Abstract
Ribonucleases III are double-stranded RNA (dsRNA) endonucleases required for the processing of a large number of prokaryotic and eukaryotic transcripts. Although the specificity of bacterial RNase III cleavage relies on antideterminants in the dsRNA, the molecular basis of eukaryotic RNase III specificity is unknown. All substrates of yeast RNase III (Rnt1p) are capped by terminal tetraloops showing the consensus AGNN and located within 13–16 bp to Rnt1p cleavage sites. We show that these tetraloops are essential for Rnt1p cleavage and that the distance to the tetraloop is the primary determinant of cleavage site selection. The presence of AGNN tetraloops also enhances Rnt1p binding, as shown by surface plasmon resonance monitoring and modification interference studies. These results define a paradigm of RNA loops and show that yeast RNase III behaves as a helical RNA ruler that recognizes these tetraloops and cleaves the dsRNA at a fixed distance to this RNA structure. These results also indicate that proteins belonging to the same class of RNA endonucleases require different structural elements for RNA cleavage.
RNase III enzymes are double-stranded RNA (dsRNA) endonucleases found in bacteria and in eukaryotes. Bacterial RNase III was discovered 3 decades ago (1). It is involved in the processing of a large number of bacterial and bacteriophage RNA substrates (reviewed in ref. 2). RNase III is thought to be a significant regulator of gene expression by controlling the decay of a subset of mRNAs (including its own mRNA) or the translation of these mRNAs (2–4). This function reflects the specificity of the enzyme, which cuts only a very specific subset of bacterial and viral RNAs. Until recently, the determinants of bacterial RNase III specificity remained controversial (2). This situation was due to the lack of obvious conserved structural features in the various RNase III substrates. By comparing cleavage site sequences and by introducing specific mutations in model substrates, Zhang and Nicholson (5) were able to show that some specific base pairs at given positions relative to the cleavage site are not found in natural sites and are not allowed for efficient cleavage in vitro. Thus, bacterial RNase III cleavage specificity relies on antideterminants in the double-stranded structure.
Eukaryotic RNase III was identified more recently on the basis of sequence similarities with the bacterial enzyme (6–8). Since its discovery, Saccharomyces cerevisiae RNase III (Rnt1p) has been shown to process the precursors of stable RNAs such as rRNA, small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs). The biological function of cleavage by Rnt1p differs with respect to the different classes of RNA targets. Processing of rRNA substrates by yeast RNase III removes the 3′ external transcribed spacer (ETS; refs. 8 and 9); 3′ end maturation of yeast snRNAs involves RNase III (7, 10–13), which cleaves the 3′ extension of snRNAs precursors downstream from the mature snRNA sequences. The remaining extension is removed by the exosome (13), which contains 3′ → 5′ exonucleases. In the case of snoRNAs, Rnt1p is required for 5′ end processing of uncapped, nonintronic snoRNAs as well as for liberating snoRNAs from a common precursor when they are encoded in polycistronic arrays (14–16). In contrast to the prokaryotic enzyme, the determinants of eukaryotic RNase III-specific cleavage are poorly understood. The recent identification of a large number of substrates for this enzyme allowed us to compare the structural features of various cleavage sites for the S. cerevisiae enzyme. Using this compilation, we have identified a conserved class of tetraloops found at the end of all dsRNAs substrates of Rnt1p. This finding suggested that these tetraloops are required for Rnt1p processing and that the enzyme acts as a dsRNA ruler to select the scissile phosphodiester bond within 13 to 16 bp of the terminal tetraloop of the RNA substrate. In this study, we address the question of the importance of these tetraloops for Rnt1p processing and test the model that cleavage site selection occurs by measuring the distance of the dsRNA from the tetraloop.
