Abstract
The inflammatory bowel diseases, Crohn's disease and ulcerative colitis, are chronic inflammatory disorders of the gastrointestinal tract. The causes of these diseases remain unknown; however, prevailing theories suggest that chronic intestinal inflammation results from a dysregulated immune response to ubiquitous bacterial antigens. While a substantial body of data has been amassed describing the role of the adaptive immune system in perpetuating and sustaining inflammation, very little is known about the early signals, prior to the development of inflammation, that initiate and direct the abnormal immune response. To this end, we characterized the gene expression profile of A/JCr mice with Helicobacter hepaticus-induced typhlitis at month 1 of infection, prior to the onset of histologic disease, and month 3 of infection, after chronic inflammation is fully established. Analysis of the gene expression in ceca of H. hepaticus infected mice revealed 25 up-regulated and 3 down-regulated genes in the month-1 postinoculation group and 31 up-regulated and 2 down-regulated genes in the month-3 postinoculation group. Among these was a subset of immune-related genes, including interferon-inducible protein 10, monokine induced by gamma interferon, macrophage-induced protein 1 alpha, and serum amyloid A1. Semiquantitative real-time reverse transcriptase PCR confirmed the increased expression levels of these genes, as well as elevated expression of gamma interferon. To our knowledge, this is the first report profiling cecal gene expression in H. hepaticus-infected A/JCr mice. The findings of altered gene expression prior to the development of any features of pathology and the ensuing chronic disease course make this an attractive model for studying early host response to microbe-induced inflammatory bowel disease.
The inflammatory bowel diseases (IBD) Crohn's disease (CD) and ulcerative colitis (UC) are recognized as important causes of gastrointestinal disease with combined prevalence rates of 175 to 400 cases per 100,000 persons in North America (2, 31, 54). CD is characterized by chronic relapsing inflammation of the gastrointestinal tract that may affect any region of the intestine from the oropharynx to the perianal area. Inflammation is commonly transmural and is sometimes accompanied by granuloma formation. In contrast to CD, UC lesions are confined to the colon and inflammation, associated with ulceration and hemorrhage in severe disease, is limited to the mucosa (48). While the clinical and histopathological presentations of these disorders are distinctive, it is theorized that they are triggered by similar pathogenic mechanisms. Present theories suggest that the IBDs are multifactorial and that development of persistent intestinal inflammation results from a dysregulated immune response generated, at least in part, against normally harmless microbial antigens (13, 29, 43). Once triggered, the ensuing immune response is channeled down a determinative pathway where inflammation is either associated with elevated interleukin-12 (IL-12), gamma interferon (IFN-γ), or tumor necrosis factor alpha (TNF-α), as in CD (32, 44), or with excessive production of IL-5, as in UC (18). Much of our present understanding about the immune signals that drive this inflammation is derived from the study of chronically inflamed tissues and from animal models of IBD at the time of disease onset (56). While these studies have provided incontrovertible evidence that a dysregulated immune response and abnormal cytokine production pattern are pivotal to the development of IBD, very little is known about the antecedent signals, i.e., those that occur prior to the development of inflammation.
The prolonged preinflammatory period and the ensuing chronic inflammatory changes in the context of an intact immune system make Helicobacter hepaticus infection in A/JCr mice an attractive model for studying gene dysregulation during the early and late stages of microbe-induced IBD. H. hepaticus is a gram-negative, microaerophilic, urease-positive, spiral rod that has been associated with intestinal disease in A/JCr mice (16). H. hepaticus infection in these mice is typically subclinical, and enteritis develops only after months of infection (15, 16, 61). Furthermore, inflammation is characterized by predominantly mononuclear cell infiltrates in the lamina propria, hyperplasia of mucosa-associated lymphoid tissue with follicle formation, and mild mucosal epithelial hyperplasia (15, 61).
In this work, we hypothesize that cecal gene dysregulation associated with H. hepaticus infection occurs prior to development of any pathological features of typhlitis and is augmented during the inflammatory phase of disease. To begin addressing this hypothesis, we focused on three specific aims. We first identified the time points that correlate to early and late stages of disease: prior to the onset of pathological lesions and after typhlitis is fully established. Second, we surveyed the gene expression profile in ceca of H. hepaticus-infected A/JCr mice in early and late disease. Third, we selected a subset of genes from the cDNA array data and used real-time reverse transcriptase PCR (RT-PCR) to quantify their expression. This study demonstrated that H. hepaticus infection in A/JCr mice is clinically and histologically silent during the 1st month of infection; by month 3, intestinal inflammation is clearly established. Analysis of the cDNA array gene expression profiles revealed 25 up-regulated and 3 down-regulated genes in the month-1 postinfection group and 31 up-regulated and 2 down-regulated genes in the month-3 postinfection group. Among these was a subset of immune-related genes, including CXC chemokine IFN-inducible protein of 10 kDa (IP-10), monokine induced by IFN-γ (MIG), the CC chemokine macrophage inflammatory protein 1α (MIP-1α), and serum amyloid A1 (SAA1). Semiquantitative real-time RT-PCR confirmed the increased expression levels of these genes as well as elevated expression of IFN-γ.
MATERIALS AND METHODS
Bacteria and cultivation.
A Helicobacter isolate (strain MU-94) was obtained from an endemically infected mouse colony by using a previously described culture technique (15, 30). The isolate was identified as H. hepaticus by ultrastructural morphology, biochemical characteristics, and sequence analysis of the 16S rRNA gene (49). For inoculation, H. hepaticus cultures were grown in brucella broth (Becton Dickinson, Franklin Lakes, N.J.) supplemented with 5% fetal calf serum (Sigma-Aldrich Co., St. Louis, Mo.). Cultures were agitated with a stir bar in a 250-ml Erlenmeyer flask and were incubated for 24 h at 37°C in a microaerobic environment with 90% N2-5% H2-5% CO2.
Animals and sample collection.
Three-week-old female A/JCr mice were obtained from the Frederick Cancer Research and Development Center (FCRDC, Frederick, Md.). Mice tested negative for Helicobacter infection by a generic PCR assay that amplifies a 374-bp region of the 16S rRNA gene from all known murine Helicobacter spp. (49). Mice were separated into four groups (two experimental and two control) of 10 mice each. Experimental mice were gavaged with 108 H. hepaticus organisms suspended in brucella broth, and control mice were sham inoculated with sterile brucella broth. Helicobacter infection was confirmed by fecal PCR assay at 2 weeks postinoculation and when mice were euthanatized for sample collection. At months 1 and 3 postinoculation, groups of infected and control mice were euthanatized by an inhaled overdose of CO2. The cecum was collected and was equally divided into two longitudinal sections. One cecal section was snap-frozen and stored at −80°C for gene expression analysis, and the remaining cecal strip was placed in 10% neutral buffered formalin for histologic evaluation.
Histopathology and intestinal lesion scoring.
