Abstract
Recent studies revealed a striking difference in the genomic organization of classic cadherin genes and one family of “nonclassic cadherin” genes designated protocadherins. Specifically, the DNA sequences encoding the ectodomain repeats of classic cadherins are interrupted by multiple introns. By contrast, all of the encoded ectodomains of each member of the protocadherin gene clusters are present in one large exon. To determine whether large ectodomain exons are a general feature of protocadherin genes we have investigated the genomic organization of several additional human protocadherin genes by using DNA sequence information in GenBank. These genes include protocadherin 12 (Pcdh12), an ortholog of the mouse vascular endothelial cadherin-2 gene; hFmi1 and hFmi2, homologs of the Drosophila planar cell polarity gene, flamingo; hFat2, a homolog of the Drosophila tumor suppressor gene fat; and the Drosophila DN-cadherin and DE-cadherin genes. Each of these genes was found to be a member of the protocadherin subfamily, based on amino acid sequence comparisons of their ectodomains. Remarkably, all of these protocadherin genes share a common feature: most of the genomic DNA sequences encoding their ectodomains are not interrupted by an intron. We conclude that the presence of unusually large exons is a characteristic feature of protocadherin genes.
Cadherins are a superfamily of transmembrane glycoproteins that play a critical role in morphogenesis during development and maintenance of neuronal connections in the adult brain (1–4). The cadherin superfamily has been divided into classic and nonclassic types (5). The extracellular domains of both types of cadherins consist of variable numbers of characteristic repeat sequences (ectodomains) that function in calcium-dependent cell adhesion. The cytoplasmic domains of classic cadherins are highly conserved and interact with catenin proteins. Catenins link cadherins with the cytoskeleton, and they also function in the regulation of gene expression (2). By contrast, the cytoplasmic domains of nonclassic cadherins do not interact with catenins.
The protocadherins constitute a subfamily of nonclassic cadherins. Their extracellular domains contain six or more cadherin ectodomain repeats, and their cytoplasmic domains differ within the subgroup (6). The third and fifth ectodomain repeats (EC3 and EC5) of classic cadherins have characteristic amino acid sequences, and these sequences are not present in protocadherins (6). In addition, classic cadherins have five ectodomains (EC1–5) that are preceded by a prosegment and a signal peptide. By contrast, protocadherins lack the prosegment.
The genomic organization of classic cadherins, including E-cadherin (7, 8), N-cadherin (9), P-cadherin (10), and VE-cadherin (11), is well characterized. In most cases, the coding region of each ectodomain is interrupted by two introns, and the positions of the introns are conserved through vertebrate evolution. In contrast to classic cadherins, which have as many as 12 introns in the sequences encoding the ectodomains, the corresponding regions of the two types of protocadherin genes thus far analyzed contain no introns (12, 13). For example, three clusters of human protocadherin genes (Pcdhα, β, and γ) have been identified on human chromosome 5. Each of the clusters contains 15 or more large (at least 2,400 nt in length), tandemly linked exons that encode all six ectodomain repeats and the transmembrane domain. The cytoplasmic domains of the Pcdhα and γ protocadherins are encoded by three small exons located downstream from the respective tandem array of exons encoding the extracellular domains. Each of the large exons in the Pcdhα and γ gene clusters is independently spliced to the first exon encoding the intracellular domain in each gene cluster (13). Thus, the Pcdhα and γ gene clusters encode multiple protocadherins that differ in their extracellular domain sequences, but have identical sequences in their intracellular domains. Based on this organization, the large exons encoding the extracellular domains were designated variable region exons, whereas the three small exons encoding the intracellular domains were designated constant region exons (13).
Other genes that can be classified as protocadherin genes have been shown to encode multiple ectodomains. For example, the mouse vascular endothelial cadherin-2 (VE-cad-2) protein contains six ectodomains (14), whereas the fly Flamingo protein contains nine (15, 16). Similarly, the fly Fat protein contains 34 extracellular cadherin ectodomains (17). To determine whether the ectodomains in these and other protocadherins are interrupted by introns, we have analyzed their genomic organization by using DNA sequences in the GenBank database. A common feature of these protocadherin genes is that multiple ectodomains are encoded by a single, large exon.
Materials and Methods
Sequence Analyses.