Materials and Methods
The template for in vitro transcription of the snR47 model hairpin substrate was obtained by annealing a forward oligonucleotide carrying a T7 promoter and the beginning of the hairpin sequence (GCGAATTCTAATACGACTCACTATAGGTAGGAGAAAGGATATTGAACATG) and the reverse primer carrying the wild-type sequence (GATAAGGAAAAAGATACCACGTTCTTCTGAACATGTTCAATATCC) or the corresponding mutant sequences and by extending them with Taq or T4 DNA polymerase. Gel-purified RNAs were 5′ end labeled with [γ-32P]ATP as described (14). To ensure folding of the RNAs, they were heated for 3 min at 85°C in 50 mM Tris⋅HCl, pH 7.6/200 mM KCl/0.1 mg⋅m−1 wheat-germ tRNA/5 mM MgCl2 and then cooled down to 55°C (0.2°C⋅s−1) and to 23°C (0.1°C⋅s−1) in a Peltier thermocycler. The potential of wild-type and GAAA mutant RNAs to form intermolecular duplex was assayed by running these RNAs in the absence of protein on native gels, and after denaturation and renaturation of these RNAs at various concentrations, no duplex formation was detected on native gels (data not shown).
Rnt1p cleavage was done at 23°C by using a recombinant 6-His-tagged version of Rnt1p cloned into pET16B (Novagen), purified by FPLC on Hi-Trap Chelating columns (Amersham Pharmacia). Cleavage with structure-specific RNases was performed in the same conditions during 20 min in 5 μl with 0.5 units of ribonuclease T1 (Roche Molecular Biochemicals), 0.1 unit of ribonuclease T2 (GIBCO/BRL) for probing of single-stranded regions, or 0.15 units of ribonuclease V1 (Amersham Pharmacia) for probing of double-stranded regions. Reactions were stopped by adding 5 μl of urea loading buffer [50% (wt/vol) urea/1× TBE/25 mM EDTA, pH 8.0/0.05% xylene cyanol; TBE = 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3], and RNAs were fractionated on 10% polyacrylamide sequencing gels.
Binding of RNAs to immobilized Rnt1p was monitored by surface plasmon resonance, as described (17), in 25 mM Hepes, pH 7.4/140 mM NaCl with nonlabeled RNAs at concentrations varying from 50 nM to 10 μM. His6–Rnt1p was immobilized to CM5 surfaces via lysine residues to carbodiimide-activated carboxyl groups on the dextran layer in the BIAcore chip. After each cycle of binding and chase, the remaining RNAs were washed with 25 mM Hepes, pH 7.4/1 M NaCl. For modification interference experiments, 1–5 pmol of 5′ end-labeled substrate was modified with diethylpyrocarbonate (DEPC) or hydrazine as described (18) to ensure less than one modification per molecule. Phosphorothioates were incorporated during in vitro transcription by T7 RNA polymerase. Binding was performed in 50 mM Tris⋅HCl, pH 7.6/200 mM KCl/2.5 mM EDTA, pH 8.0/0.2 mg⋅ml−1 tRNA/20% (vol/vol) glycerol with 50 to 100 nM of His6–Rnt1p and 10 nM labeled RNAs to ensure ≈50% of binding. These values corresponded to the range of the Kd determined by a gel shift experiment with unmodified RNA. After a 20-min incubation at 4°C, samples were loaded onto an 8% acrylamide:bisacrylamide (37.5:1)/0.25× TBE gel (20 × 20 cm, 1.5 mm thick). Migration was performed at 5°C for 2 to 3 h (500 V). Free and bound RNAs were excised from polyacrylamide gels and eluted overnight in 400 μl of 0.2 M NaCl/25 mM EDTA, pH 8.0/1% sodium dodecyl sulfate. After elution, 5 μg of proteinase K was added; RNAs were extracted with phenol-chloroform-isoamylic alcohol (50:49:1); and ethanol was precipitated. Aniline cleavage of DEPC- and hydrazine-modified bases was performed as described (18). Iodine cleavage of phosphorothioate-substituted RNAs was done for 3 min at 95°C in 98% (vol/vol) formamide/25 mM EDTA, pH 8.0/250 μM iodine/0.05% xylene cyanol. Cleaved RNAs were fractionated on 12% sequencing gels.