Formalin-fixed ceca from H. hepaticus-infected and control mice were embedded in paraffin, cut in 5-μm-thick sections, and processed for staining with hematoxylin and eosin. To objectively assess the severity of intestinal disease in H. hepaticus-infected mice, we adapted the scoring system described by Mohammadi and coworkers for analysis of chronic gastritis in H. felis-infected mice (34). Briefly, lesions were scored for intensity of inflammation (0 = none, 1 = mild, 2 = moderate, and 3 = severe), longitudinal extent (1 = one or two small foci, 2 = patchy, and 3 = diffuse), and vertical extent of inflammation (1 = basal mucosal inflammation, 2 = full-thickness mucosal inflammation, and 3 = transmural inflammation). In addition, lesions were scored for hyperplasia by using the following criteria: hyperplasia is defined as the presence of basophilic staining “crypt” epithelial cells in at least the lower two-thirds of the gland or at least doubling of the height of the mucosal epithelium. Focal hyperplasia was given a score of 1, and diffuse hyperplasia was given a score of 2. The scores for intensity of inflammation, longitudinal and vertical extents of inflammation, and hyperplasia were added together to give a total score for each animal. Because the minimum inflammation score using this system is 3 (mild, focal, and basal), inflammation and hyperplasia were not on comparable scales. To rectify this bias, lesion scores of >2 were adjusted by subtracting 2 from the total score to give a total adjusted score. The total adjusted score was used to compare lesions in H. hepaticus-infected and sham-infected control mice.
RNA extraction from cecal tissue.
Frozen ceca were thawed in a solution of phenol and guanidine isothiocyanite (TRIzol reagent; Invitrogen, Carlsbad, Calif.). The ceca were homogenized by using a polypropylene pestle that was driven by a Pellet Pestle Motor (Kimble/Kontes, Vineland, N.J.). Total RNA was isolated according to the manufacturer's protocol (TRIzol reagent; Invitrogen), and the extracted RNA was dissolved in 100 μl of water treated with diethyl pyrocarbonate (Sigma-Aldrich Co.). Because contamination by genomic DNA is potentially problematic when working with cDNA arrays, the extracted RNA sample was DNase digested (RNase-Free DNase Set; Qiagen, Valencia, Calif.) and was repurified by using a silica-gel membrane column (RNeasy; Qiagen). The quantity and quality of RNA were assessed by measuring the absorbance at 260 nm and the relative intensity of 28S and 18S rRNA bands on a 1.5% agarose denaturing gel (NorthernMax; Ambion, Austin, Tex.) stained with ethidium bromide, respectively.
Complex probe preparation and hybridization to cDNA expression arrays.
The Atlas Mouse 1.2II cDNA expression array (Clontech, Palo Alto, Calif.), which interrogates expression of 1,176 genes, was used to profile gene expression of cecal tissue from H. hepaticus-infected mice at months 1 and 3 of infection. This panel includes genes that play key roles in many different biological processes, including cytokines, growth factors, transcription factors, cell cycle regulators, and nine housekeeping genes. The gene expression profile for each time point was compiled from the statistical analysis of 18 (nine uninfected controls; nine H. hepaticus-infected animals) biological replicates.
Complex probe preparation.
The cDNA complex probes were prepared by using the Array-Advantage AA cDNA labeling and hybridization kit (Ambion). Briefly, 32P-radiolabeled cDNA probes were prepared by reverse transcribing 10 μg of total RNA with 400 U of Moloney murine leukemia virus RT and 1× gene-specific primers (Atlas Mouse 1.2II cDNA expression array; Clontech) in the presence of 30 μCi of [32P]dATP (Perkin-Elmer Life Sciences, Boston, Mass.) and 1× deoxyneucleoside triphosphate (dNTP) mix (Strip-EZ dNTP; Ambion). Unincorporated [32P]dATP was removed by filtration through NucAway Spin Columns (Ambion). Incorporation of [32P]dATP into cDNA was measured in a scintillation counter, and typical activity was estimated as 1× 106 to 5 × 106 cpm.
Hybridization.
The probe sets were hybridized to Atlas Mouse 1.2II cDNA expression arrays (Clontech) according to the Array-Advantage AA (Ambion) instructions. The Atlas Mouse 1.2II cDNA expression arrays were prehybridized for 1 h at 50°C in 10 ml of ULTRArray hybridization buffer (Ambion). Heat-denatured herring sperm DNA was added to the hybridization buffer at the end of the prehybridization period, and the array membrane was incubated for an additional 5 min. Heat-denatured complex probe cDNA (1 × 106 to 5 × 106 cpm) was then added to the hybridization buffer and was incubated at 52°C for 16 h. The membranes were washed twice at 52°C for 30 min with low-stringency buffer containing 2× SSC (1× SSC is 0.15 M NacCl plus 0.015 M sodium citrate) and 1% sodium dodecyl sulfate and then twice at 52°C for 30 min with high-stringency buffer containing 0.5× SSC and 1% sodium dodecyl sulfate. The Atlas Arrays were exposed to a phosphor screen (Bio-Rad Laboratories, Hercules, Calif.) for 24 and 72 h and were imaged with the Personal Molecular Imager FX System (Bio-Rad). Gene signal intensities in ceca of H. hepaticus-infected and sham-infected mice were measured and tabulated for each address on the array by using AtlasImage software (Clontech).
Confirmation of differentially expressed genes.
Semiquantitative real-time RT-PCR was used to measure the transcriptional activation of five proinflammatory genes, i.e., SAA1, IP-10, MIG, MIP-1α, and IFN-γ. These genes were chosen from among the differentially expressed genes discovered in the cDNA expression array analysis studies and were based on published reports that demonstrate their association with chronic human inflammatory diseases and animal models of IBD (1, 3, 10, 14, 17, 20, 50, 52, 60).
Reverse transcription.
Five micrograms of total RNA was reverse transcribed with Moloney murine leukemia virus RT and oligo(dT) primers according to the manufacturer's protocol (Superscript; Invitrogen). The cDNA was diluted with Tris (10 mM, pH 8.5) to a final concentration of 20 ng/μl.
Primer sequences and plasmids.
The primer sequences for IFN-γ (40), MIG (46), IP-10 (9), and hypoxanthine-guanine phosphoribosyltransferase (HPRT) (40) have been previously reported in the literature. The primer sequences for MIP-1α and SAA1 were designed from published mRNA sequences by using OMIGA software (Accelrys Inc., San Diego, Calif.). Sequences for all primer pairs are shown in Table 1. Quantification was done by comparing the fluorescence of experimental samples to that of plasmid standards containing known concentrations of the cloned amplified product. Standards were generated from linearized plasmids containing cloned amplicons of selected primer sets by using the Zero Blunt Topo PCR-cloning kit (Invitrogen).
TABLE 1.
Sense and antisense primer pairs used for PCR amplification
| Gene name | Sense (5′-3′) | Antisense (5′-3′) | Fragment size (bp) |
|---|---|---|---|
| HPRT (40) | GTA ATG ATC AGT CAA CGG GGG AC | CCA GCA AGC TTG CAA CCT TAA CCA | 165 |
| IP-10 (9) | CCT ATC CTG CCC ACG TGT TGA G | CAT CCT GCA GGA GGA GTA GCA G | 301 |
| MIG (46) | GGG CAA GTG TCC CTT TCC TTC | GGG CTC TAG GCT GAC CCA AAT | 198 |
| IFN-γ (40) | TGG AGG AAC TGG CAA AAG GAT GGT | TTG GGA CAA TCT CTT CCC CAC | 336 |
| MIP-1α | GCT CAA CAT CAT GAA GGT CTC C | TGC CGG TTT CTC TTA GTC AGG | 222 |
| SAA1 | TCA CGA GGC TTT CCA AGG | TGG TCA GCA ATG GTG TCC | 219 |
Real-time RT-PCR.