Iterative blast searches of the GenBank database were carried out by using the sequences of the Pcdh gene clusters. Numerous entries with significant E values were identified. A systematic analysis of these entries revealed four additional human Pcdh genes and provided information necessary to determine their exon/intron organization. The genomic sequences were downloaded and compared with orthologous full-length cDNA and numerous human expressed sequence tag sequences by using the GCG package and blast 2 program. The splice sites were identified manually, based on the alignment of coding sequences at both the nucleotide and amino acid levels. The Pcdh12 gene sequence was derived from GenBank accession numbers AC005740, AC012625, and AC010264, whereas the hFmi1 gene sequence was derived from AC005923 and AC005903. The partial cDNA sequence of hFmi1 previously was reported as hMEGF2 (18). The hFmi2 gene sequence was derived from AL021392, AL031597, and AL031588. While this manuscript was in preparation, hFmi2 was designated hCelsr1 by the Sanger Center. The hFat2 gene sequence was derived from GenBank accession numbers AC011337, AC011374, and AC008670. The chromosomal locations of Pcdh12 and hFat2 genes were derived from the web site www-hgc.lbl.gov/biology/bacmap/2.gif, and those of hFmi1 and hFmi2 genes were from the corresponding GenBank entries. The organization of human cadherin 16 (Ksp-cadherin) gene was determined by comparing the cDNA (AF016272) and genomic DNA (AC009084 and AC008716) sequences. The organization of fly fmi (starry night) gene was determined by comparing the cDNA (AB028498 and AF172329) and genomic DNA (AC015222 and AC009343) sequences. The organization of fly fat gene was determined by comparing the cDNA (M80537) and genomic DNA (AC004380 and AC004371) sequences. The organization of fly DN-cadherin gene was determined by comparing the cDNA (AB002397) and genomic DNA (AC018021, AC018194, AC018219, AC009202, and AC009457) sequences. The organization of fly DE-cadherin gene was determined by comparing the cDNA (D28749) and genomic DNA (AC005268) sequences. The corresponding positions of introns within the protocadherin proteins were determined manually based on the translation of the exon sequences.
Evolutionary Analyses.
For the members of human Pcdh clusters, the variable region-coding sequences were translated. For other protocadherin proteins, the corresponding amino acids were used. For Pcdh12, hFat2, hFmi1, and hFmi2, the exon 1 sequences were translated. The polypeptides were aligned by using the pileup program of the GCG package with default parameters. An evolutionary tree was reconstructed by using paup (Phylogenetic Analysis Using Parsimony), version 4.0.0, with distance as an optimality criterion. The beginning and end positions of the alignment to be analyzed were determined according to the shortest sequence. Gaps in the alignment were ignored. The robustness of the tree partitions was evaluated by using the bootstrap method with a neighbor-joining search. The sequences are from the GenBank database: human Pcdhα1 (AF152479); human Pcdhβ1 (AF152488); human Pcdhγ-a1 (AF152507); mouse CNR1 (D80916); mouse Pcdh 2c (U88550); rat Pcdh3 (L43592); rat Pcdh4 (AB004276); chicken Pcdh2 (AB004689); human Pcdh1 (L11370); human Pcdh7 (AB006755); human Pcdh8 (AF061573); and human Pcdh68 (AF029343).
Results and Discussion
The Intron/Exon Organization of Classic Cadherin and Protocadherin Genes.
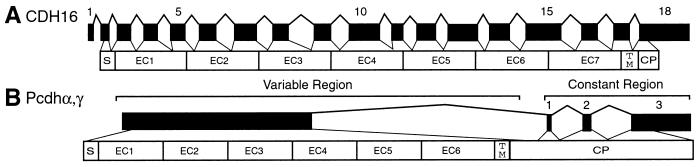
Studies of the genomic organization of a number of classic cadherin genes revealed that the DNA sequences encoding individual ectodomains are interrupted by two introns (7–11). An example of this is provided by human Ksp-cadherin (CDH16) gene (Fig. 1A). The Ksp-cadherin gene, which is expressed only in kidney cells, maps to chromosome 16q21–16q22 (19), a region that contains at least seven other cadherin genes (CDH1, CDH3, CDH5, CDH8, CDH11, CDH13, and CDH15). The DNA sequences encoding each of the ectodomains of the Ksp-cadherin gene (Fig. 1A) and all of the other genes characterized thus far in this cluster are interrupted by two introns. This intron/exon organization is typical of classic cadherin genes (7–11). By contrast, the six ectodomains of every member of the human protocadherin α, β, and γ proteins are encoded in a single, unusually large exon (Fig. 1B). To date, the genomic organization of only one other Pcdh gene (Pcdh8) has been determined (12), and all of its six ectodomains are encoded by a single, large exon. We have identified several additional human protocadherin genes and analyzed their genomic organizations to determine whether single exons encoding multiple ectodomains are a common feature of Pcdh genes.
