Abstract
Cullins assemble the largest family of ubiquitin ligases by binding with ROC1 and various substrate receptors. CUL4 function is linked with many cellular processes, but its substrate-recruiting mechanism remains elusive. We identified a protein motif, the DWD box (DDB1-binding WD40 protein), and demonstrated the binding of 15 DWD proteins with DDB1–CUL4A. We provide evidence supporting the critical function of the DWD box and DDB1’s role as the linker mediating DWD protein association with CUL4A. A database search predicts that about one-third of WD40 proteins, 90 in humans, contain the DWD box, suggesting a potentially large number of DWD–DDB1–CUL4–ROC1 E3 ligases.
Keywords: Ubiquitin, cullin, WD40, DDB1, CUL4, DWD
The ubiquitin–proteasome pathway regulates the concentration and conformation of many cellular proteins in response to changes in physiological conditions. This pathway consists of a cascade of three activities performed by E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) enzymes (Hochstrasser 1996; King et al. 1996; Hershko and Ciechanover 1998). A critical step in this process is how individual protein substrates are recruited to specific E3 ligases. The RING family represents the major family of E3 ligases. Members either contain an intrinsic RING finger domain (as in MDM2 and BRCA1) or bind in trans with a small RING finger protein, such as ROC1 (also known as Rbx1 and Hrt1) by the cullins, to recruit and activate an E2 (Jackson et al. 2000; Petroski and Deshaies 2005).
A remarkable aspect of cullin E3 ligases is that each cullin can assemble into many distinct cullin-RING-dependent ligases (CRLs) by interacting with a conserved motif present in multiple proteins (Petroski and Deshaies 2005). To recruit specific substrates, CUL1 utilizes an N-terminal domain to bind with a linker protein, SKP1 (Feldman et al. 1997; Skowyra et al. 1997; N. Zheng et al. 2002), which does not interact with other cullins (Michel and Xiong 1998). SKP1 uses a separate domain to bind with a conserved protein motif, the F box, which, via its additional protein–protein interaction modules, recruits various substrates, often phosphorylated, to the CUL1–ROC1 catalytic core. To bring specific substrates to CUL2- and CUL5-dependent ligases, a heterodimeric linker complex containing elongins B and C binds simultaneously to an analogous N-terminal domain in CUL2 and CUL5 and to two similar protein motifs, the VHL box and SOCS box. VHL and SOCS proteins, via their additional protein–protein interaction modules, target various substrates differentially to the CUL2–ROC1 or CUL5–ROC2 catalytic cores (Kamura et al. 1998, 2001, 2004; Stebbins et al. 1999; Zhang et al. 1999). Omitting a linker, CUL3 utilizes its N-terminal domain to bind to proteins with a conserved 100-residue protein motif known as a BTB domain, which, via additional protein–protein interaction domains, then target various substrates to the CUL3–ROC1 catalytic core (Furukawa et al. 2003; Geyer et al. 2003; Pintard et al. 2003; Xu et al. 2003). The presence of multiple substrate receptors—mammals express >60 F-box, 40 SOCS, and 200 BTB proteins—suggests that cullins may form the largest family of E3 ligase complexes and control the ubiquitination of a wide variety of substrates. The substrate recruiting mechanism of CUL4, which has two closely related paralogs, CUL4A and CUL4B, in mammals and presents as a single gene in Schizosaccharomyces pombe, Caenorhabditis elegans, and Drosophila, has remained elusive. Various reports have suggested that CUL4 may assemble multiple ligases to target many different substrates. One protein that has frequently been found in association with these various different CUL4 complexes is DDB1, first identified as a damaged DNA-binding protein (Chu and Chang 1988). The emerging picture is that DDB1 functions as a critical factor for CUL4–ROC ligases and possibly acts as a linker to bridge a substrate receptor(s) to CUL4. This study is directed toward understanding the substrate recruiting mechanism and the architecture of DDB1–CUL4 ligases.