Results
Although frequently observed on Rnt1p substrates, the presence of an AGNN tetraloop structure at the tip of the RNA substrate did not seem to be absolutely required for Rnt1p processing, because some substrates seemed to lack such structures (8, 14, 15). To check the absence of AGNN tetraloops in these substrates, we reexamined their potential secondary structures. After refolding these RNAs with mfold (19) and reassessment of the physiological relevance of one of these substrates (the 5′ external transcribed spacer in the precursor of rRNA; ref. 9), it seemed that all Rnt1p physiological substrates but one bear an AGNN tetraloop (Table 1), the U1 snRNA 3′ sequence showing a single nucleotide variation in the consensus (UGUU). The first 2 nt of these tetraloops are strongly conserved as AG; the remaining 2 bases show no conservation, albeit cytosines were almost absent from the third and fourth position. This small AGNN consensus sequence may reflect a requirement for the substrate RNA stability and/or folding, as for the GNRA or UNCG tetraloops. Alternatively, this tetraloop may act as a recognition signal for Rnt1p. The latter provocative hypothesis was challenged by results obtained with Escherichia coli RNase III, which showed that the terminal tetraloop of the T7 R1.1 substrate is not required for in vitro cleavage (20), but was supported by the observation that the distance between the terminal loops and the cleavage sites is strongly constrained (between 13 and 16 bp; Table 1). To test for a function of AGNN tetraloops in substrate recognition by yeast RNase III, we synthesized a model substrate derived from the 5′ extension of the snR47 snoRNA precursor, which contains only the stem loop region where Rnt1p cleavage occurs (Fig. 1). This substrate was cleaved efficiently in vitro in the presence of recombinant Rnt1p (Fig. 1), at the same site observed for the full-length substrate (as shown by primer extension mapping; data not shown). Mutations were introduced in this model substrate to change the conserved sequence of the tetraloop and to vary the distance between the terminal tetraloop and the cleavage site (Fig. 1). 5′ end-labeled wild-type and mutant substrates were incubated in the presence of recombinant His6–Rnt1p to assess the influence of these mutations on in vitro cleavage. A single substitution in the loop (AGAA → CGAA) was sufficient to decrease Rnt1p activity, as shown by the increased amount of uncut precursor (Fig. 1). Changing the AGAA tetraloop into a stable tetraloop, GAAA (Fig. 2) or UUCG (data not shown), abolished Rnt1p cleavage, although weak residual activity could be detected at prolonged incubation times with high amounts of protein (data not shown). The same result was obtained with a single mutation, AGAA → ACAA, which completely inhibited cleavage (data not shown). We next varied the distance between the natural cleavage site and the tetraloop by inserting or deleting base pairs between these two elements. Deletion of 1 bp between the tetraloop and the cleavage site induced the activation of a second cleavage in addition to the normal cleavage site. This second cleavage was shifted 1 base downstream from the normal cleavage (Fig. 1). Inserting 1 bp between the tetraloop and the cleavage site was sufficient to shift the major cleavage site by 1 base upstream from the normal cleavage (Fig. 1). Insertions or deletions of 2 bp or insertion of 3 bp resulted in corresponding shifts of the cleavage sites (Fig. 1). These results show that changing the distance between the tetraloop and the cleavage site is sufficient to change the location of the cleavage site accordingly, showing that the distance between the tetraloop and the cleavage site is a primary determinant of cleavage site selection.
Table 1.