Figure 1.
Comparison of the genomic organization of human cadherin 16 (A) and members of the human Pcdhα and γ gene clusters (B). Exons are indicated by solid rectangles, with the encoded protein shown below. CDH16, human cadherin 16; S, signal peptide; EC1–7, cadherin ectodomain 1–7; TM, transmembrane segment; CP, cytoplasmic domain.
Genomic Organization of the Human Pcdh12 Gene.
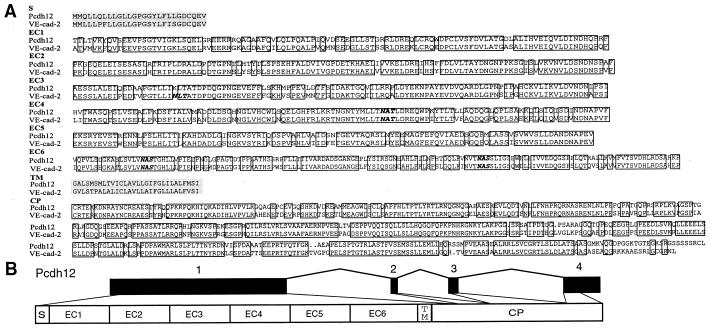
We have identified a human Pcdh gene (Fig. 2), which maps to the 5q31 region, approximately 0.3 Mb telomeric to the Pcdhα, β, and γ gene clusters. This human Pcdh gene, which we designate Pcdh12, encodes a protein of 1,184 aa, with 81% sequence identity with the mouse VE-cad-2 protein (14) (Fig. 2A). The high degree of sequence identity between Pcdh12 and VE-cad-2 strongly suggests that they are orthologs. The VE-cad-2 gene was cloned originally from heart microvascular endothelial cells and has been shown to localize at intercellular junctions of endothelial cells, suggesting that these proteins may play an important role in cell–cell interactions at interendothelial junctions (14).
Figure 2.
(A) An alignment of the human Pcdh12 and mouse vascular endothelial cadherin-2 (VE-cad-2) proteins. Identical residues are boxed. Putative N-glycosylation sites are indicated by bold italic letters. The two shaded regions correspond to the signal sequence and transmembrane domain, respectively. (B) Genomic organization of human Pcdh12 gene.
Like other protocadherins, the Pcdh12 protein has two regions of hydrophobic amino acids, a putative N-terminal signal peptide and a transmembrane domain (Fig. 2A). Between these hydrophobic sequences, there are six ectodomain repeats that have features of protocadherins (Fig. 2A). Specifically, they have the conserved motifs DXDXGXN, AXDXGXPXL, and VXVXVXDXNDNAPXF (Fig. 3). In addition, like all protocadherin genes characterized thus far, Pcdh12 does not have a prosegment between the signal peptide and ectodomains (Fig. 2). The cytoplasmic domain of Pcdh12 bears no similarity to that of any other cadherins in the database. Thus, it is likely that Pcdh12 functions through a yet-to-be-identified signaling pathway. Based on all of these sequence characteristics, we conclude that Pcdh12 is a member of the Pcdh gene family.
Figure 3.
Sequence alignment of human protocadherin and mouse N-cadherin ectodomain repeat sequences. Conserved amino acid sequences are indicated by white letters on a black background. Asterisks near the beginning of Pcdh12 EC6 and in the middle of hFmi2 EC4 represent 30 and 17 aa, respectively, which are not shown. Inclusion of these sequences would disrupt the alignment. Consensus sequences are shown at the bottom.