Results and Discussion
CUL4A associates with a potentially large number of cellular proteins
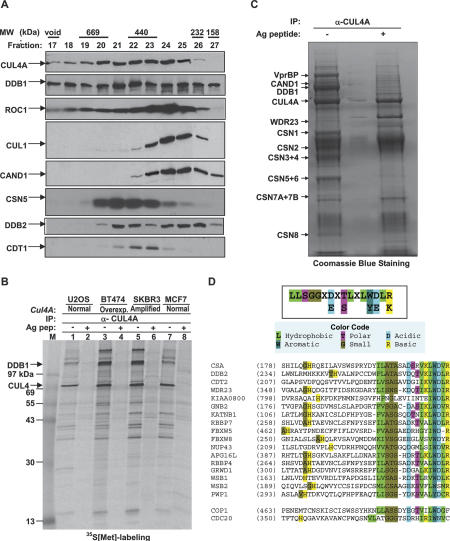
To begin searching for the substrate targeting mechanism of CUL4, we first examined CUL4A complexes in vivo by size-exclusion chromatography to gain an initial view of their size distribution. This study revealed that CUL4A exists in a number of different complexes, varying in size from ∼250 kDa to >700 kDa (Fig. 1A). Notably, two proteins that are critical partners of CUL4A, DDB1 and ROC1, also exist in many different complexes. In comparison, the distribution of most CUL1 complexes is limited to a much narrower size range—smaller than 440 kDa, very similar to that of CAND1, a negative regulator of CRLs that binds to an unneddylated form of cullins and blocks the binding of the SKP1 linker with CUL1 (Liu et al. 2002; J. Zheng et al. 2002; Goldenberg et al. 2004). The distribution of CSN5, the catalytic subunit of the COP9/signalosome, exhibits an almost mutually exclusive pattern from that of CUL1 and CAND1, but yet overlaps significantly with CUL4A. These observations are consistent with the finding that CUL4A molecules abundantly associate with the COP9/signalosome (Fig. 1C; Groisman et al. 2003), but only a small portion is associated with CAND1, suggesting that CUL4A assembles into many different and active complexes in vivo.
Figure 1.
Human CUL4 associates with potentially multiple WD40 repeat proteins. (A) Clarified lysate from HeLa cells was fractionated over a Superdex 200 gel filtration chromatography column, resolved by SDS-PAGE, and Western blotted with indicated antibodies. (B) 35S-labeled, clarified lysates from four cell lines were immunoprecipitated with an anti-CUL4A antibody and resolved by SDS-PAGE, followed by autoradiography. Identity of CUL4A and DDB1 were subsequently confirmed by mass spectrometric analysis. (C) Immunopurification and mass spectrometric analysis of CUL4A complexes. CUL4A immunocomplexes were precipitated from human BT474 cells and resolved by SDS-PAGE followed by staining with Coomassie brilliant blue. Bands identified by mass spectrometry are indicated. (D) Identification of a putative DDB1-binding motif in WD40 proteins, the DWD box. The DWD box is predicted to be present in >100 human WD40 repeat proteins. Only three reported in the literature (DDB2, CSA, and COP1) and 15 demonstrated in this study are included. CDC20 does not contain a DWD box and does not detectably interact with DDB1–CUL4A.
To identify proteins that interact with CUL4A in these different complexes, we analyzed CUL4A complexes by 35S-IP in various tissues (data not shown) and cell lines, including two breast cancer lines (SKBR3 and BT474) known to overexpress CUL4A (Chen et al. 1998), as well as two other cell lines that do not overexpress CUL4A. These analyses revealed a large number of potential CUL4A-interacting proteins as determined by antigen peptide competition (Fig. 1B). We then scaled up immunopurification of the CUL4A complex from BT474 cells and analyzed it by mass spectrometry. We have thus far identified 13 CUL4A-associated proteins (Fig. 1C). These include 11 proteins whose association with CUL4A had been previously identified (ROC1, DDB1, CAND1, and the eight COP9/signalosome subunits) and two novel proteins, WDR23 and VprBP. Coomassie blue staining indicates that CUL4A associates nearly stochiometrically with DDB1, but with the COP9/signalsome at a much lower level. Together, these results support the notion that CUL4A may associate with multiple different cellular proteins to assemble various E3 complexes and that DDB1 is a critical, if not essential, component of CUL4A ligases.