Selected differentially expressed genes were quantified by using real-time RT-PCR (LightCycler; Roche Diagnostic). PCRs and melting curves were performed in a 20-μl volume in glass capillaries that contained a 0.5 μM concentration of each primer, 3 mM MgCl2, QuantiTect SYBR Green PCR Master Mix containing dNTP mix, HotStart Taq DNA polymerase, reaction buffer, and SYBR green I (Qiagen), and cDNA. To quantify the number of copies of specific cDNA, a standard curve was created by using known concentrations (101 to 105 copies) of the pCR-Blunt II-TOPO (Invitrogen) plasmid containing the amplicon of interest. The PCRs were incubated at 95°C for 15 min to activate the polymerase. Forty cycles consisting of a 15-s denaturing at 94°C; 20-s annealing at 55°C (MIP-1α), 57°C (MIG), 59°C (SAA1) or 60°C (IP-10 and IFN-γ); and 30-s extension at 72°C were used to amplify the genes of interest. The ramp rate was 3°C/s for annealing and 20°C/s for all other steps. Fluorescence was monitored at the end of each extension phase. Following amplification, melting curves were generated to verify PCR product identity. HPRT levels were used to normalize H. hepaticus-infected and sham-infected samples for comparison.
Statistical analysis. (i) Lesion scoring.
Cecal tissue sections from uninfected control (n = 10) and H. hepaticus-infected (n = 10) mice, at months 1 and 3 of infection, were evaluated by a comparative pathologist who assigned scores without prior knowledge of the mouse infection status. A median lesion score for each group of animals was determined, and these scores were used for statistical comparisons. Because these data are ordinal data, they were statistically compared by using the nonparametric one-way analysis of variance (ANOVA) on ranks (Kruskal-Wallis). The statistical software package, SigmaStat (SPSS, Inc., Chicago, Ill.), was used for these statistical analyses.
(ii) cDNA expression arrays.
The gene expression profiles, at months 1 and 3 of infection, were compiled from the statistical analysis of 18 (nine uninfected controls and nine H. hepaticus-infected mice) biological replicates. Gene signal intensities were normalized by comparing the mean global signal intensity of an individual array to the mean global signal intensity of all arrays. Normalized gene expression levels in H. hepaticus-infected mice were compared to those of controls and were analyzed for statistical significance by using Significance Analysis of Microarray (SAM) software (59).
(iii) Confirmation of differentially expressed genes.
Semiquantitative RT-PCR was used to measure SAA1, IP-10, MIG, MIP-1α, and IFN-γ mRNA levels in cecal tissue of uninfected control (n = 10) and H. hepaticus-infected (n = 10) mice at months 1 and 3 of infection. The significance of differences between mean expression levels of these selected genes was determined by using a one-way ANOVA (Student-Newman-Keuls).
RESULTS
Mice and infection status.
Helicobacter infection was confirmed by fecal PCR assay at 2 weeks postinoculation and when mice were euthanatized for sample collection. All of the sham-inoculated control mice remained PCR negative for Helicobacter sp. throughout the study. Helicobacter DNA was detected in the feces of inoculated mice within 2 weeks of infection, and mice remained colonized for the duration of the study.
Histologic examination of ceca and cecal lesion scores.
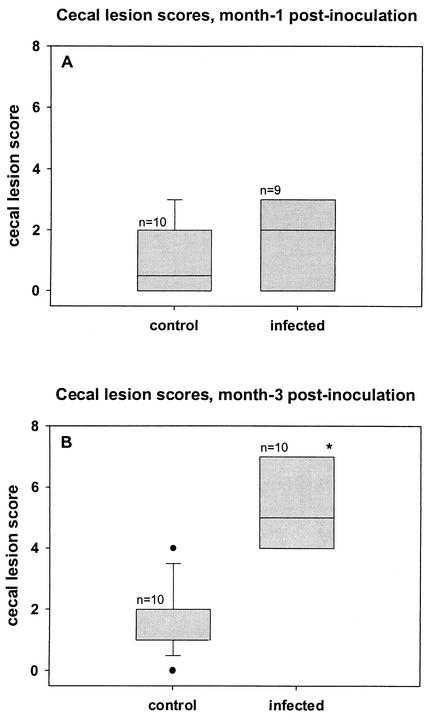
Cecal lesions were scored in uninfected controls (n = 10) and H. hepaticus-infected (n = 10) mice at months 1 and 3 of infection. Mild focal inflammation was observed in the ceca of two mice in each control group and three mice in the month-1 infection group (Fig. 1A to C). The difference between the median cecal lesion score of control and month-1 infection groups was not statistically significant (Fig. 2A and B). In contrast, cecal lesions in the 3-month infection group were characterized by patchy-to-diffuse mononuclear cell infiltrates in the lamina propria with mild-to-moderate mucosal epithelial hyperplasia (Fig. 1D). Furthermore, the median cecal lesion score was significantly elevated (P < 0.05) compared to that of the age-matched control and month-1 infection groups (Fig. 2A and B).
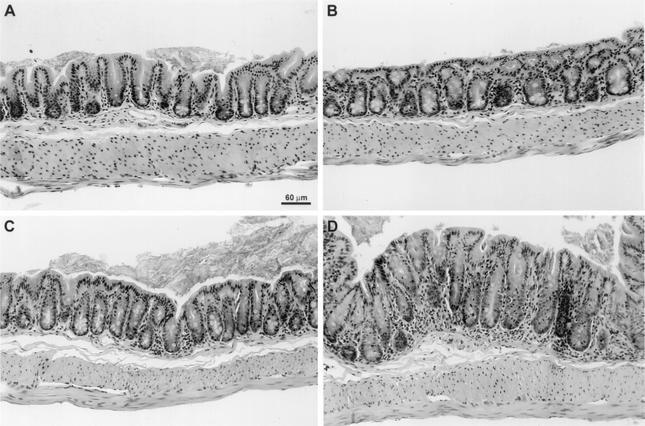
FIG. 1.
Photomicrograph of cecal sections from H. hepaticus-infected (B and D) and -naïve control (A and C) mice at months 1 (A and B) and 3 (C and D) postinoculation. (A, B, and D) No histologic lesions were identified in cecal sections from naïve control and month-1 postinfection groups. (C) Histologic lesions in the month-3 postinfection group were characterized by moderate mononuclear cell infiltrates and mild mucosal hyperplasia. Hematoxylin and eosin stain; bar = 60 μm.
FIG. 2.
The ceca were scored for intensity, longitudinal extent, and vertical extent of inflammation as well as hyperplasia at months 1 (A) and 3 (B) postinfection. (A) Median cecal lesion scores of naïve control and H. hepaticus-infected mice at month 1 postinfection were not statistically different. (B) Median cecal lesion scores of H. hepaticus-infected mice at month 3 postinfection were significantly increased compared to those of their age-matched cohorts. *, P < 0.05; Kruskal-Wallis, one-way ANOVA on ranks.
cDNA array gene expression profile.
The Atlas Mouse 1.2II cDNA expression array (Clontech) used in this study contains 1,176, 200- to 600-bp, cDNA fragments that have been amplified from a region of the mRNA that lacks repetitive or highly homologous sequences. These cDNA fragments represent a diverse set of genes with a variety of functions, including cell surface antigens, transcription factors, cell cycle proteins, cell adhesion receptors/proteins, immune system proteins, extracellular transport and carrier proteins, oncogenes, tumor suppressors, stress response proteins, membrane channel and transporter proteins, extracellular matrix proteins, trafficking/targeting proteins, apoptosis-associated proteins, RNA processing, cell signaling, and extracellular communication proteins, to name a few. This array format, with a broad representation of genes, was chosen to enhance the likelihood of identifying genes that are known to play a role in IBD as well as discovering new genes that are not a part of our present understanding of the disease. The cDNA array data presented in this study were derived from the analysis of whole cecal tissue rather than from uniform cell sets. While it may be argued that some cell-specific signals may be diluted or completely lost by analyzing whole tissue, profiling gene expression in complex heterogeneous cecal tissue has allowed us to identify dysregulated genes in the context of an intact immune system.
