC (upon transduction with the beta-galactosidase expression vector) was not detected at this level of sensitivity but only on longer exposures (Fig. 4). (B) The cells were harvested at 24 and 48 h postinfection as indicated, followed by the quantification of virus yield. This was determined by freeze-thawing the cells, infecting fresh cell monolayers with serial dilutions of the lysate, and staining infected cells by immunofluorescence, with antibodies to E2A-72 kDa. The number of infectious units obtained per cell, determined in at leastthree independent experiments, is shown (columns) along with the standard deviation (bars). (C) The cells were harvested after 6, 8, and 11 h postinfection as indicated, followed by the preparation of total RNA and analysis by agarose gel electrophoresis (the positions of 28S and 18S rRNAs are indicated). The mRNAs of E1A and E2A were amplified by RT-PCR for the indicated numbers of temperature cycles. (D) The cells were harvested at the indicated time points postinfection, followed by immunoblot detection of the E2A-72-kDa DNA binding protein. (E) At the indicated time points postinfection, a portion of the adenovirus genome was amplified by semiquantitative PCR for 15 or 20 cycles. Ad, adenovirus; β-Gal, beta-galactosidase.
MYC affects the proper formation of adenovirus replication centers.
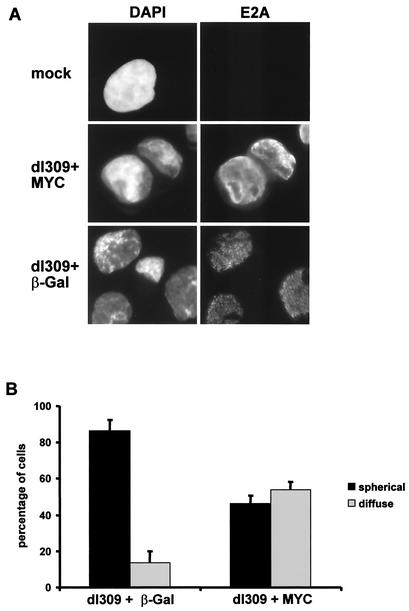
To further address the mechanism by which MYC interferes with adenovirus replication, we analyzed the morphology of intranuclear virus replication centers. Twenty-four hours after infection of HeLa cells with a combination of wild-type adenovirus and expression vectors for beta-galactosidase or MYC, the cells were fixed and stained with an antibody to the adenovirus E2A-72-kDa protein, a well-characterized marker of the replication centers of virus DNA (30). When cells were coinfected with dl309 and the beta-galactosidase expression vector, the usual pattern of spheric replication centers was observed in the vast majority of the cells (Fig. 7A). In contrast, the staining pattern of E2A was considerably altered in the presence of overexpressed MYC. In these cells, E2A was either distributed diffusely within the cell nuclei, as is usually observed at a stage of infection before the replication of virus DNA, or found in structures that were no longer spheric but had an irregular shape. In contrast, cells displaying the regular spheric replication centers were less frequent (Fig. 7A). To obtain a more quantitative picture of these changes, the cells were counted and scored according to the presence or absence of 10 or more spheric replication centers in one nucleus. Several independent experiments consistently revealed that the number of cells containing a diffuse distribution of E2A was increased about threefold in the presence of MYC (Fig. 7B). We conclude that MYC is capable of directly or indirectly antagonizing the proper formation of adenovirus replication centers.
FIG. 7.
Effect of MYC overexpression on the formation of adenovirus replication centers. (A) HeLa cells were infected with a combination of dl309 and expression vectors as in Fig. 6. After 24 h, the cells were immunostained with an antibody to the E2A-72-kDa DNA binding protein. The cells were counterstained with DAPI. (B) After treatment as for panel A, at least 200 cells were scored and divided into two categories: (i) spherical, indicating that at least 10 distinct spherical formations stained with an anti-E2A antibody were observed in the nucleus of a cell, and (ii) diffuse, indicating that fewer than nine distinct spherical formations or an entirely diffuse staining pattern of E2A was observed. β-Gal, beta-galactosidase.
DISCUSSION
We have shown that a limited number of cellular genes are differentially expressed in response to adenovirus infection. Most of these genes are negatively regulated, among them myc. While myc mRNA levels were consistently reduced by adenovirus infection, this was not observed for MYC protein levels. The reason appears to be that MYC protein is stabilized in infected cells, and this effect can be attributed to adenovirus E1A. One interpretation of these results is that E1A stabilizes MYC as a side effect of its growth-stimulating activities, whereas mechanisms were selected during evolution of the virus to hold down myc expression at the RNA level. Consistently, we found that excess MYC antagonizes virus replication.