Identification of a DWD box conserved in DDB1-binding WD40 proteins
One intriguing finding from our preliminary studies of CUL4A-interacting proteins is the identification of two WD40 repeat proteins, WDR23 and VprBP. While neither protein has been functionally characterized or linked with CUL4–DDB1, the significance of this identification is appealing when considering that four previously identified CUL4A-interacting proteins—DDB2, CSA, COP1, and CDT2—also contain WD40 repeats (Groisman et al. 2003; Wertz et al. 2004; Holmberg et al. 2005; Liu et al. 2005). The WD40 repeat is loosely defined at the primary sequence level by a Gly–His dipeptide 10–20 residues N-terminal from a Trp–Asp (WD) dipeptide, and is typically ∼40 residues in length. WD40 proteins form a propeller-like structure, typically with seven blades, each composed of four anti-parallel β-sheets. There are >700 human WD40 repeat protein entries in the current PFAM database (http://www.sanger.ac.uk/Software/Pfam/index.shtml), corresponding to 320 unique genes after eliminating duplicates and truncated forms. As not all WD40 repeat proteins associate with DDB1 or CUL4 even when overexpressed (see below), it is likely that only a subset of WD40 repeat proteins interact with the DDB1–CUL4A ligase.
To identify the signature motif shared by DDB1-binding WD40 proteins, we carried out extensive primary sequence alignment and computer modeling. The WD40 repeats were first compared using NCBI-Blast searches and the CLUSTAL algorithm, then adjusted manually and continuously refined as more DDB1-binding proteins were experimentally identified (see below). This analysis identified a 16-residue stretch that we refer to as the DWD box (DDB1-binding and WD40 repeat) (Fig. 1D) and is defined by three features. (1) The first 14 residues in the DWD box correspond to the second half of a WD40 repeat that is more conserved than other WD40 repeats. These 14 residues include three highly conserved residues, Asp7, Trp13, and Asp14; five hydrophobic residues (Ile, Leu, or Val) at positions 1, 2, 10, 12, and 15; and three small residues (Ala, Gly, Ser, or Thr) at position 3, 4, and 5. (2) Arg16 following the WD dipeptide is a signature residue of the DWD box. Notably, the Arg residue in the DWD box of DDB2 is mutated in several human patients with xeroderma pigmentosum group E (XP-E), and reduces its binding with DDB1 (see Fig. 3, below; Rapic-Otrin et al. 2003). The crystal structure of the heterotrimeric G protein β1 subunit, a DWD protein (see below), has been solved and shows that several conserved residues, including the Arg16, within the DWD box are located on the protein surface (Sondek et al. 1996), suggesting that these residues might participate in binding with other proteins. (3) A DWD protein usually contains one and sometimes two DWD boxes, but rarely three.
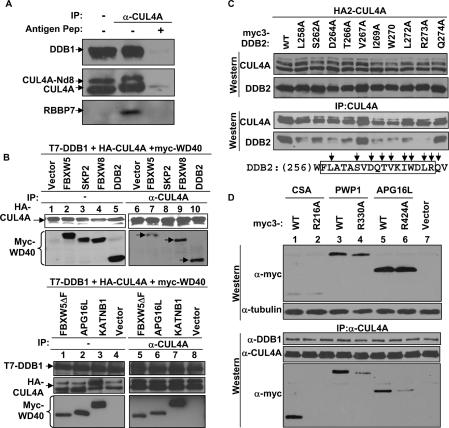
Figure 3.
Testing the DWD hypothesis. (A) CUL4A associates with endogenous RBBP7. 293T cells were lysed with 0.1% NP-40 lysis buffer and immunoprecipitated with an anti-CUL4A antibody, with or without molar excess of antigen peptide. The immunocomplexes were resolved by SDS-PAGE and Western blotted with indicated antibodies. (B) F-box-and-WD40 proteins FBXW5 and FBXW8 were characterized for binding with DDB1–CUL4A. SKP2 (F-box protein without WD40 repeats) and DDB2 were included as a negative and a positive control, respectively. A mutant of FBXW5 deleting the F box was tested for binding with DDB1–CUL4A along with two additional DWD proteins, APG16L and KATNB1, that do not contain an F box. (C) Individual residues of the DWD box of DDB2 protein were changed to alanine and tested for binding with CUL4A by IP-Western analysis. (D) The conserved arginine of the DWD box is critical for binding with DDB1–CUL4A. The arginines of three DWD proteins were changed to alanine and tested for binding with CUL4A by IP-Western analysis.