Gene expression was measured in the ceca of 18 (nine uninfected control and nine H. hepaticus-infected) mice at months 1 and 3 of infection. Gene signal intensities were normalized by comparing the mean global signal intensity of an individual array to the mean global signal intensity of all arrays. Normalized gene expression levels were analyzed for statistical significance by using SAM software by Chu, Tibshirani, Tusher, and Narasimhan at Stanford University (7, 55, 59). This method of statistical analysis was chosen because it offers a better balance between the total number of hypotheses rejected and the probability of making a type I error than do more conservative multiple-hypothesis tests like Bonferroni's or Westfall and Young's. The SAM algorithm calculates a score (d) for each gene based on the changes in gene expression relative to the standard deviation of repeated measures. A random data set was constructed by permutations of the data and was compared to the actual data set to determine the percentage of genes identified as significant by chance and to determine which genes had significantly altered expression. The cutoff for significance was determined by a tuning parameter (Δ), set by the user, to optimize the number of significant genes while maintaining a conservative false-discovery rate (FDR). Tables 2 and 3 show genes with significantly altered expression at months 1 and 3 postinfection, respectively. Analysis of the gene expressions profile in the cecum of H. hepaticus-infected mice at 1 month postinfection, with an FDR of 6.49%, demonstrated 25 up-regulated and 3 down-regulated genes. In the 3-month postinfection group, with an FDR of 6.45%, 31 up-regulated and 2 down-regulated genes were discovered. These genes have a wide range of functions, including acute-phase response proteins (SAA1), cell adhesion receptors/proteins (integrin beta-1 binding protein), intracellular kinase network members (mitogen-activated protein kinase kinase 1), intracellular signal transducers (IL-6 signal transducer gp130 and IFN-γ-inducible protein 47 kDa), proteosomal proteins (proteosome subunit beta type 4), cell surface antigens (CD38 antigen and CD3 antigen), and transcription factors (IFN-dependent positive-acting transcription factor 3 gamma). From these data, we identified a subset of immune-related genes that were up-regulated early in the immune response and have been associated with chronic human inflammatory diseases. These include CXC chemokines IP-10, MIG, and MIP-1α.
TABLE 2.
Differentially expressed genes at month 1 of infectionc
| Gene name | GenBank no. | Function | Score (d)a | Change (n fold) | q (%)b |
|---|---|---|---|---|---|
| Up-regulated genes | |||||
| SAA1 | M11131 | Acute-phase response | 6.82 | 1.64 | 3.78 |
| CD38 antigen | L11332 | Synthesis/hydrolysis of cyclic ADP-ribose | 6.56 | 1.78 | 3.78 |
| MHC-II Ia-associated invariant chain | X05430 | Antigen presentation | 5.98 | 1.97 | 3.78 |
| Cytochrome P450 IIJ6 | U62295 | Electron transport | 5.54 | 1.35 | 3.78 |
| Integrin beta 1-binding protein 1 | AJ001373 | Cell adhesion | 5.42 | 1.38 | 3.78 |
| IFN-dependent positive-acting transcription factor 3 gamma | U51992 | Transcription factor | 5.21 | 1.67 | 3.78 |
| IP-10 | M86829 | Lymphocyte activation/inflammatory | 5.20 | 1.50 | 3.78 |
| Mitogen-activated protein kinase kinase | L02526 | Signal transduction | 5.15 | 1.33 | 3.78 |
| IL-6 signal transducer (gp130) | X62646 | Signal transduction | 5.02 | 1.66 | 3.78 |
| MIP-1α | M23447 | Chemotaxis/cell activation | 4.92 | 1.63 | 3.78 |
| Proteasome subunit beta type 4 | U65636 | Protein catabolism | 4.91 | 1.36 | 3.78 |
| Lymphocyte antigen 57 | U65636 | B-cell receptor signaling | 4.89 | 1.40 | 3.78 |
| IFN-γ-inducible protein 47 kDa | M63630 | Gene regulation/protein trafficking | 4.72 | 2.11 | 3.78 |
| MIG | M34815 | Lymphocyte activation/inflammatory | 4.62 | 3.45 | 3.78 |
| Neurotrophic factor family receptor alpha 3 | AB008833 | Tyrosine kinase transmembrane receptor | 4.48 | 2.96 | 3.78 |
| Protein tyrosine phosphatase receptor type C | M14342 | T-cell activation | 4.08 | 1.57 | 3.78 |
| Drebrin-like | U58884 | Actin binding | 3.92 | 1.28 | 3.78 |
| Stefin A3 | M92419 | Proteinase inhibitor | 3.42 | 1.16 | 3.78 |
| Arachidonate 15-lipoxygenase | L34570 | Leukotriene biosynthesis | 3.32 | 1.49 | 3.78 |
| Solute carrier family 7 member 5 | AB017189 | Amino acid transport | 3.25 | 1.48 | 3.78 |
| Basic keratin complex 2 gene 8 | X12789 | Cytoskeleton organization | 3.21 | 1.15 | 3.78 |
| CD3 antigen gamma polypeptide | Y00635 | T-cell receptor signaling | 3.16 | 1.61 | 6.49 |
| Caveolin; 22-kDa caveolar protein | U07645 | Scaffolding protein | 3.01 | 1.63 | 6.49 |
| DNA methyltransferase 3A | AF068625 | DNA methylation | 2.99 | 1.30 | 6.49 |
| Opioid receptor sigma 1 | AF004927 | Ergosterol biosynthesis | 2.84 | 1.30 | 6.49 |
| Down-regulated genes | |||||
| Eukaryotic translation initiation factor 4 gamma 2 | U76112 | Regulation of translation | 5.10 | 0.74 | 3.78 |
| Antioxidant protein 2 | AF004670 | Redox regulation | 4.78 | 0.76 | 3.78 |
| Meprin 1 alpha | M74897 | Metalloprotease | 4.36 | 0.73 | 3.78 |
d, change in gene expression relative to the standard deviation of repeated measures.
q, the probability that a gene appearing to be differentially expressed is not actually differentially expressed.
The median false significant number is 1.82; the median FDR is 6.49.
TABLE 3.