Compilation of tetraloop sequences at Rnt1p cleavage sites
| Substrate | RNA class | Sequence | Distance from cleavage site, nt | Distance from mature RNA, nt | Refs. |
|---|---|---|---|---|---|
| 3′ETS | rRNA | AGGA | 15 | +29 (25S) | 8, 9 |
| U1 | snRNA | UGGU | 15–18 | +97 | 12 |
| U2 | snRNA | AGUU | 13–14 | +43 | 11 |
| U4 | snRNA | AGUU | 14–15 | +148 | 13 |
| U5 | snRNA | AGUC | 13–14 | +40 (U5L) | 10 |
| snR36 | snoRNA | AGUA | 14–15 | −94 | 15 |
| snR40 | snoRNA | AGUU | 14–15 | −89 | 15 |
| snR43 | snoRNA | AGUG | 13–14 | −62 | 15 |
| snR46 | snoRNA | AGGA | 14–15 | −115 | 15 |
| snR47 | snoRNA | AGAA | 14–15 | −57 | 15 |
| snR73–72 | snoRNA | AGUU | 16 | −54 (snR72) | 16 |
| snR75–74 | snoRNA | AGUU | 14 | −66 (snR74) | 16 |
| snR78–77 | snoRNA | AGUA | 15 | −90 (snR77) | 16 |
| snR79 | snoRNA | AGGA | 15–16 | −76 | 15 |
| snR41–70 | snoRNA | AGUA | 14 | −67 (snR41) | 15 |
| snR70–51 | snoRNA | AGUU | 15–16 | −42 (snR51) | 15 |
| snR190–U14 | snoRNA | AGUU | 14 | −62 (snR190) | 14 |
The sequence of the terminal tetraloops and the distance between the first nucleotide of the loop and the upstream cleavage site were compiled from refs. 8–16. The distance in nucleotides of the first nucleotide of the tetraloop to the first nucleotide of the corresponding mature RNA is indicated: “+” and “−” meaning that the tetraloops are located downstream or upstream from the mature RNA, respectively. The tetraloop sequence shown for the snR79 snoRNA cleavage site differs from that of ref. 15, because an alternative tetraloop could be found at a distance that fits the consensus better. For the cleavage site separating snR190 from U14 (14) and the site between snR41 and snR70 (15), we found by refolding these RNAs at the vicinities of the cleavage sites that potential AGNN tetraloops could cap a short helix that coaxially stacks on the long helix where cleavage occurs, in a conformation reminiscent of the snR40 cleavage site (15). The A0 site on the 5′ ETS of rRNA is not shown on the list, because recent results are opening the question of whether the in vitro cleavage of the 5′ ETS substrate is physiological (ref. 9 and M. Ares, personal communication). The distance for the A1 site at the 3′ ETS is different from that described in ref. 8 and indicated in ref. 9, in agreement with the unpublished observation that in vitro cleavage with FPLC-purified recombinant GST-Rnt1p and a shortened version of the 3′ ETS occurs 15 bp from the tetraloop (M. Ares, S. Abou Elela, and R. Nagel, personal communication).
Figure 1.
Single-turnover cleavage of wild-type and mutant model substrates by recombinant Rnt1p. 5′ end-labeled RNAs (20 fmol) were incubated for 2 min with recombinant His6–Rnt1p (800 fmol) or in buffer with no enzyme (Mock), and products were fractionated on a 10% sequencing gel. The arrows labeled WT, −1, −2, +1, +2, and +3 indicate the positions of the major cleavage products for the wild-type substrate and for the corresponding insertion-deletion mutants, respectively. The radioactive symbol indicates the 5′ end label of the RNA.
Figure 2.