Multiple DWD proteins associate with DDB1–CUL4A
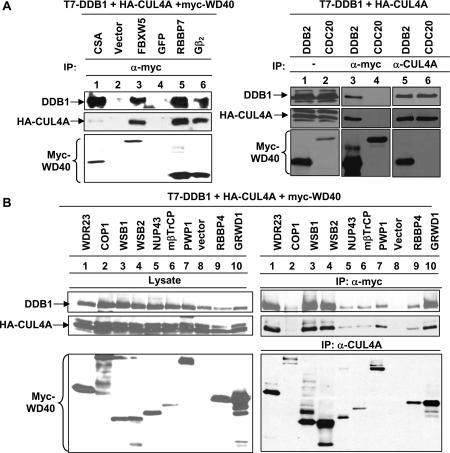
To experimentally test the DWD hypothesis, we randomly chose and expressed a number of putative DWD proteins and examined their binding with CUL4A– DDB1 by coupled IP-Western assays. Considering DDB1 as a possible linker, we triply transfected plasmids expressing CUL4A, DDB1, and individual WD40 proteins. Association between WD40 proteins and DDB1–CUL4A was determined by either Western blotting of the WD40 protein in the CUL4A immunocomplex or, conversely, blotting either CUL4A or DDB1 protein in the WD40 protein immunocomplex (Fig. 2A,B). DDB2 and CSA, the two best-characterized DWD proteins, interact with DDB1 strongly and were included as positive controls. We have identified thus far 15 novel human WD40 proteins that positively bind with DDB1–CUL4A. Twelve are associated with CUL4A–DDB1 in a readily detectable manner when compared with DDB2 and CSA, including VprBP and WDR23 (Fig. 1C); FBXW5, RBBP7, and Gβ2 (Fig. 2A); WSB1, WSB2, PWP1, and GRWD1 (Fig. 2B); and FBXW8, APG16L, and KATNB1 (Fig. 3B). Three WD40 proteins, NUP43, mβTrCP, and RBBP4, weakly interact with DDB1 and CUL4A (Fig. 2B). COP1 contains conserved WD40 repeats, but not the signature Arg16, and only associates with DDB1–CUL4A weakly in comparison with other WD40 proteins (Fig. 2B). It is currently not clear whether COP1 directly interacts with DDB1 or is bridged by another protein as suggested by the finding that a mutation outside the WD40 repeats in COP1 disrupted its binding with DET1 as well as with DDB1 (Wertz et al. 2004). CDC20, a WD40 protein that is missing several conserved residues, including the signature Arg16 (Fig. 1D), did not detectably interact with DDB1–CUL4A in a reciprocal IP-Western assay (Fig. 2A), suggesting that only a subset of, but not all, WD40 proteins interact with DDB1–CUL4A. Thus, together with three previously reported DDB1-binding WD40 proteins—DDB2, CSA, and COP1—a total of 18 DWD proteins have been demonstrated to bind with DDB1–CUL4 experimentally.
Figure 2.
Multiple WD40 repeat proteins bind with CUL4A– DDB1. (A,B) WD40 proteins were cloned into pcDNA3-myc3 expression vectors and cotransfected into 293T cells with plasmids expressing HA-CUL4A and T7-DDB1. Cell lysates were immunoprecipitated and Western blotted as indicated.
DWD box is important for DWD proteins binding to DDB1–CUL4A
To provide evidence supporting the DWD hypothesis, we first determined the in vivo association of endogenous CUL4A with a newly identified DWD protein, RBBP7, to which an antibody is available. RBBP7 was readily detected in the CUL4A immunocomplex and was competed off by molar excess of antigen peptide (Fig. 3A).
Finding that at least two F-box-containing, WD40 repeat proteins, FBXW5 (Fig. 2A) and FBXW8 (Fig. 3A), interact with DDB1–CUL4A is somewhat unexpected given that F-box proteins interact with CUL1. We determined whether the F box is required for the binding of WD40 proteins with DDB1–CUL4A. Binding assays showed that the F-box protein SKP2, which does not contain WD40 repeats, did not interact with CUL4A, and that deletion of the F box from FBXW5 did not seem to affect its binding with CUL4A (Fig. 3A). Two additional WD40 proteins that do not contain an F box, APG16L and KATNB1, were readily detected in the CUL4A immunocomplex in the same binding assay. We therefore conclude that WD40 repeats, but not the F box, are required for interacting with DDB1–CUL4A.
To directly test the DWD hypothesis, we performed site-directed mutagenesis of the DWD box of DDB2 (Fig. 3C). Nine residues in the DWD box were changed to alanine and individual mutant proteins were coexpressed with CUL4A. The expression of both CUL4A and DDB2 mutants were verified by direct Western blotting, and DDB2–CUL4A association was determined by IP-Western analysis. DDB2–CUL4A association was nearly completely disrupted by the mutation of the signature Arg at position 16 (corresponding to Arg273 in DDB2); substantially reduced by the mutations of five conserved residues—Leu at position 2 (corresponding to Leu258 in DDB2), Asp at position 7 (Asp264), Ile at position 10 (Ile269), Trp at position 13 (Trp270), and Leu at position 15 (Leu272); and reduced to a lesser extent by the mutation of Ser at position 3 (Ser262). Mutation of Val at position 11 (Val267), which is not highly conserved among the DWD proteins, and another residue, Gln274, outside the DWD box did not appreciably affect DDB2–CUL4A binding.