As illustrated in Fig. 2B, the genomic organization of the Pcdh12 gene is similar to that of the human Pcdhα, β, and γ gene clusters. In both cases, the extracellular and transmembrane domains are encoded by a single, large exon. However, in contrast to the human Pcdhα and γ gene clusters, where there are multiple variable region exons upstream from a single set of constant region exons, the Pcdh12 gene has only a single exon encoding all six ectodomains and the transmembrane domain. Similar to the organization of the constant region of human Pcdhα and γ gene clusters, the cytoplasmic domain of the Pcdh12 protein also is encoded by three small exons (Fig. 2B). However, the exons are larger than the corresponding constant region exons of human Pcdhα and γ gene clusters and their sequences are divergent. Thus, they encode a distinct intracellular domain.
Genomic Organization of Two Human Flamingo Genes.
A search of the DNA sequence database revealed two novel human Pcdh genes (Fig. 4) that appear to be the homologs of the recently cloned Drosophila planar cell polarity gene fmi (starry night) (15, 16). The Drosophila fmi gene plays a critical role in tissue polarity (15, 16, 20). The Fmi protein localizes to the proximal and distal cell boundaries in wing epithelia and plays an essential role in regulating planar cell polarization during prehair morphogenesis (15). The fmi gene also controls the planar polarity of sensory bristles (20). In addition, the Fmi protein may play a role in axon outgrowth (15) and dendrite formation (21) in the nervous system.
Figure 4.
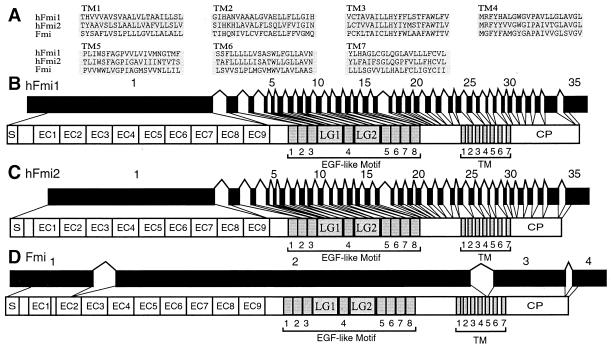
Alignment of the seven transmembrane segments of two human and the fly Fmi proteins (A) and genomic organization of the human flamingo 1 (B) and flamingo 2 (C) genes in comparison with that of the fly flamingo gene (D). hFmi1–2, human Flamingo 1–2; Fmi, fly Flamingo; LG, laminin A-G domain.
There are at least three human Fmi homologs, and the organization of two of them is reported here. The third was identified as partial cDNA sequence (Kiaa0279), and its genomic organization is not known (22). The hFlamingo1 (hFmi1) gene localizes on the 3p21 region of human chromosome 3, whereas the hFlamingo2 (hFmi2) gene localizes to the 22q13 region of human chromosome 22. There is a 56% amino acid sequence identity between hFmi1 and hFmi2 proteins, and the genomic organizations of the two genes are essentially identical (see below). Thus, the fmi genes constitute a subfamily of the cadherin superfamily, and the hFmi1 and hFmi2 genes are paralogs of the subfamily. The hFmi1 protein displays 92% sequence identity to the rat MEGF2 protein (18), whereas hFmi2 protein displays 82% sequence identity to the mouse Celsr1 protein (23). In contrast to all other members of the cadherin superfamily, the striking feature of these cadherins is the presence of seven transmembrane domains. A comparison of the transmembrane sequences of the human and fly Fmi protein is shown in Fig. 4A. The transmembrane region is homologous to a subfamily of G protein-coupled receptors.
Both of the human Fmi proteins have nine protocadherin ectodomain repeats (Fig. 4 B and C). However, they do not have the characteristic leucine residue within the middle region of the ectodomain repeats, which is conserved among members of the protocadherin clusters. Instead, they have an isoleucine residue that is conserved in the ectodomains of mouse N-cadherin protein (Fig. 3). In addition, both human Fmi proteins lack a prosegment between the signal peptide and ectodomains. However, they have eight epidermal growth factor (EGF)-like motifs and two laminin A-G domains (Fig. 4 B and C). The laminin A-G domain is present in other cadherin-like proteins, such as fly Fat (17), DE-cadherin (24) and DN-cadherin (25), and sea urchin G-cadherin (26). This domain is also present in other neural proteins, such as laminin A, agrin, merosin, slit, crumbs, and neurexin.