Several cellular genes have been reported earlier to be modified by adenovirus infection. In agreement with our results, the amount of myc mRNA was downregulated in KB cells infected by adenovirus type 2 (32), a system similar to the one described here. The same study suggested that major alterations occur in the general transcription patterns of adenovirus-infected cells. Based on our findings, however, these alterations appear to be more limited than expected, with the caveats discussed below.
One previous study used arrays to look at changes of cellular transcription brought about by adenovirus (14). However, this report analyzed a smaller number of genes (588 total, 98 detectable) in murine liver after transduction with a nonreplicative adenovirus vector, a system that has little in common with our study. Correspondingly, differentially expressed genes were identified that are mostly involved in the interferon response. We propose that these genes may have been induced as an indirect effect of virus infection and the subsequent immune response, not necessarily as a direct consequence of immediate virus-cell interaction.
In general, the choice of target cells certainly affects the selection of genes that are dysregulated by adenovirus infection. For instance, primary cells can be expected to respond more extensively to transforming adenovirus proteins, such as the E1 and E4 gene products, than transformed cells that already carry alterations in cellular growth control pathways, e.g., the Rb and p53 pathways. The study presented here was mainly aiming at elucidating the alterations of cellular gene expression during the late phase of infection, when replication centers are formed and the early-to-late switch of viral transcription is complete. Accordingly, it is very possible that different numbers and species of genes would have been found dysregulated at different time points postinfection. Further, it should be kept in mind that the number of cellular genes examined represents no more than one-sixth of the total estimated to comprise the human genome. Finally, the cDNA array methodology, which estimates changes in steady-state RNA concentrations, is inherently limited in its ability to detect inhibition of gene expression: substantial decreases in RNA levels will occur only when the mRNA has a relatively short half-life. All these considerations argue that more than the genes found in this screen can be expected to be differentially regulated by adenovirus infection.
Currently, it is unknown by what mechanism(s) adenovirus infection suppresses the expression of the genes identified in our screen. The elimination of suppression by an inhibitor of DNA replication suggests that the onset of the late phase is a prerequisite for the regulation of these genes. Perhaps the virus replication centers absorb transcription factors that would otherwise allow the efficient transcription of the identified cellular genes. If this is true, however, it is surprising that not more cellular genes are affected. Thus, if any cellular transcription factors are removed from target genes by adenovirus infection to an extent that reduces their overall activity, then such transcription factors appear to be essential for the function of relatively few cellular promoters. If productive adenovirus infection leaves most cellular genes transcriptionally active, it appears that the continuing cellular transcription does not negatively influence adenovirus replication. This may be explained, at least in part, by the known ability of adenovirus to block cellular gene expression on the translation level. The E1B-55 kDa and E4orf6 proteins mediate this host cell shutdown phenomenon through their impact on mRNA export and translation (2, 3, 15, 31, 33), although some cellular mRNA species can escape this block and still yield protein (45).
Direct effects of E1A on the transcription and mRNA levels of myc have been previously reported, sometimes resulting in upregulation (16, 23, 39), sometimes in repression of myc (9, 32, 42). It should be noted that most of these studies were confined to promoter analysis in transient reporter assays. Such assays can sometimes yield misleading results, since they do not accurately reflect the situation of a chromosomally integrated, chromatin-packed gene. Moreover, reporter constructs frequently contain only a subset of the regulatory elements relevant to control gene expression, due to their limited size. In any case, E1A alone did not appear to be sufficient to downregulate myc mRNA in our study (Fig. 3B).
We have identified E1A as a factor that mediates the stabilization of MYC. The question remains by what mechanism(s) this occurs. Initial mapping studies revealed that several activities of E1A might cooperate to achieve this effect (our unpublished observations). Since E1A interacts with the transcriptional coactivator and acetyltransferase p300, it is a possibility that sequestration of p300 by E1A might alter MYC stability. This concept is supported by the finding that depletion of p300 upregulates MYC (21). However, in our hands, an E1A mutant lacking the capability to bind p300 was still partially able to stabilize MYC (data not shown). Another common binding factor used by both proteins is the complex of p400 and TRRAP (12, 28). Given the striking analogies between the biological activities of E1A and MYC, we favor the hypothesis that E1A might affect the stability of MYC by competing with the interaction of MYC with one or several partner proteins. Such interaction partners might, for instance, affect the ubiquitination of MYC.