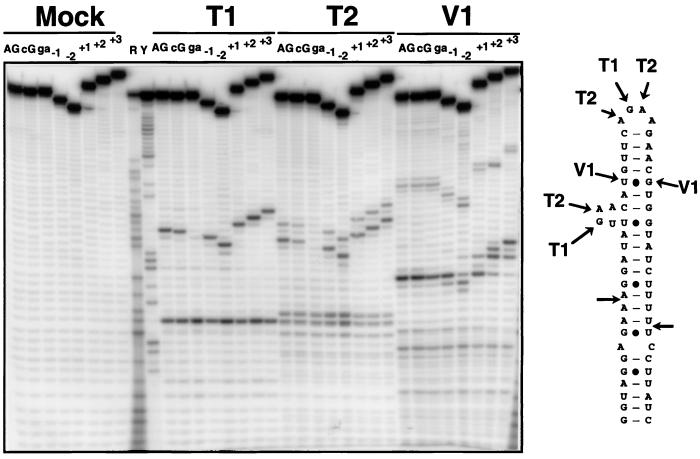
RNase probing of 5′ end-labeled wild-type and mutant model substrates. Shown are the cleavage products of wild-type and various mutant substrates with RNase T1, T2, and V1 or with buffer alone (Mock). The cleavage sites of these RNases on the wild-type substrate are indicated on the secondary structure. R and Y are size markers obtained by iodine cleavage of RNAs substituted with purine (R) or pyrimidine (Y) phosphorothioates. AG, wild-type substrate; cG, CGAA tetraloop mutant; ga, GAAA tetraloop mutant.
Loss of cleavage in the tetraloop mutants could be due to perturbations in the folding of mutant RNAs. To rule out this hypothesis, we compared the secondary structure probing patterns obtained with the wild-type and mutant substrates with the structure-specific ribonucleases T1, T2, and V1. The cleavage patterns observed for the wild-type and the mutant substrates were strikingly similar, taking into account the differences in size of these substrates (Fig. 2). The only exceptions were the T1 and T2 cleavages observed in the terminal tetraloop, which were inhibited in the case of the GAAA tetraloop mutant (Fig. 2). This result suggests that, in the wild-type substrate, the terminal AGAA tetraloop is in a single-stranded conformation and exposed to cleavage and therefore accessible to proteins. Changing it into a strongly internally structured GAAA tetraloop (21, 22) results in protection against cleavage by ribonucleases T1 and T2. These results allowed us to conclude that the mutations that inhibit Rnt1p cleavage do not strongly affect the overall RNA structure, at least at the level of the secondary structure. The results obtained with the T1 and T2 also confirmed some elements of the predicted secondary structure of the snR47 model substrate, because regions expected to be single-stranded (the terminal tetraloop and the asymmetrical loop) were attacked by these nucleases. Although the cleavage patterns obtained with the V1 endonuclease were more difficult to interpret, cleavage of some nucleotides by this enzyme outside of the regions cleaved by the T1 and T2 confirmed the double-stranded status of these positions.
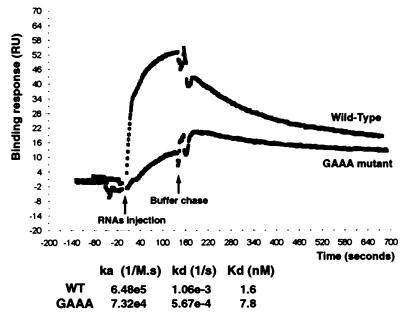
To investigate whether AGNN tetraloops are required for the binding of Rnt1p to its RNA substrate uncoupled from cleavage, we examined the binding of the wild-type and of the GAAA tetraloop mutant RNAs in the absence of Mg2+, which is required for cleavage. Binding of the RNA substrates was monitored in real time by surface plasmon resonance with a BIAcore system. The recombinant Rnt1p protein was immobilized to a dextran surface, and a continuous flow of RNA solution at various concentrations (50 nM to 10 μM) was applied to monitor in real time the association of the RNAs to the protein. The dissociation phase was monitored by chasing the RNAs with buffer alone. Fig. 3 shows the results obtained at 50 nM RNA and shows only one data set, because the high-salt washing regeneration procedure used between two binding assays at different RNA concentrations progressively incapacitated the immobilized protein. Consequently, it was realistically impossible to generate a meaningful binding curve amenable to simple Scatchard-type analysis. Therefore, the purpose of these experiments was not to obtain thermodynamically valid rate constants but rather to compare the observed rate constants for a wild type and a mutant under similar conditions. Under these conditions, the association phase (ka) is faster (≈10 times) with the wild-type substrate than with a mutant carrying a GAAA mutation, and the dissociation phase (kd) is twice as fast with the wild-type than with the GAAA mutant (Fig. 3). At this concentration, the resulting dissociation constant (Kd) was five times lower for the RNA bearing a wild-type AGAA tetraloop (1.6 nM) than for the mutant (7.8 nM). The ka/kd ratios were found to be consistently higher with the mutant than with the wild type at other RNA concentrations analyzed (data not shown). This assay suggested that the presence of AGNN tetraloops enhances binding of dsRNA by recombinant Rnt1p but that most of the binding observed is probably mediated by the dsRNA-binding domain found in the protein (8). However, in the case of the GAAA mutant, this binding is likely to be nonproductive, as shown by the lack of cleavage of this substrate (Fig. 1).