We have determined the genomic organization of both human Fmi genes. The intron/exon structures are highly conserved (Fig. 4 B and C) but are different from the genomic organization of the fly fmi gene, which was reported recently (16) and is shown in Fig. 4D for comparison. Both hFmi1 and hFmi2 have 34 introns. All splice sites conform to the GT-AG rule, and almost all introns appear in the homologous positions in the two proteins. In addition, the corresponding exons have the same size or differ only by several codons (data not shown). However, the intron sizes are very different. Almost all introns of hFmi2 are larger than the corresponding introns of hFmi1 (data not shown). The first two introns of hFmi2 gene are especially large, 70,281 and 24,295 nt in length, respectively.
The first exons of both hFmi1 and hFmi2 are very large, at least 4,029 and 3,544 nt in length, respectively. Moreover, the large exons encode the signal peptide and all nine ectodomain repeats in both hFmi1 and hFmi2 proteins (Fig. 4 B and C). Thus, the intron/exon organization of both genes is similar to that of Pcdh genes, where the first exon encodes the signal peptide and all six ectodomain repeats.
Genomic Organization of Human and Fly fat Genes.
The fly fat gene encodes a tumor suppressor that controls cell proliferation in imaginal discs (17). The Fat protein is a large, cadherin-like protein containing 34 cadherin ectodomains (17). The sequence features of the ectodomain repeats are more similar to those of protocadherins than to those of classic cadherins (6, 17). The cytoplasmic domain of all Fat proteins contains sequences homologous to the β-catenin-binding region of the classic cadherins.
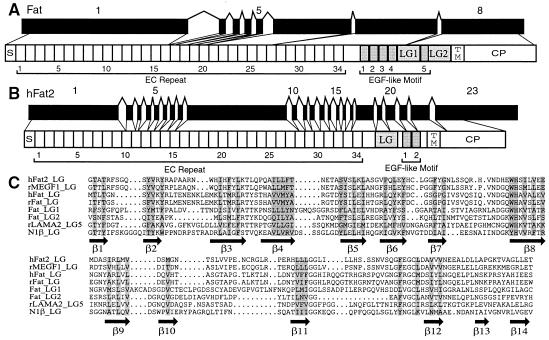
The genomic organization of fly fat gene is shown in Fig. 5A. Interestingly, the genomic DNA sequences encoding these ectodomains display variable intron/exon organization. The first exon encodes ectodomains 1–15 (at least 5,616 nt in length, Fig. 5A); the next five ectodomains are interrupted by a single intron, as is ectodomain 27. However, ectodomains 21–26 and 28–34 are encoded by large exons. Although some of the ectodomain sequences are interrupted by introns, neither the number nor the position of these introns corresponds to that of the introns highly conserved among the classic cadherins.
Figure 5.
Genomic organization of fly fat (A) and human Fat 2 (B) genes and a structure-based sequence alignment of the laminin A-G domains in Fat homologs (C). Sequences were aligned by using the pileup program of the GCG package and then manually adjusted based on recent structural information of the LNS domain of neurexin 1β (28) and LG5 domain of laminin α2 chain (29). Secondary structure elements of the 14 β-sheets are indicated below the alignment. Conserved core residues are shaded. rMEGF1, rat multiple EGF-like motifs containing protein; rLAMA2, rat laminin α2 chain; N1β, neurexin 1β.
A human Fat homolog was cloned previously and localized to chromosome 4q34-q35 (27). However, its genomic organization is not known. Here, we identified a novel human Fat homolog, designated hFat2 (Fig. 5B), which is located on chromosome 5q33 region, approximately 6–7 Mb telomeric to the Pcdhα, β, and γ gene clusters. The hFat2 gene encodes a protein of 4,349 aa and contains a signal peptide, 34 cadherin ectodomains, a laminin A-G domain, two EGF-like motifs, a transmembrane domain, and a cytoplasmic domain (Fig. 5B). All of the ectodomain sequences are protocadherin-like. A comparison of the sequences of the laminin A-G domains in different Fat homologs is shown in Fig. 5C. The laminin A-G domain consists of 14 regularly spaced β-sheets, which are found in all Fat homologs, the brain-specific, cell-surface protein neurexin (28), and the extracellular protein laminin (29).