To further test the DWD hypothesis, we introduced alanine substitutions targeting the signature arginine residue in the DWD box of three additional DWD proteins, CSA, PWP1, and APG16L. The Arg-to-Ala substitution nearly completely disrupted the binding of DDB1–CUL4A with CSA, and substantially reduced the binding of DDB1–CUL4A with PWP1 and APG16L (Fig. 3D). Together, these bioinformatic analysis and binding and mutagenesis experiments demonstrate that the DWD box not only correlates with but also is directly involved in mediating the binding of a subset of WD40 proteins with DDB1–CUL4. Whether the DWD motif is sufficient for binding with DDB1 has not been tested due to the unique structure of WD40 proteins. Each WD40 propeller contains seven blades, and every blade is made of four β-sheets: three from one WD40 repeat and one from the next repeat. The DWD box corresponds to the second and third β-sheets of one blade. These features suggest that while the DWD box is required for binding to DDB1, other residues are likely involved as well and also make it unfeasible to map the binding sequence by deletion analysis without causing conformational collapse of the entire blade.
DDB1 links the DWD protein DDB2 to CUL4A
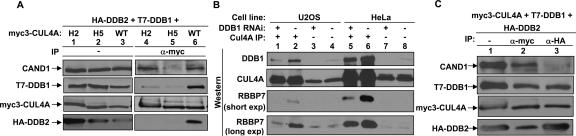
The results from our binding assays are most consistent with a model where DDB1 functions as a linker to bridge the DWD proteins to CUL4A. To directly test this model, we examined the interaction of DDB2 with CUL4A in more detail. Two hydrophobic helical surfaces in the N-terminal tip of CUL1, H2 and H5, bind with hydrophobic and polar residues from SKP1 to form a large interface. The N-terminal regions of other cullins form similar H2 and H5 helices, which contain residues that are invariably conserved in orthologs, but are different in paralogs (N. Zheng et al. 2002). We have previously shown that the substitution of residues in the H2 (86LYQAV90 → 86AAAAA90) or H5 (139WQDH142 → 39AADA142) helices of CUL4A substantially reduced DDB1–CUL4A binding (Fig. 3B; Hu et al. 2004). Binding assays demonstrated that mutation of either the H2 or H5 helix of CUL4A almost completely abolished CUL4A–DDB2 interaction (Fig. 4A). We noted that the steady-state level of DDB2 was higher when coexpressed with either the H2 or H5 mutant of CUL4A, a finding that is consistent with the possibility that DDB2 is degraded by the DDB1–CUL4A–ROC1 ligase, either as a direct substrate or as a substrate receptor that is degraded after delivering the substrate. Infection of a retrovirus expressing short hairpin RNA (shRNA) targeting DDB1 reduced the steady-state level of DDB1 by >85% in U2OS cells and about half in HeLa cells, did not change the steady-state level of either CUL4A or RBBP7, but substantially reduced the in vivo association of CUL4A–RBBP7 in both U2OS and HeLa cells (Fig. 4B). Together, these results support a model that DDB1 acts as a linker bridging the binding of DWD proteins and CUL4.
Figure 4.
DDB1 bridges WD40 repeat proteins to CUL4. (A) Intact H2 and H5 helices of CUL4A are required for binding with DDB1 and DDB2. 293T cells were cotransfected with plasmids expressing indicated proteins. The steady-state level and protein–protein interactions were determined by direct immunoblotting and IP-Western, respectively. (B) Silencing DDB1 reduced CUL4A–RBBP7 association. DDB1 was knocked down by the infection of a retrovirus expressing shRNA targeting DDB1. The steady-state level and CUL4A–RBBP7 association were determined by direct Western and IP-Western analysis. (C) CAND1 and DDB2 form mutually exclusive complexes with CUL4A. 293T cells were cotransfected with plasmids expressing HA-DDB2, T7-DDB1, and myc3-CUL4A. Cell lysates were immunoprecipitated and blotted with indicated antibodies.