The inhibitory effect of excess MYC on adenovirus replication might suggest that mechanisms to negatively regulate myc mRNA levels were selected during the evolution of adenovirus. However, it is still unknown how MYC interferes with adenovirus replication. We have determined that MYC only marginally affects early adenovirus gene expression but antagonizes viral DNA replication. This correlates with inhibited formation of the virus replication centers. MYC was previously implicated in the direct regulation of cellular DNA replication, e.g., through the association with enzymes of DNA replication (36) or through binding to p21/CDKN1A, replacing the proliferating cell nuclear antigen (PCNA) (19). Interestingly, MYC enhances, rather than decreases, the replication of simian virus 40 DNA by cellular polymerases (6). However, adenovirus uses a virus-encoded polymerase to replicate its DNA, and therefore, the requirements for the replication of its genome might be entirely distinct from those for cellular DNA or the genome of simian virus 40. Nonetheless, it is conceivable that MYC might interact with components of the DNA replication machinery used by the virus and interfere with their function.
Our data reveal a complex interplay between adenovirus infection and the expression of myc. Not only is myc among the relatively few genes that show a profoundly altered amount of mRNA upon infection, but MYC is further regulated at the level of protein stability and, in turn, is capable of affecting virus replication.
Acknowledgments
We thank C. Lenz-Bauer for excellent technical assistance; M. Eilers and M. Krause for invaluable help with cDNA arrays; V. Böhm for help with plasmid cloning and virus titrations; T. Shenk for adenoviruses dl309, dl338, and dl355; P. Hearing for adenovirus E4inorf3; and H. Hermeking and B. Vogelstein for an adenovirus vector expressing MYC.
This work was supported by the German Research Foundation and the P. E. Kempkes Foundation.
Footnotes
We dedicate this paper to Hans-Dieter Klenk on the occasion of his 65th birthday in appreciation of his generous and continuous support.
REFERENCES
- 1.Amati, B., S. R. Frank, D. Donjerkovic, and S. Taubert. 2001. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta 1471:M135-M145. [DOI] [PubMed] [Google Scholar]
- 2.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell, Jr. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltz, G. A., and S. J. Flint. 1979. Inhibition of HeLa cell protein synthesis during adenovirus infection. Restriction of cellular messenger RNA sequences to the nucleus. J. Mol. Biol. 131:353-373. [DOI] [PubMed] [Google Scholar]
- 4.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 5.Breckenridge, D. G., and G. C. Shore. 2000. Regulation of apoptosis by E1A and Myc oncoproteins. Crit. Rev. Eukaryot. Gene Expr. 10:273-280. [DOI] [PubMed] [Google Scholar]
- 6.Classon, M., M. Henriksson, J. Sumegi, G. Klein, M. L. Hammarskjold, and M. L. Hammaskjold. 1987. Elevated c-myc expression facilitates the replication of SV40 DNA in human lymphoma cells. Nature 330:272-274. [DOI] [PubMed] [Google Scholar]
- 7.Contente, A., A. Dittmer, M. C. Koch, J. Roth, and M. Dobbelstein. 2002. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 30:315-320. [DOI] [PubMed] [Google Scholar]
- 8.de Stanchina, E., M. E. McCurrach, F. Zindy, S. Y. Shieh, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 12:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dion, P. A., J. Fleurent, D. Roberge, and J. M. Weber. 1992. Reduction of c-myc expression correlated with E1a expression but not with the transformed phenotype. Virus Res. 26:231-240. [DOI] [PubMed] [Google Scholar]
- 10.Eisenman, R. N. 2001. Deconstructing myc. Genes Dev. 15:2023-2030. [DOI] [PubMed] [Google Scholar]
- 11.Fessler, S. P., and C. S. Young. 1998. Control of adenovirus early gene expression during the late phase of infection. J. Virol. 72:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 13.Gallimore, P. H., and A. S. Turnell. 2001. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene 20:7824-7835. [DOI] [PubMed] [Google Scholar]
- 14.Haberberger, T. C., K. Kupfer, and J. E. Murphy. 2000. Profiling of genes which are differentially expressed in mouse liver in response to adenoviral vectors and delivered genes. Gene Ther. 7:903-909. [DOI] [PubMed] [Google Scholar]
- 15.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiebert, S. W., M. Lipp, and J. R. Nevins. 1989. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc. Natl. Acad. Sci. USA 86:3594-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 19.Kitaura, H., M. Shinshi, Y. Uchikoshi, T. Ono, S. M. Iguchi-Ariga, and H. Ariga. 2000. Reciprocal regulation via protein-protein interaction between c-Myc and p21(cip1/waf1/sdi1) in DNA replication and transcription. J. Biol. Chem. 275:10477-10483. [DOI] [PubMed] [Google Scholar]