Differentially expressed genes at month 3 of infectionc
| Gene name | GenBank no. | Function | Score (d)a | Change (n fold) | q (%)b |
|---|---|---|---|---|---|
| Up-regulated genes | |||||
| Metallothionein I activator | S63758 | DNA-dependent RNA polymerase | 7.60 | 1.95 | 2.76 |
| Karyopherin (importin) beta 1 | D45836 | Nuclear protein import | 6.79 | 1.18 | 2.76 |
| Alpha-2-macroglobulin | M93264 | Proteinase inhibitor | 5.59 | 1.76 | 2.76 |
| SAA3 | X03479 | Acute phase response | 5.36 | 1.63 | 2.76 |
| SRY box containing gene 7 | AB023419 | Transcription factor | 5.29 | 1.84 | 2.76 |
| SAA1 | M11131 | Acute phase response | 5.24 | 1.46 | 2.76 |
| Tumor necrosis factor superfamily member 11 | AF019048 | Augmentation of dendritic cell stimulation of T cells | 5.07 | 1.50 | 2.76 |
| IFN-γ-inducible protein 47 kDa | M63630 | Gene regulation/protein trafficking | 4.95 | 1.83 | 2.76 |
| Peptidoglycan recognition protein | AF076482 | Binding of peptidoglycan/innate immunity | 4.49 | 1.34 | 2.76 |
| MHC-II Ia-associated invariant chain | X05430 | Antigen presentation | 4.41 | 2.78 | 2.76 |
| Coagulation factor II (thrombin) receptor-like 1 | Z48043 | Trypsin receptor | 4.34 | 1.22 | 2.76 |
| Insulin-like growth factor binding protein 4 precursor | X81582 | Cell growth | 4.23 | 1.39 | 2.76 |
| Caveolin; 22-kDa caveolar protein | U07645 | Scaffolding protein | 4.16 | 1.88 | 2.76 |
| Fibroblast growth factor 9 | D38258 | Cell growth/tissue repair | 4.15 | 1.76 | 2.76 |
| Macrophage migration inhibitory factor | Z23048 | Macrophage regulation | 4.15 | 1.47 | 2.76 |
| Muscarinic acetylcholine receptor Mm3 | S74908 | Cholinergic receptor | 4.12 | 1.43 | 2.76 |
| Solute carrier family 34 member 1 | L33878 | Phosphate transportation | 4.08 | 1.67 | 2.76 |
| Coagulation factor VIII | L05573 | Blood coagulation factor | 4.04 | 1.38 | 2.76 |
| CD38 antigen | L11332 | Synthesis/hydrolysis of cyclic ADP-ribose | 4.04 | 2.41 | 2.76 |
| Solute carrier family 8 member 1 | U70033 | Calcium transport | 4.03 | 1.55 | 2.76 |
| Complement component 1 q subcomponent beta polypeptide | M22531 | Complement activation, classical pathway | 3.96 | 2.40 | 2.76 |
| Calbindin-D9K | AF028071 | Calcium binding | 3.90 | 3.74 | 2.76 |
| Coagulation factor XIII beta subunit | D10071 | Blood coagulation factor | 3.85 | 1.34 | 2.76 |
| Myelin-associated glycoprotein | M31811 | Cell adhesion | 3.84 | 1.29 | 2.76 |
| Nonmuscle myosin light chain 3 | U04443 | Cytoskeleton organization | 3.80 | 1.40 | 2.76 |
| MIP-1α | M23447 | Chemotaxis/cell activation | 3.61 | 2.27 | 4.84 |
| Thyroid-stimulating hormone receptor | U02601 | Control of thyroid cell metabolism | 3.59 | 1.47 | 4.84 |
| Growth differentiation factor 5 | U08337 | Bone growth | 3.33 | 1.24 | 4.84 |
| Apolipoprotein H | D10056 | Heparin binding | 3.32 | 1.39 | 4.84 |
| MIG | M34815 | Lymphocyte activation/inflammatory | 3.19 | 1.74 | 6.45 |
| Low-affinity immunoglobulin G Fc receptor II beta | M16367 | Antibody-dependent cellular cytotoxicity | 3.12 | 1.77 | 6.45 |
| Down-regulated genes | |||||
| Alcohol dehydrogenase 1 complex | M11307 | Alcohol dehydrogenase, zinc dependent | 6.45 | 0.65 | 2.76 |
| Mitochondrial aldehyde dehydrogenase 2 | U07235 | Aldehyde dehydrogenase (NAD+) | 5.90 | 0.47 | 2.76 |
d, change in gene expression relative to the standard deviation of repeated measures.
q, the probability that a gene that appears to be differentially expressed is not actually differentially expressed.
The median false significant number is 2.32; the median FDR is 6.45.
Confirmation of select genes by real-time RT-PCR.
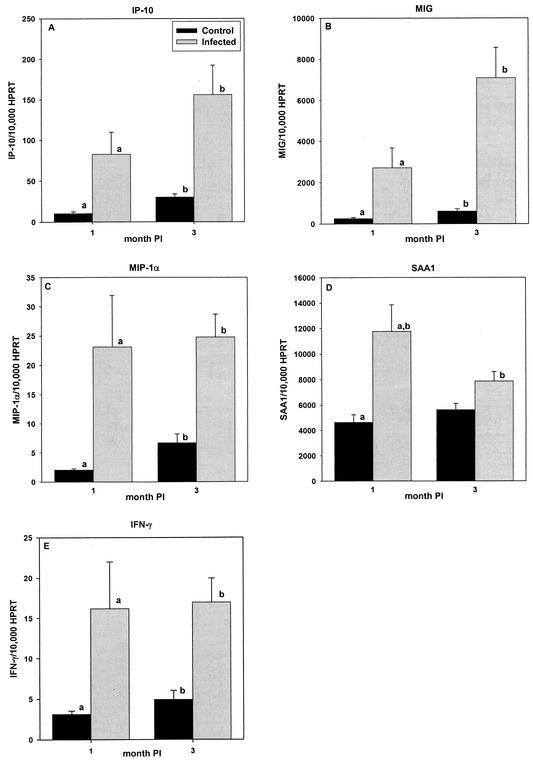
Semiquantitative RT-PCR was used to measure IP-10, MIG, MIP-1α, and SAA1 mRNA levels in ceca of mice chronically infected with H. hepaticus. We selected this subset of genes from the cDNA array data based on our present understanding of the ontogeny of mucosal inflammation and on the likelihood that these genes are involved in initiating and perpetuating the immune response. Curiously, our cDNA array studies did not detect expression of IFN-γ, an important inflammatory mediator that is known to be dysregulated in human and animal models of IBD (1, 8, 26, 27, 39, 42). Accordingly, a semiquantitative RT-PCR assay was developed to measure expression of this gene. H. hepaticus-infected mice (n = 10/group) showed statistically significant (P < 0.05) elevations in MIP-1α, MIG, IP-10, and IFN-γ gene expression at months 1 and 3 postinfection (Fig. 3A to C and E to F) when compared to sham-inoculated controls (n = 10/group). The level of SAA1 was significantly (P < 0.05) elevated at month 1 but not month 3 postinfection, compared with the level in age-matched controls (Fig. 3D); furthermore, expression of SAA1 at month 1 was significantly higher (P < 0.05) than at month 3 of infection.
FIG. 3.
The mean number of MIP-1α (A), MIG (B), IP-10 (C), SAA1 (D), and IFN-γ (E) mRNA molecules relative to the number of HPRT mRNA molecules in the ceca of naïve control and H. hepaticus-infected A/JCr mice at months 1 and 3 postinoculation (PI). Data represent mean plus or minus standard error of the mean of 10 mice per group. Letters (a and b) represent statistically significant (P < 0.05) difference according to Student-Newman-Keuls, one-way ANOVA.
DISCUSSION
In this study, we characterized gene expression in the cecum of A/JCr mice with H. hepaticus-induced typhlitis as it progressed from early to late disease. Understanding the dysregulated host immune response ensuing from H. hepaticus infection is an important step toward unraveling the complex immune circuitry that leads to chronic, progressive, mucosal inflammation. By studying the molecular details of this interaction in the context of the heterogeneous and changing cell population that makes up the intestinal mucosa, we can identify host genes that are critical to development of IBD. Gene expression profiling is an attractive approach to aid in identifying those genes that are involved in disease pathogenesis, because it makes possible the interrogation of thousands of genes simultaneously, including those genes not intuitively thought to be altered. Our array studies showed that H. hepaticus infection in female A/JCr mice induced significant changes in the expression levels of ∼2.5% of the 1,176 genes analyzed. Some of these genes have previously been described in human and animal models of IBD. These genes include SAA1 (17, 41), major histocompatibility complex class II (MHC-II) Ia-associated invariant chain (25, 35), CXC chemokines IP-10 and MIG (14, 52), IL-6 signal transducer gp130 (21), and macrophage migration inhibitory factor (6, 36, 37, 42). We also identified a subset of genes that are associated with the immune system but have not been specifically associated with IBD. This subset includes TNF superfamily member 11, known to augment dendritic cell stimulation of T cells by induction of IL-6, IL-1, IL-12, and IL-15 from dendritic cells (22); IFN-γ-inducible protein 47 kDa, which plays a role in IFN-γ-mediated clearance of protozoa and bacterial infection by regulating protein expression and trafficking (5, 57, 58); and MIP-1α, a CC class chemokine that has been shown to up-regulate cell-mediated immune response by preferentially recruiting Th1 lymphocytes to sites of inflammation and by promoting the development of IFN-γ-producing T cells by an IL-12-independent pathway (23, 24).