The genomic DNA encoding hFat2 protein spans a region of about 60 kb and contains 22 introns (Fig. 5B). All introns are conventional GT-AG introns except intron 4, which is a rare GC-AG intron (30). As with the other protocadherin-like genes, multiple ectodomains (EC1–9 and EC16–27) are encoded in a single, large exon. However, the DNA sequences encoding the other ectodomains (EC10–15 and EC28–33) are interrupted by introns. Thus, similar to the fly Fat gene, the human gene has a mixed intron/exon organization.
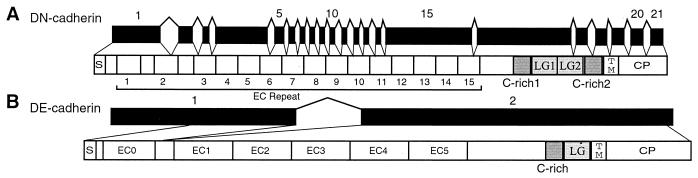
Genomic Organization of Drosophila DN-Cadherin and DE-Cadherin Genes.
The fly DN-cadherin gene is required for axon patterning in the central nervous system (25). This gene encodes a cadherin protein containing 15 ectodomain repeats, two cysteine-rich segments (C-rich), two laminin A-G domains, a transmembrane domain, and a cytoplasmic domain. Analysis of the genomic DNA encoding this protein reveals a mixed intron/exon organization. The coding region of some ectodomains is interrupted by one or two introns whereas other multiple ectodomains are encoded by a single exon (Fig. 6A).
Figure 6.
Genomic organization of fly DN-cadherin (A) and DE-cadherin (B) genes. C-rich, cysteine-rich domain.
The fly DE-cadherin originally was classified as a homolog of vertebrate classic cadherins because its cytoplasmic domain interacts with catenins (24). However, the amino acid sequences of its ectodomains are protocadherin-like (6), and the coding region of the DE-cadherin gene is interrupted by only a single intron (Fig. 6B). The exon 1 of DE-cadherin encodes the signal peptide and the N-terminal cadherin ectodomain (EC0) repeat, whereas exon 2 encodes the remaining five cadherin ectodomain (EC1–5) repeats, the transmembrane domain, and the cytoplasmic domain (Fig. 6B). The first cadherin ectodomain repeat probably is removed by proteolysis during its maturation (24). Thus, the mature form of the DE-cadherin protein is encoded by a single, large exon.
Phylogenetic Analysis of Protocadherin Genes.
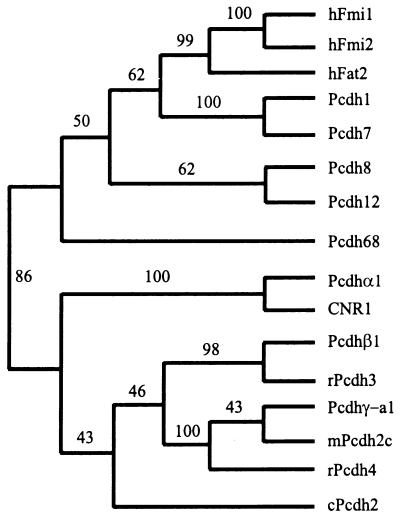
An evolutionary tree of the amino acid sequences encoded by the large exon 1 of the human Pcdh genes described above and those encoded by single variable region exons in the Pcdhα, β, and γ gene clusters is presented in Fig. 7. The tree shows that the protocadherins encoded by the Pcdhα, β, and γ gene clusters constitute a large subfamily of cadherin-like proteins. All of the other protocadherins are in a separate group. Consistent with their genomic organization, the two human Fmi paralogs are in one small branch and are closely related to the hFat2 protein. The large protocadherin proteins are evolutionarily closer to seven-ectodomain-containing protocadherins (Pcdh1 and Pcdh7) than to six-ectodomain-containing protocadherins (Pcdh8, Pcdh12, Pcdh68, and members of the Pcdh clusters). These results strongly suggest that the large protocadherin genes evolved from a primordial protocadherin-type gene. Thus, it seems very likely that a basic evolutionary unit of these protocadherin genes is a large exon encoding multiple cadherin ectodomains.
Figure 7.
Evolutionary relationships between the ectodomains of different cadherin-like proteins. The evolutionary tree was reconstructed by using the neighbor-joining method of the paup program. The tree is unrooted. The bootstrap nodes are labeled with the percentage support for that partition based on 1,000 bootstrap replicates.