- 20.Koch, P., J. Gatfield, C. Löber, U. Hobom, C. Lenz-Stöppler, J. Roth, and M. Dobbelstein. 2001. Efficient replication of adenovirus despite the overexpression of active and non-degradable p53. Cancer Res. 61:5941-5947. [PubMed] [Google Scholar]
- 21.Kolli, S., A. M. Buchmann, J. Williams, S. Weitzman, and B. Thimmapaya. 2001. Antisense-mediated depletion of p300 in human cells leads to premature G1 exit and up-regulation of c-MYC. Proc. Natl. Acad. Sci. USA 98:4646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipp, M., R. Schilling, and G. Bernhardt. 1989. Trans-activation of human MYC: the second promoter is target for the stimulation by adenovirus E1a proteins. Oncogene 4:535-541. [PubMed] [Google Scholar]
- 24.Liu, H. T., R. Baserga, and W. E. Mercer. 1985. Adenovirus type 2 activates cell cycle-dependent genes that are a subset of those activated by serum. Mol. Cell. Biol. 5:2936-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutz, P., and C. Kedinger. 1996. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J. Virol. 70:1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutz, W., J. Leon, and M. Eilers. 2002. Contributions of Myc to tumorigenesis. Biochim. Biophys. Acta 1602:61-71. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda, S., J. Rouault, J. Magaud, and C. Berthet. 2001. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 497:67-72. [DOI] [PubMed] [Google Scholar]
- 28.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 29.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 99:6274-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosahl, T., and W. Doerfler. 1992. Alterations in the levels of expression of specific cellular genes in adenovirus-infected and -transformed cells. Virus Res. 26:71-90. [DOI] [PubMed] [Google Scholar]
- 33.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2111-2148. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 34.Smith, R. R., R. J. Huebner, W. P. Rowe, W. E. Schatten, and L. B. Thomas. 1956. Studies on the use of viruses in the treatment of carcinoma of the cervix. Cancer 9:1211-1218. [DOI] [PubMed] [Google Scholar]
- 35.Smith-Mungo, L. I., and H. M. Kagan. 1998. Lysyl oxidase: properties, regulation and multiple functions in biology. Matrix Biol. 16:387-398. [DOI] [PubMed] [Google Scholar]
- 36.Studzinski, G. P., U. T. Shankavaram, D. C. Moore, and P. V. Reddy. 1991. Association of c-myc protein with enzymes of DNA replication in high molecular weight fractions from mammalian cells. J. Cell. Physiol. 147:412-419. [DOI] [PubMed] [Google Scholar]
- 37.Tauber, B., and T. Dobner. 2001. Adenovirus early E4 genes in viral oncogenesis. Oncogene 20:7847-7854. [DOI] [PubMed] [Google Scholar]
- 38.Tauber, B., and T. Dobner. 2001. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 278:1-23. [DOI] [PubMed] [Google Scholar]
- 39.Thalmeier, K., H. Synovzik, R. Mertz, E. L. Winnacker, and M. Lipp. 1989. Nuclear factor E2F mediates basic transcription and trans-activation by E1a of the human MYC promoter. Genes Dev. 3:527-536. [DOI] [PubMed] [Google Scholar]
- 40.Tribouley, C., P. Lutz, A. Staub, and C. Kedinger. 1994. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 68:4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Dam, H., R. Offringa, A. M. Smits, J. L. Bos, N. C. Jones, and A. J. van der Eb. 1989. The repression of the growth factor-inducible genes JE, c-myc and stromelysin by adenovirus E1A is mediated by conserved region 1. Oncogene 4:1207-1212. [PubMed] [Google Scholar]
- 43.Weigel, S., and M. Dobbelstein. 2000. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J. Virol. 74:764-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wienzek, S., and M. Dobbelstein. 2001. Viral and cellular factors that target the promyelocytic leukemia oncogenic domains strongly activate a glucocorticoid-responsive promoter. J. Virol. 75:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, U. C., W. Huang, and S. J. Flint. 1996. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J. Virol. 70:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [Online.] http://nar.oupjournals.org/cgi/content/full/30/4/e15. [DOI] [PMC free article] [PubMed]
- 47.Zaika, A., M. Irwin, C. Sansome, and U. M. Moll. 2001. Oncogenes induce and activate endogenous p73 protein. J. Biol. Chem. 276:11310-11316. [DOI] [PubMed] [Google Scholar]
- 48.Zantema, A., and A. J. van der Eb. 1995. Modulation of gene expression by adenovirus transformation. Curr. Top. Microbiol. Immunol. 199:1-23. [DOI] [PubMed] [Google Scholar]