Figure 3.
Binding of wild-type (WT) and mutant snR47 substrates by immobilized Rnt1p. The binding and the dissociation of the RNAs were followed by monitoring the response (RU, arbitrary units) on a BIAcore system. Shown is a representative profile for the wild-type and the GAAA mutant substrates at a concentration of 50 nM. The association rate ka and dissociation rate kd are indicated, as well as the resulting dissociation constant Kd for both RNAs. The ka reported in the figure is the apparent second order rate constant (M−1⋅s−1) and depends on the RNA concentration, whereas the kd was independent of RNA concentration.
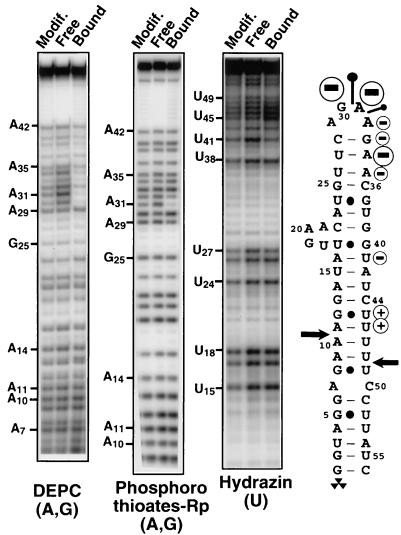
To look more thoroughly for RNA components required for Rnt1p binding, we examined the interference of specific chemical modifications of the RNA substrate (18) on Rnt1p binding by using native gel electrophoresis. 5′ end-labeled RNAs were chemically modified and incubated with Rnt1p, and after incubation, bound and free RNA populations were fractionated on native gels. After gel purification, chemically modified positions were cleaved, and the pattern of modifications found in the RNAs present in the different subpopulations was visualized by fractionation on denaturing polyacrylamide gels. Modifications that inhibit enzyme binding are expected to be enriched in the population of free RNAs and underrepresented in the bound RNAs population. This procedure allowed us to evaluate the effects of various chemical modifications of the RNA substrate on the protein binding. Purines were modified by DEPC, and uridines were modified by hydrazine (18). We also searched for nonbridging phosphate oxygens involved in Rnt1p binding by incorporating pro-Rp phosphorothioates. This modification interference approach allowed us to identify a subset of bases and phosphate oxygens whose modification strongly interferes with Rnt1p binding (Fig. 4). The strongest effects of base modifications on Rnt1p binding were found in the second and the third nucleotides (G30 and A31) of the tetraloop and on the second nucleotide of the stem that follows the tetraloop (A34). Modest inhibitory effects were also found at nucleotides A32, G33, and A35 located in the tetraloop or close to it. In contrast, no interference effects were found in the area of the cleavage site (Fig. 4). Incorporation of a pro-Rp phosphorothioate on the phosphate following the conserved guanosine of the tetraloop (5′ to A31) strongly inhibited binding; a weaker inhibitory effect was found on the following phosphate (5′ to A32). In conclusion, most of the bases and nonbridging Rp phosphate oxygens required for Rnt1p binding to its RNA substrates are localized on the terminal tetraloop or at its vicinity on the 3′ side of the helix. This result contrasts with the double-stranded region near the cleavage site, where single base modifications showed no effect on Rnt1p binding, at least in the conditions of binding with no divalent cations available. However, this negative result should be interpreted with caution. A recent structural study showed that the interaction of a dsRNA with a dsRNA-binding domain is the result of many individual interactions (23), and single modifications within the RNA stem may not be sufficient to affect binding of the dsRNA. Also, DEPC modification mostly affects the major groove, and interactions involving the minor groove of the dsRNA may have been missed in our study, because we did not use minor groove-specific modifications.