A number of studies have clearly established the critical role of the adaptive immune system in IBD. However, data from the majority of these studies are derived from tissue with chronic inflammation. Thus, there is very little known about the interface between the innate and adaptive immune responses and the identity of the early response genes and about how these genes are triggered and how the early response genes drive the immune response toward a Th1 or Th2 dominant cytokine profile. The pathological changes in the cecum of H. hepaticus-infected female A/JCr mice are delayed until after the 1st month of infection. This preinflammatory period is opportune for the study of early gene dysregulation associated with initiation of typhlitis. Despite the lack of histologic lesions at month 1 of infection, our cDNA array studies identified 25 up-regulated and 3 down-regulated genes. Furthermore, many of the genes altered at month 1 of infection are also dysregulated at the height of pathology. Of the genes discovered in the cDNA array studies, the CXC chemokines IP-10 and MIG, CC chemokine MIP-1α, and SAA1 are thought to play an important role in recruiting inflammatory cells and thereby shaping the ensuing immune response. By using real-time RT-PCR, we validated the cDNA array findings and quantified the expression level for these selected genes. Our real-time RT-PCR studies showed that the CXC chemokines MIG and IP-10 and the CC chemokine MIP-1α were up-regulated at months 1 and 3 postinfection. SAA1 was up-regulated at month 1 but not month 3 postinfection.
Human IBD can generally be divided into two major groups, CD and UC, based upon the clinical and pathological presentation. Studies in patients with CD demonstrate that mucosal inflammation is associated with a Th1 cytokine profile, whereas, although the data are controversial, UC is thought to have a Th2 predominance (12, 18, 45, 51, 53). Accordingly, most animal models of mucosal inflammation are classified as Th1 or Th2 phenotypes, based on the nature of T-cell-mediated inflammation. While the mucosal cytokine profile of H. hepaticus-infected A/JCr mice has not been clearly defined, studies of the systemic immune response of A/JCr mice (61) and the mucosal immune response of IL-10 knockout mice (28) to H. hepaticus infection, along with the gene expression data presented herein, suggest that the dysregulated mucosal immune response to H. hepaticus infection is characteristic of a Th1-like phenotype. Our real-time RT-PCR data demonstrate that IFN-γ, a hallmark cytokine for the Th1 paradigm, is up-regulated by month 1 and that enhanced expression continues to month 3 of infection. IFN-γ specifically induces the transcription of a number of genes, including IFN-γ-inducible protein 47 kDa (19), IFN-dependent positive-acting transcription factor 3 gamma (33), MHC-II Ia-associated invariant chain (4), and most notably the CXC chemokines IP-10 and MIG identified in our array studies. Whereas IP-10 can be induced in response to other proinflammatory signals, including IFN-β, IFN-α lipopolysaccharide, TNF-α, IL-1, IL-6, IL-12, and environmental antigens, independent of IFN-γ, the synthesis of MIG is induced exclusively by IFN-γ (11). Support for the role of these chemokines in the pathogenesis of IBD comes from studies that use animal models with a Th1-mediated immune response similar to that seen in CD. MIG is up-regulated in chronically inflamed colons of IL-10−/− mice, and reversal of colitis with anti-IL-12 monoclonal antibody is associated with inhibition of expression (52). Moreover, IP-10 and MIG mRNA levels are increased in recombinase-activated gene-2 knockout mice (Rag-2−/−) reconstituted with CD4+CD45RBhigh. In contrast, these chemokines are not elevated in Rag-2−/− mice reconstituted with a combination of CD4+CD45RBhigh and CD4+CD45RBlow or in mice treated with IL-10 or anti-IL-12 monoclonal antibody. Curiously, Uguccioni et al. observed elevated expression of IP-10 in biopsy specimens from patients with UC (60). The significance of this finding is unknown, but its ubiquitous expression may be reflective of the fact that IP-10 is induced by a variety of signals, including IL-1 and IL-6, known to be up-regulated in patients with UC as well those with CD (38, 47).
In summary, this report demonstrates that H. hepaticus infection in A/JCr mice is associated with significant alterations in gene expression. Moreover, gene dysregulation begins prior to the development of any features of pathology and persists during the chronic stage of IBD. Consistent with findings from previously reported Helicobacter studies, our expression data showing increased IFN-γ, IP-10, and MIG mRNA levels suggest that the mucosal immune response to H. hepaticus infection is consistent with a Th1 phenotype.
Acknowledgments
We thank Kim Mullinax, Laurie Roesel, and Lisa Zell for their assistance with animal necropsy and sample preparation.
This work was supported by National Institutes of Health training grant NIH-T32-RR07004 and Research Animal Diagnostic Laboratory (RADIL) research funds.