We previously demonstrated that like SKP1–CUL1, the DDB1–CUL4A association is also negatively regulated by CAND1 (Hu et al. 2004). The model that DDB1 acts as a linker to bring other DWD proteins to CUL4 predicts that CAND1 and DWD proteins would interact with CUL4 in a mutually exclusive manner. To test this prediction, we determined the interactions among these proteins. As expected, CAND1, DDB1, and the DWD protein DDB2 were readily detected in the CUL4A immunocomplex (Fig. 4C). In the same assay, both DDB1 and CUL4A, but not CAND1, were detected in the DDB2 immunocomplex (Fig. 4C), further supporting the model that DDB1 acts as a linker bridging the binding of DWD protein and CUL4.
A large number of DWD proteins are present in different organisms
A remarkable feature of the cullin family of E3 ligases is the assembly of multiple E3 ligases by each individual cullin. Despite its linkage with multiple cellular pathways and many putative substrates, the substrate recruiting mechanism for CUL4 has remained elusive. The evidence presented here supports the model that a subset of WD40 proteins, defined by the presence of a DWD box, bind to DDB1, and that DDB1 functions as a linker connecting the DWD proteins to CUL4. A DWD protein could either be a substrate itself or function as a substrate receptor to recruit other protein(s) for ubiquitination by the CUL4–ROC catalytic core.
Database searches predict that there may be as many as 90 DWD proteins in humans (Supplementary Fig. 1), 74 in mice (Supplementary Fig. 2), 75 in Drosophila (Supplementary Fig. 3), 36 in C. elegans (Supplementary Fig. 4), 71 in Arabidopsis (Supplementary Fig. 5), 33 in S. pombe (Supplementary Fig. 6), and 20 in Saccharomyces cerevisiae (Supplementary Fig. 7). These observations suggest that CUL4–ROC1 could potentially form a large family of distinct ubiquitin ligases with individual DWD proteins and target the ubiquitination of many substrate proteins. Among the DWD proteins that have been experimentally tested positive for binding with DDB1– CUL4A are two F-box proteins, FBXW5 and FBXW8, and two SOCS proteins, WSB1 and WSB2 (Fig. 2). FBXW5 and WSB1 have been shown to bind with CUL1 and CUL2/5, respectively (Kamura et al. 2001; Arai et al. 2003). Whether these receptor proteins bind simultaneously with both CUL1 and CUL4 or CUL2/5 and CUL4, and whether cullin heterodimers more efficiently promote substrate polyubiquitination is an interesting possibility that is yet to be tested. The presence of 20 WD40 proteins in the S. cerevisiae genome with the potential to bind with DDB1 is equally intriguing since there is no obvious homolog of either DDB1 or CUL4 in budding yeast. Two obvious candidate linkers for binding with these putative DWD proteins are CPSF160 (the cleavage and polyadenylation specificity factor A, 160-kDa subunit) and SAP130 (spliceosome-associated protein 130). Both CPSF160 and SAP130 are present in budding yeast and exhibit a low-level similarity with DDB1 at the primary sequence level, but contain similar β-propellers, as predicted by computational modeling (Neuwald and Poleksic 2000) and reinforced by comparison with the DDB1 crystal structure (Li et al. 2006).
Materials and methods
Antibodies, immunopurification, and mass spectrometric analysis Antibodies to HA (12CA5, Boehringer-Mannheim), Myc (9E10, NeoMarker), T7 (Novagen), and Flag (M2, Sigma) were purchased commercially. Rabbit polyclonal antibodies to CUL4A, DDB1, and CDT1 have been described (Hu et al. 2004). A rabbit polyclonal antibody against CUL4B was produced by injection of a synthetic peptide antigen to residues 34–62 of CUL4B (AAQEVRSATDGNTSTTPPTSAKKRKLNSS). A rabbit polyclonal antibody against DDB2 was the kind gift of Dr. Altaf Wani (Ohio State University, Columbus, OH). To purify the endogenous CUL4A complex, BT474 cells from 47 150-mm plates were lysed with a 0.5% NP-40 lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% NaCl, 50 mM NaF), and lysates were pooled (300 mg total). Clarified lysates were immunoprecipitated with affinity-purified anti-CUL4A antibody (2 μg/mg lysate, ±10 μg/mg antigen peptide). Immunocomplexes were resolved by SDS-PAGE and stained with Coomassie blue, and the protein bands were digested with trypsin and subjected to mass spectrometric analysis at the University of North Carolina Proteomics Core Facility.