Figure 4.
Interference of DEPC, phosphorothioate, and hydrazine modifications of the snR47 substrate on Rnt1p binding. RNAs modified by DEPC, hydrazine, or by incorporation of purine phosphorothioates (thios A and G) were incubated with recombinant Rnt1p protein, and bound and free RNAs were gel purified. After cleavage of DEPC- or hydrazine-modified RNAs by aniline and of phosphorothioate-modified RNAs by iodine, the corresponding cleaved RNAs populations were fractionated on 12% polyacrylamide gels. The strongest effects of modifications are indicated on the secondary structure of the snR47 stem loop. “−” and “+” signs indicate inhibitory and stimulatory effects of base modifications on Rnt1p binding, respectively. Pins indicate the location of the nonbridging Rp phosphate oxygens where substitution by sulfur inhibits Rnt1p binding. The size of the circles and of the pins is roughly proportional to the inhibitory effect, as quantitated by PhosphorImager analysis. No inhibitory effects of phosphorothioate incorporation at pyrimidines were found (data not shown). Cytosines and the 5′ first 5 nt of the substrate were not mapped.
Discussion
The finding that cleavage by eukaryotic RNase III requires a single-stranded structure found in a conserved class of tetraloops was unexpected, given that bacterial RNase III cleavage does not require terminal tetraloops (20) but, rather, depends on antideterminants in the dsRNA structure (5). Therefore, despite the sequence conservation between the bacterial and the yeast enzyme, they require different RNA elements for efficient cleavage. Our results from in vitro cleavage, modification interference, and binding studies indicate that Rnt1p recognizes the single-stranded terminal tetraloop and the adjacent nucleotides and will cleave the RNA within the stem at a constrained distance relative to the tetraloop. The effects of mutations on Rnt1p binding were significantly weaker than the effects of the same mutations on cleavage activity. This result suggests that the major quantitative binding determinant relies on the dsRNA structure. It remains that the presence of an AGNN tetraloop is strictly required for productive binding (i.e., binding leading to a cleavage event), and binding studies with gel shift or plasmon surface resonance experiments cannot discriminate between nonproductive and productive binding events.
Our findings indicate that yeast RNase III behaves as a helical RNA ruler by measuring the distance from the tetraloop in a helical context. Bacterial RNase III has also been proposed to cut at a more or less fixed distance, but the docking element is in this case the 3′ end of the region of continuous RNA complementarity (24). Also, the bacterial enzyme cuts at a shorter distance (between 10 to 14 bp; about one helix turn) than the yeast enzyme (between 14 to 16 bp to the tetraloop, about one and a half helix turns). These differences indicate that the angle in the RNA helix between the cleavage site and the docking site is significantly different. These observations provide additional arguments that indicate that the bacterial and the yeast enzymes require different structural RNA elements for RNA cleavage. The existence of RNA molecular rulers has been previously described in more complex endonucleolytic systems. For example, the distance from the anticodon stem loop determines the site of cleavage in tRNA precursors by the multimeric tRNA intron endonuclease (25, 26). Histone mRNA 3′ processing also occurs at a fixed distance from the downstream element recognized by a ribonucleoprotein, the U7 snRNP (27).