Editor: F. C. Wang
REFERENCES
- 1.Akagi, S., E. Hiyama, Y. Imamura, Y. Takesue, Y. Matsuura, and T. Yokoyama. 2000. Interleukin-10 expression in intestine of Crohn disease. Int. J. Mol. Med. 5:389-395. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, C., J. Blanchard, P. Rawsthorne, and A. Wajda. 1999. Epidemiology of Crohn's disease and ulcerative colitis in a central Canadian province: a population-based study. Am. J. Epidemiol. 149:916-924. [DOI] [PubMed] [Google Scholar]
- 3.Boorsma, D. M., P. J. van Beek, C. Nieboer, T. J. Stoof, R. Willemze, C. P. Tensen, and T. L. Sorensen. 2001. Expression of specific chemokines and chemokine receptors in the central nervous system of multiple sclerosis patients. J. Pathol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 4.Cao, Z. A., B. B. Moore, D. Quezada, C. H. Chang, and P. P. Jones. 2000. Identification of an IFN-gamma responsive region in an intron of the invariant chain gene. Eur. J. Immunol. 30:2604-2611. [DOI] [PubMed] [Google Scholar]
- 5.Collazo, C. M., G. S. Yap, G. D. Sempowski, K. C. Lusby, L. Tessarollo, G. F. Woude, A. Sher, and G. A. Taylor. 2001. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J. Exp. Med. 194:181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong, Y. P., A. C. Abadia-Molina, A. R. Satoskar, K. Clarke, S. T. Rietdijk, W. A. Faubion, E. Mizoguchi, C. N. Metz, M. A. Sahli, T. ten Hove, A. C. Keates, J. B. Lubetsky, R. J. Farrell, P. Michetti, S. J. van Deventer, E. Lolis, J. R. David, A. K. Bhan, and C. Terhorst. 2001. Development of chronic colitis is dependent on the cytokine MIF. Nat. Immunol. 2:1061-1066. [DOI] [PubMed] [Google Scholar]
- 7.Efron, B., R. Tibshirani, J. D. Storey, and V. Tusher. 2001. Empirical Bayes analysis of a microarray experiment. J. Am. Stat. Assoc. 96:1151-1160. [Google Scholar]
- 8.Egger, B., M. Bajaj-Elliott, T. T. MacDonald, R. Inglin, V. E. Eysselein, and M. W. Buchler. 2000. Characterisation of acute murine dextran sodium sulphate colitis: cytokine profile and dose dependency. Digestion 62:240-248. [DOI] [PubMed] [Google Scholar]
- 9.Enk, A. H., and S. I. Katz. 1992. Early molecular events in the induction phase of contact sensitivity. Proc. Natl. Acad. Sci. USA 89:1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdman, S., J. G. Fox, C. A. Dangler, D. Feldman, and B. H. Horwitz. 2001. Typhlocolitis in NF-kappa B-deficient mice. J. Immunol. 166:1443-1447. [DOI] [PubMed] [Google Scholar]
- 11.Farber, J. M. 1997. Mig and IP-10: CXC chemokines that target lymphocytes. J. Leukoc. Biol. 61:246-257. [PubMed] [Google Scholar]
- 12.Farrell, R. J., and M. A. Peppercorn. 2002. Ulcerative colitis. Lancet 359:331-340. [DOI] [PubMed] [Google Scholar]
- 13.Fiocchi, C. 1998. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology 115:182-205. [DOI] [PubMed] [Google Scholar]
- 14.Fort, M., R. Lesley, N. Davidson, S. Menon, F. Brombacher, M. Leach, and D. Rennick. 2001. IL-4 exacerbates disease in a Th1 cell transfer model of colitis. J. Immunol. 166:2793-2800. [DOI] [PubMed] [Google Scholar]
- 15.Fox, J. G., X. Li, L. Yan, R. J. Cahill, R. Hurley, R. Lewis, and J. C. Murphy. 1996. Chronic proliferative hepatitis in A/JCr mice associated with persistent Helicobacter hepaticus infection: a model of helicobacter-induced carcinogenesis. Infect. Immun. 64:1548-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox, J. G., L. Yan, B. Shames, J. Campbell, J. C. Murphy, and X. Li. 1996. Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect. Immun. 64:3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukushima, K., H. Ogawa, T. Kitayama, T. Yamada, H. Naito, Y. Funayama, S. Matsuno, and I. Sasaki. 2002. Epithelial induction of serum amyloid A in experimental mucosal inflammation. Dig. Dis. Sci. 47:1438-1446. [DOI] [PubMed] [Google Scholar]
- 18.Fuss, I. J., M. Neurath, M. Boirivant, J. S. Klein, C. de la Motte, S. A. Strong, C. Fiocchi, and W. Strober. 1996. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn's disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J. Immunol. 157:1261-1270. [PubMed] [Google Scholar]
- 19.Gilly, M., and R. Wall. 1992. The IRG-47 gene is IFN-gamma induced in B cells and encodes a protein with GTP-binding motifs. J. Immunol. 148:3275-3281. [PubMed] [Google Scholar]
- 20.Grimm, M. C., and W. F. Doe. 1996. Chemokines in inflammatory bowel disease mucosa—expression of RANTES, macrophage inflammatory protein (MIP)-1-alpha, MIP-1-beta, and gamma-interferon-inducible protein-10 by macrophages, lymphocytes, endothelial cells, and granulomas. Inflamm. Bowel Dis. 2:88-96. [PubMed] [Google Scholar]
- 21.Holub, M. C., E. Mako, T. Devay, M. Dank, C. Szalai, A. Fenyvesi, and A. Falus. 1998. Increased interleukin-6 levels, interleukin-6 receptor and gp130 expression in peripheral lymphocytes of patients with inflammatory bowel disease. Scand. J. Gastroenterol. Suppl. 228:47-50. [PubMed] [Google Scholar]
- 22.Josien, R., B. R. Wong, H. L. Li, R. M. Steinman, and Y. Choi. 1999. TRANCE, a TNF family member, is differentially expressed on T cell subsets and induces cytokine production in dendritic cells. J. Immunol. 162:2562-2568. [PubMed] [Google Scholar]
- 23.Karpus, W. J., and K. J. Kennedy. 1997. MIP-1alpha and MCP-1 differentially regulate acute and relapsing autoimmune encephalomyelitis as well as Th1/Th2 lymphocyte differentiation. J. Leukoc. Biol. 62:681-687. [PubMed] [Google Scholar]
- 24.Karpus, W. J., N. W. Lukacs, K. J. Kennedy, W. S. Smith, S. D. Hurst, and T. A. Barrett. 1997. Differential CC chemokine-induced enhancement of T helper cell cytokine production. J. Immunol. 158:4129-4136. [PubMed] [Google Scholar]
- 25.Koretz, K., F. Momburg, H. F. Otto, and P. Moller. 1987. Sequential induction of MHC antigens on autochthonous cells of ileum affected by Crohn's disease. Am. J. Pathol. 129:493-502. [PMC free article] [PubMed] [Google Scholar]
- 26.Kullberg, M. C., D. Jankovic, P. L. Gorelick, P. Caspar, J. J. Letterio, A. W. Cheever, and A. Sher. 2002. Bacteria-triggered CD4+ T regulatory cells suppress Helicobacter hepaticus-induced colitis. J. Exp. Med. 196:505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kullberg, M. C., A. G. Rothfuchs, D. Jankovic, P. Caspar, T. A. Wynn, P. L. Gorelick, A. W. Cheever, and A. Sher. 2001. Helicobacter hepaticus-induced colitis in interleukin-10-deficient mice: cytokine requirements for the induction and maintenance of intestinal inflammation. Infect. Immun. 69:4232-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kullberg, M. C., J. M. Ward, P. L. Gorelick, P. Caspar, S. Hieny, A. Cheever, D. Jankovic, and A. Sher. 1998. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect. Immun. 66:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linskens, R. K., X. W. Huijsdens, P. H. Savelkoul, C. M. Vandenbroucke-Grauls, and S. G. Meuwissen. 2001. The bacterial flora in inflammatory bowel disease: current insights in pathogenesis and the influence of antibiotics and probiotics. Scand. J. Gastroenterol. Suppl. 234:29-40. [DOI] [PubMed] [Google Scholar]
- 30.Livingston, R. S., L. K. Riley, E. K. Steffen, C. L. Besch-Williford, R. R. Hook, Jr., and C. L. Franklin. 1997. Serodiagnosis of Helicobacter hepaticus infection in mice by an enzyme-linked immunosorbent assay. J. Clin. Microbiol. 35:1236-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loftus, E. V., Jr., P. Schoenfeld, and W. J. Sandborn. 2002. The epidemiology and natural history of Crohn's disease in population-based patient cohorts from North America: a systematic review. Aliment. Pharmacol. Ther. 16:51-60. [DOI] [PubMed] [Google Scholar]