Plasmids, cell culture, and cell transfection
Plasmids expressing human CUL4A, DDB1, and CDT1 were as previously described (Ohta et al. 1999; Liu et al. 2002; Furukawa et al. 2003; Hu et al. 2004). Plasmids expressing DDB2, CSA, and all other WD40 proteins were produced by amplifying cDNA from either human HeLa or thymus (kind gift of Dr. Lishan Su, University of North Carolina, Chapel Hill, NC) cDNA libraries and subcloning into pcDNA3-based mammalian expression vectors. Mutations were introduced by site-directed mutagenesis using the QuickChange Kit (Stratagene) and verified by DNA sequencing. All human cells were cultured in DMEM containing 10% FBS in a 37°C incubator with 5% CO2, except 293T cells, which were cultured in DMEM containing 10% newborn calf serum. Cell transfections were carried out using a calcium-phosphate buffer.
Gel filtration chromatography
To examine the elution profile of CUL4A and associated proteins, HeLa cells were lysed with the 0.5% NP-40 lysis buffer, and clarified lysate was resolved through a Superdex-200 gel filtration column (GE/Amersham). Fractions (0.5 mL) were collected, and 50 μL of each was resolved via SDS-PAGE and immunoblotted with antibodies as indicated. High-molecular-weight standards (GE/Amersham) were resolved through the same column, and the peak fraction for each was determined.
35S-IP
About 80% confluent 100-mm tissue culture dishes of cells were washed with 1× PBS and then incubated in DMEM without methionine or cysteine (ICN, Inc.) + 10% dialyzed FBS, for 30 min. 35S-methionine (NEN) (0.5 mCi) was added to each dish and incubated for 4 h to label newly synthesized proteins. The cells were then washed twice with 5 mL of cold 1× PBS and lysed with 1 mL of 0.5% NP-40 lysis buffer per plate. Clarified lysates were divided in half, and each half was immunoprecipitated with 10 μg of anti-CUL4A antibody, ±50 μg of antigen peptide. The immunocomplexes were immobilized on Protein-A agarose beads and resolved via SDS-PAGE. The gel was then dried and exposed to film.
RNA interference (RNAI)
A duplex oligonucleotide encoding human DDB1-specific shRNA (5′-CCGGCAGCATTGACTTACCAGGCATCTTCCTGTCAATGCCTGGT AAGTCAATGCTGTTTTTG-3′) was ligated into the PMKO.1 vector (Addgene plasmid 8452). Retrovirus production was carried out according to a standard protocol and then used to infect U2OS and HeLa cells. The infected cell lines were then selected by puromycin (2 μg/mL) for 4 d before analyzing protein expression and complex formation.
Note added in proof
After submission of this paper, similar findings were published by the laboratories of Drs. Johannes Walter and Wade Harper (Jin et al. 2006) and Drs. Ning Zhang and Randall Moon (Angers et al. 2006).
Acknowledgments
We thank Brenda Temple for assisting in computer modeling, Altaf Wani for providing the DDB2 antibody, and Ning Zheng and members of the Xiong laboratory for discussion throughout this work. C.M.M. is supported by a DOD Breast Cancer Predoctoral Fellowship. This study is supported by an NIH grant (GM067113) to Y.X.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1483206.