Evolution of Cadherin-Like Genes.
Phylogenetic analyses suggest that there are four paralogous subfamilies, E-, N-, P-, and R-cadherins, of vertebrate classic cadherin proteins (31) and that these genes duplicated early in evolution (32). The conservation of intron positions among classic cadherins suggests that there was a constraint on unequal crossing-over among these genes (9). Through systematic analyses of protocadherin genomic organization, we found that a single exon encoding multiple cadherin ectodomains is a characteristic feature of protocadherin genomic organization. In addition, some protocadherins have unusually large exons encoding more than nine ectodomains. These large exons could have been generated by unequal crossing-over between the six-ectodomain-encoding protocadherin-type exons lacking an intron.
Most human genes have multiple small exons and large introns. For example, the average size of vertebrate exons is approximately 134 nt (33) and the average size of human first exons is about 284 nt (34). The small exon size is thought to be a consequence of “exon definition,” the mechanism by which splice sites are recognized by cross-exon interactions between splicing components bound to the splice sites flanking each exon. A surprising characteristic of the organization of the protocadherin genes is that multiple cadherin ectodomain repeats are encoded by a single, large exon. However, it is interesting to note that in most cases the large exon is the 5′-most exon in the pre-mRNA. Previous studies have shown that the cap-binding complex can promote the recognition of the first 5′ splice site in pre-mRNA (35). Thus, the cap-binding complex may play a more prominent role in the processing of the large 5′-most exons of the protocadherin pre-mRNAs.
The tandem organization of the Pcdhα, β, and γ gene clusters on the human chromosome 5q31 region suggests that the Pcdh clusters evolved from duplications of a large exon of a primordial protocadherin that encodes the extracellular and transmembrane domains (13). For example, it is possible that the first round of duplication generated the Pcdhγ-c genes and the ancestor of the Pcdhγ-a/b genes. The resulting genes then were duplicated to form the ancestors of Pcdhγ and Pcdhα gene clusters. The present organization of human Pcdh clusters appears to be a consequence of extensive duplication of the variable regions of each cluster. The protocadherins encoded by these gene clusters are thought to play an important role in the establishment and maintenance of specific neural cell–cell connections in the vertebrate brain (3, 13). The difference in the genomic organizations of classic cadherin and protocadherin genes suggests that the two types of cadherin-like genes diverged from each other very early in evolution and then evolved as separate subfamilies.
Acknowledgments
We thank members of the Maniatis laboratory for useful suggestions and C. Nabholz and B. Tasic for critical comments on the manuscript. We are grateful to J.-F. Cheng for invaluable discussions on the chromosomal locations of Pcdh12 and hFat2 genes. This work was supported by National Institutes of Health Grant GM42231 (to T.M.). Q.W. was supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation (DRG-1559).
Abbreviations
- VE-cad-2
vascular endothelial cadherin-2
- CDH
cadherin
- Pcdh
protocadherin
- Fmi
flamingo
- EC
ectodomain
- EGF
epidermal growth factor
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF231022 (hFat2), AF231023 (hFmi1), AF231024 (hFmi2), and AF231025 (Pcdh12)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060027397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060027397
References
- 1.Takeichi M. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 2.Gumbiner B M. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro L, Colman D R. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 4.Tepass U. Curr Opin Cell Biol. 1999;11:540–548. doi: 10.1016/s0955-0674(99)00025-3. [DOI] [PubMed] [Google Scholar]