- 32.MacDonald, T. T., G. Monteleone, and S. L. Pender. 2000. Recent developments in the immunology of inflammatory bowel disease. Scand. J. Immunol. 51:2-9. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto, M., N. Tanaka, H. Harada, T. Kimura, T. Yokochi, M. Kitagawa, C. Schindler, and T. Taniguchi. 1999. Activation of the transcription factor ISGF3 by interferon-gamma. Biol. Chem. 380:699-703. [DOI] [PubMed] [Google Scholar]
- 34.Mohammadi, M., R. Redline, J. Nedrud, and S. Czinn. 1996. Role of the host in pathogenesis of Helicobacter-associated gastritis: H. felis infection of inbred and congenic mouse strains. Infect. Immun. 64:238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Momburg, F., K. Koretz, A. Von Herbay, and P. Moller. 1988. Nonimmune human cells can express MHC class II antigens in the absence of invariant chain—an immunohistological study on normal and chronically inflamed small intestine. Clin. Exp. Immunol. 72:367-372. [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami, H., S. M. Akbar, H. Matsui, N. Horiike, and M. Onji. 2002. Macrophage migration inhibitory factor activates antigen-presenting dendritic cells and induces inflammatory cytokines in ulcerative colitis. Clin. Exp. Immunol. 128:504-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami, H., S. M. Akbar, H. Matsui, and M. Onji. 2001. Macrophage migration inhibitory factor in the sera and at the colonic mucosa in patients with ulcerative colitis: clinical implications and pathogenic significance. Eur. J. Clin. Investig. 31:337-343. [DOI] [PubMed] [Google Scholar]
- 38.Murata, Y., Y. Ishiguro, J. Itoh, A. Munakata, and Y. Yoshida. 1995. The role of proinflammatory and immunoregulatory cytokines in the pathogenesis of ulcerative colitis. J. Gastroenterol. 30(Suppl. 8):56-60. [PubMed] [Google Scholar]
- 39.Niessner, M., and B. A. Volk. 1995. Altered Th1/Th2 cytokine profiles in the intestinal mucosa of patients with inflammatory bowel disease as assessed by quantitative reversed transcribed polymerase chain reaction (RT-PCR). Clin. Exp. Immunol. 101:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Garra, A., R. Chang, N. Go, R. Hastings, G. Haughton, and M. Howard. 1992. Ly-1 B (B-1) cells are the main source of B cell-derived interleukin 10. Eur. J. Immunol. 22:711-717. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa, H., K. Fukushima, I. Sasaki, and S. Matsuno. 2000. Identification of genes involved in mucosal defense and inflammation associated with normal enteric bacteria. Am. J. Physiol. Gastrointest. Liver Physiol. 279:G492-G499. [DOI] [PubMed] [Google Scholar]
- 42.Ohkawara, T., J. Nishihira, H. Takeda, S. Hige, M. Kato, T. Sugiyama, T. Iwanaga, H. Nakamura, Y. Mizue, and M. Asaka. 2002. Amelioration of dextran sulfate sodium-induced colitis by anti-macrophage migration inhibitory factor antibody in mice. Gastroenterology 123:256-270. [DOI] [PubMed] [Google Scholar]
- 43.Papadakis, K. A., and S. R. Targan. 1999. Current theories on the causes of inflammatory bowel disease. Gastroenterol. Clin. N. Am. 28:283-296. [DOI] [PubMed] [Google Scholar]
- 44.Papadakis, K. A., and S. R. Targan. 2000. Role of cytokines in the pathogenesis of inflammatory bowel disease. Annu. Rev. Med. 51:289-298. [DOI] [PubMed] [Google Scholar]
- 45.Parronchi, P., P. Romagnani, F. Annunziato, S. Sampognaro, A. Becchio, L. Giannarini, E. Maggi, C. Pupilli, F. Tonelli, and S. Romagnani. 1997. Type 1 T-helper cell predominance and interleukin-12 expression in the gut of patients with Crohn's disease. Am. J. Pathol. 150:823-832. [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu, B., K. A. Frait, F. Reich, E. Komuniecki, and S. W. Chensue. 2001. Chemokine expression dynamics in mycobacterial (type-1) and schistosomal (type-2) antigen-elicited pulmonary granuloma formation. Am. J. Pathol. 158:1503-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reimund, J. M., C. Wittersheim, S. Dumont, C. D. Muller, R. Baumann, P. Poindron, and B. Duclos. 1996. Mucosal inflammatory cytokine production by intestinal biopsies in patients with ulcerative colitis and Crohn's disease. J. Clin. Immunol. 16:144-150. [DOI] [PubMed] [Google Scholar]
- 48.Riddell, R. H. 2000. Pathology of idiopathic inflammatory bowel disease, p. 427-452. In J. B. Kirsner (ed.), Inflammatory bowel disease, 5th ed. W. B. Saunders, Philadelphia, Pa.
- 49.Riley, L. K., R. R. Hook, Jr., C. L. Besch-Williford, and C. L. Franklin. 1999. Identification of murine helicobacters by PCR and restriction enzyme analyses. Clin. Diagn. Lab. Immunol. 6:745-750.10473529 [Google Scholar]
- 50.Robinson, N., A. Patel, H. Varsani, P. Woo, and D. D. Patel. 2000. CXCR3 and CCR5 ligands in rheumatoid arthritis synovium. Arthritis Rheum. 43:765-774. [DOI] [PubMed] [Google Scholar]
- 51.Romagnani, P., F. Annunziato, M. C. Baccari, and P. Parronchi. 1997. T cells and cytokines in Crohn's disease. Curr. Opin. Immunol. 9:793-799. [DOI] [PubMed] [Google Scholar]
- 52.Scheerens, H., E. Hessel, R. de Waal-Malefyt, M. W. Leach, and D. Rennick. 2001. Characterization of chemokines and chemokine receptors in two murine models of inflammatory bowel disease: IL-10−/− mice and Rag-2−/− mice reconstituted with CD4+CD45RBhigh T cells. Eur. J. Immunol. 31:1465-1474. [DOI] [PubMed] [Google Scholar]
- 53.Shanahan, F. 2002. Crohn's disease. Lancet 359:62-69. [DOI] [PubMed] [Google Scholar]
- 54.Stonnington, C. M., S. F. Phillips, L. J. Melton III, and A. R. Zinsmeister. 1987. Chronic ulcerative colitis: incidence and prevalence in a community. Gut 28:402-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storey, J. D., and D. Siegmund. 2001. Approximate p-values for local sequence alignments: numerical studies. J. Comput. Biol. 8:549-556. [DOI] [PubMed] [Google Scholar]
- 56.Strober, W., I. J. Fuss, and R. S. Blumberg. 2002. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 20:495-549. [DOI] [PubMed] [Google Scholar]
- 57.Taylor, G. A., C. M. Collazo, G. S. Yap, K. Nguyen, T. A. Gregorio, L. S. Taylor, B. Eagleson, L. Secrest, E. A. Southon, S. W. Reid, L. Tessarollo, M. Bray, D. W. McVicar, K. L. Komschlies, H. A. Young, C. A. Biron, A. Sher, and G. F. Vande Woude. 2000. Pathogen-specific loss of host resistance in mice lacking the IFN-gamma-inducible gene IGTP. Proc. Natl. Acad. Sci. USA 97:751-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, G. A., R. Stauber, S. Rulong, E. Hudson, V. Pei, G. N. Pavlakis, J. H. Resau, and G. F. Vande Woude. 1997. The inducibly expressed GTPase localizes to the endoplasmic reticulum, independently of GTP binding. J. Biol. Chem. 272:10639-10645. [DOI] [PubMed] [Google Scholar]
- 59.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. (Erratum, 98:10515.) [DOI] [PMC free article] [PubMed]
- 60.Uguccioni, M., P. Gionchetti, D. F. Robbiani, F. Rizzello, S. Peruzzo, M. Campieri, and M. Baggiolini. 1999. Increased expression of IP-10, IL-8, MCP-1, and MCP-3 in ulcerative colitis. Am. J. Pathol. 155:331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whary, M. T., T. J. Morgan, C. A. Dangler, K. J. Gaudes, N. S. Taylor, and J. G. Fox. 1998. Chronic active hepatitis induced by Helicobacter hepaticus in the A/JCr mouse is associated with a Th1 cell-mediated immune response. Infect. Immun. 66:3142-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]