References
- Angers, S., Li, T., Yi, X., MacCoss, M.J., Moon, R.T., Zheng, N. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature. 2006 doi: 10.1038/nature05175. Epub September 10, 2006; doi: 10.1038/ nature05175. [DOI] [PubMed] [Google Scholar]
- Arai, T., Kasper, J.S., Skaar, J.R., Ali, S.H., Takahashi, C., DeCaprio, J.A. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc. Natl. Acad. Sci. 2003;100:9855–9860. doi: 10.1073/pnas.1733908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.C., Manjeshwar, S., Lu, Y., Moore, D., Ljung, B.M., Kuo, W.L., Dairkee, S.H., Wernick, M., Collins, C., Smith, H.S. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. [PubMed] [Google Scholar]
- Chu, G., Chang, E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- Feldman, R.M.R., Correll, C.C., Kaplan, K.B., Deshaies, R.J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Furukawa, M., He, Y.J., Borchers, C., Xiong, Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nat. Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- Geyer, R., Wee, S., Anderson, S., Yates, J., Wolf, D.A. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- Goldenberg, S.J., Cascio, T.C., Shumway, S.D., Garbutt, K.C., Liu, J., Xiong, Y., Zheng, N. Structure of the Cand1–Cul1–Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A.F., Tanaka, K., Nakatani, Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Hershko, A., Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Holmberg, C., Fleck, O., Hansen, H.A., Liu, C., Slaaby, R., Carr, A.M., Nielsen, O. Ddb1 controls genome stability and meiosis in fission yeast. Genes & Dev. 2005;19:853–862. doi: 10.1101/gad.329905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., McCall, C.M., Ohta, T., Xiong, Y. Targeted ubiquitination of CDT1 by the DDB1–CUL4A–ROC1 ligase in response to DNA damage. Nat. Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Jackson, P., Eldridge, A.G., Freed, E., Furstenthal, L., Hsu, J.Y., Kaiser, B.K., Reimann, J.D.R. The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Sato, S., Haque, D., Liu, L., Kaelin, W.G., Jr., Conaway, R.C., Conaway, J.W. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras,WD-40 repeat, and ankyrin repeat families. Genes & Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J., Arias, E.E., Chen, J., Harper, J.W., Walter, J.C. A family of diverse Cul4–Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Burian, D., Yan, Q., Schmidt, S.L., Lane, W.S., Querido, E., Branton, P.E., Shilatifard, A., Conaway, R.C., Conaway, J.W. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 2001;276:29748–29753. doi: 10.1074/jbc.M103093200. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Maenaka, K., Kotoshiba, S., Matsumoto, M., Kohda, D., Conaway, R.C., Conaway, J.W., Nakayama, K.I. VHL-box and SOCS-box domains determine binding specificity for Cul2–Rbx1 and Cul5–Rbx2 modules of ubiquitin ligases. Genes & Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R.W., Deshaies, R.J., Peters, J.-M., Kirschner, M.W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Li, T., Chen, X., Garbutt, K.C., Zhou, P., Zheng, N. Structure of DDB1 in complex with a paramyxovirus V protein: Viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Liu, J., Furukawa, M., Matsumoto, T., Xiong, Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1– SKP1 binding and SCF ligases. Mol. Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Liu, C., Poitelea, M., Watson, A., Yoshida, S.H., Shimoda, C., Holmberg, C., Nielsen, O., Carr, A.M. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4–Ddb1–CSN ubiquitin ligase. EMBO J. 2005;24:3940–3951. doi: 10.1038/sj.emboj.7600854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, J.J., Xiong, Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:435–449. [PubMed] [Google Scholar]
- Neuwald, A.F., Poleksic, A. PSI-BLAST searches using hidden markov models of structural repeats: Prediction of an unusual sliding DNA clamp and of β-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res. 2000;28:3570–3580. doi: 10.1093/nar/28.18.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., Michel, J.J., Schottelius, A.J., Xiong, Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Petroski, M.D., Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pintard, L., Willis, J.H., Willems, A., Johnson, J.L., Srayko, M., Kurz, T., Glaser, S., Mains, P.E., Tyers, M., Bowerman, B., et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- Rapic-Otrin, V., Navazza, V., Nardo, T., Botta, E., McLenigan, M., Bisi, D.C., Levine, A.S., Stefanini, M. True XP group E patients have a defective UV-damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum. Mol. Genet. 2003;12:1507–1522. doi: 10.1093/hmg/ddg174. [DOI] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K., Tyers, M., Elledge, S.J., Harper, J.W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D.G., Hamm, H.E., Sigler, P.B. Crystal structure of a G-protein βγ dimer at 2.1A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Stebbins, C.E., Kaelin, W.G., Jr., Pavletich, N.P. Structure of the VHL–ElonginC–ElonginB complex: Implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- Wertz, I.E., O'Rourke, K.M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R.J., Dixit, V.M. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.H., Vidal, M., Elledge, S.J., Harper, J.W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- Zhang, J.G., Farley, A., Nicholson, S.E., Willson, T.A., Zugaro, L.M., Simpson, R.J., Moritz, R.L., Cary, D., Richardson, R., Hausmann, G., et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J., Yang, X., Harrell, J.M., Ryzhikov, S., Shim, E.H., Lykke-Andersen, K., Wei, N., Sun, H., Kobayashi, R., Zhang, H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell. 2002;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Zheng, N., Schulman, B.A., Song, L., Miller, J.J., Jeffrey, P.D., Wang, P., Chu, C., Koepp, D.M., Elledge, S.J., Pagano, M., et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]