- 5.Uemura T. Cell. 1998;93:1095–1098. doi: 10.1016/s0092-8674(00)81452-x. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki S T. J Cell Sci. 1996;109:2609–2611. doi: 10.1242/jcs.109.11.2609. [DOI] [PubMed] [Google Scholar]
- 7.Sorkin B C, Gallin W J, Edelman G M, Cunningham B A. Proc Natl Acad Sci USA. 1991;88:11545–11549. doi: 10.1073/pnas.88.24.11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ringwald M, Baribault H, Schmidt C, Kemler R. Nucleic Acids Res. 1991;19:6533–6539. doi: 10.1093/nar/19.23.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyatani S, Copeland N G, Gilbert D J, Jenkins N A, Takeichi M. Proc Natl Acad Sci USA. 1992;89:8443–8447. doi: 10.1073/pnas.89.18.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatta M, Miyatani S, Copeland N G, Gilbert D J, Jenkins N A, Takeichi M. Nucleic Acids Res. 1991;19:4437–4441. doi: 10.1093/nar/19.16.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber P, Dalmon J, Engiles J, Breviario F, Gory S, Siracusa L D, Buchberg A M, Dejana E. Genomics. 1996;32:21–28. doi: 10.1006/geno.1996.0072. [DOI] [PubMed] [Google Scholar]
- 12.Strehl S, Glatt K, Liu Q M, Glatt H, Lalande M. Genomics. 1998;53:81–89. doi: 10.1006/geno.1998.5467. [DOI] [PubMed] [Google Scholar]
- 13.Wu Q, Maniatis T. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 14.Telo P, Breviario F, Huber P, Panzeri C, Dejana E. J Biol Chem. 1998;273:17565–17572. doi: 10.1074/jbc.273.28.17565. [DOI] [PubMed] [Google Scholar]
- 15.Usui T, Shima Y, Shimada Y, Hirano S, Burgess R W, Schwarz T L, Takeichi M, Uemura T. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 16.Chae J, Kim M, Goo J H, Collier S, Gubb D, Charlton J, Adler P N, Park W J. Development (Cambridge, UK) 1999;126:5421–5429. doi: 10.1242/dev.126.23.5421. [DOI] [PubMed] [Google Scholar]
- 17.Mahoney P A, Weber U, Onofrechuk P, Biessmann H, Bryant P J, Goodman C S. Cell. 1991;67:853–868. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama M, Nakajima D, Nagase T, Nomura N, Seki N, Ohara O. Genomics. 1998;51:27–34. doi: 10.1006/geno.1998.5341. [DOI] [PubMed] [Google Scholar]
- 19.Thomson R B, Ward D C, Quaggin S E, Igarashi P, Muckler Z E, Aronson P S. Genomics. 1998;51:445–451. doi: 10.1006/geno.1998.5402. [DOI] [PubMed] [Google Scholar]
- 20.Lu B, Usui T, Uemura T, Jan L, Jan Y N. Curr Biol. 1999;9:1247–1250. doi: 10.1016/s0960-9822(99)80505-3. [DOI] [PubMed] [Google Scholar]
- 21.Gao F B, Brenman J E, Jan L Y, Jan Y N. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N. DNA Res. 1996;3:321–329. doi: 10.1093/dnares/3.5.321. [DOI] [PubMed] [Google Scholar]
- 23.Hadjantonakis A K, Formstone C J, Little P F R. Mech Dev. 1998;78:91–95. doi: 10.1016/s0925-4773(98)00153-1. [DOI] [PubMed] [Google Scholar]
- 24.Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. Dev Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- 25.Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 26.Miller J R, McClay D R. Dev Biol. 1997;192:323–339. doi: 10.1006/dbio.1997.8740. [DOI] [PubMed] [Google Scholar]
- 27.Dunne J, Hanby A M, Poulsom R, Jones T A, Sheer D, Chin W G, Da S M, Zhao Q, Beverley P C, Owen M J. Genomics. 1995;30:207–223. doi: 10.1006/geno.1995.9884. [DOI] [PubMed] [Google Scholar]
- 28.Rudenko G, Nguyen T, Chelliah Y, Südhof T C, Deisenhofer J. Cell. 1999;99:93–101. doi: 10.1016/s0092-8674(00)80065-3. [DOI] [PubMed] [Google Scholar]
- 29.Hohenester E, Tisi D, Talts J F, Timpl R. Mol Cell. 1999;4:783–792. doi: 10.1016/s1097-2765(00)80388-3. [DOI] [PubMed] [Google Scholar]
- 30.Wu Q, Krainer A R. Mol Cell Biol. 1999;19:3225–3236. doi: 10.1128/mcb.19.5.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallin W J. Mol Biol Evol. 1998;15:1099–1107. doi: 10.1093/oxfordjournals.molbev.a026017. [DOI] [PubMed] [Google Scholar]
- 32.Pouliot Y. BioEssays. 1992;14:743–748. doi: 10.1002/bies.950141104. [DOI] [PubMed] [Google Scholar]
- 33.Sterner D A, Carlo T, Berget S M. Proc Natl Acad Sci USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M Q. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 35.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]