In addition to providing insights into the mechanisms of cleavage specificity of an eukaryotic dsRNA endonuclease, these results also define a paradigm of RNA loops, in addition to the well known GNRA and UNCG tetraloops. The phylogeny of Rnt1p substrates strongly suggests a 4-nt loop structure, because all of the substrates examined show potential Watson–Crick base pairing between at least the last 3 bases preceding the AGNN sequence and the 3 bases after it. Although this phylogenetic observation is not sufficient by itself to conclude definitely that the AGNN sequence constitutes a genuine tetraloop, it provides a strong argument in favor of a 4-nt loop conformation. Further structural studies will be required to address this point. Whatever their precise shape, these loops provide a docking site for Rnt1p, and they may also help the substrate to adopt a hairpin conformation. For example, cytosine is never found on the third position; this counterselection may be explained by the need to prevent the formation of dimers of two RNA substrates and to favor a monomeric hairpin conformation. AGNN tetraloops are not found frequently in cellular RNAs, in contrast to the GNRA or UNCG tetraloops (unpublished observations). Because of the universal conservation of this structure in Rnt1p substrates, the presence of an AGNN terminal tetraloop in a cellular RNA provides a strong predictive argument for the presence of a yeast RNase III cleavage site 14–16 bp away from this tetraloop in this transcript. This property can be used for substrate prediction of eukaryotic RNase III. During the course of a previous study aimed at identifying Rnt1p cleavage sites in snoRNA precursors, predictions of the location of the cleavage site based on the presence of these tetraloops were always confirmed experimentally subsequently. Given that the yeast genome is sequenced entirely, one could imagine screening in silico the whole genome for the presence of such structures. This task is difficult, however, because the nucleotide conservation in the loop affects only 2 nt, the remaining two being not strongly constrained (with the exception of the almost absence of cytosine); in addition, the stems capped by these tetraloops do not form perfect Watson–Crick structures; they frequently include A:C and G:U wobble pairs and several internal bulges or loops. Therefore, it is hard to put constraints on search parameters that would be strong enough to eliminate a plethora of false positives. The identification of these structures in silico is therefore not an easy task, and for the moment, identification of Rnt1p cleavage sites depends on in vivo studies.
Another question remaining is the universality of this recognition signal for eukaryotic RNases III. There are a few physiological sites identified for the Schizosaccharomyces pombe RNase III, PacIp, but these targets RNAs do not seem to present AGNN tetraloops (28, 29). However, because the structural information is very limited, the identification of more physiological substrates is required to draw any conclusions. Also, it would be interesting to identify RNA substrates of RNases III from higher eukaryotes to conclude whether AGNN tetraloops are recognition signals specific for the S. cerevisiae enzyme or whether they represent a common recognition feature of substrates of the eukaryotic subclass of RNases III.
Acknowledgments
We thank B. Felden, P. Legrain, A. Nolte, I. Pumberton, and M. Teixeira for advice and suggestions; M. Teixeira for help with protein purifi- cation; M. Ares and J. Staley for reading the manuscript; and M. Ares, S. Abou Elela, and R. Nagel for communication of their results before publication.
Abbreviations
- dsRNA
double-stranded RNA
- snRNA
small nuclear RNA
- snoRNA
small nucleolar RNA
- ETS
external transcribed spacer
- DEPC
diethylpyrocarbonate
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070043997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070043997
References
- 1.Robertson H D, Webster R E, Zinder N D. J Biol Chem. 1968;243:82–91. [PubMed] [Google Scholar]
- 2.Court D. In: Control of mRNA Stability. Brawerman G, Belasco J, editors. New York: Academic; 1993. pp. 70–116. [Google Scholar]
- 3.Apirion D, Miczak A. BioEssays. 1993;15:113–120. doi: 10.1002/bies.950150207. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson A W. In: Ribonucleases: Structures and Functions. D'Alessio G, Riordan J F, editors. New York: Academic; 1997. pp. 1–49. [Google Scholar]
- 5.Zhang K, Nicholson A W. Proc Natl Acad Sci USA. 1997;94:13437–13441. doi: 10.1073/pnas.94.25.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iino Y, Sugimoto A, Yamamoto M. EMBO J. 1991;10:221–226. doi: 10.1002/j.1460-2075.1991.tb07939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotondo G, Gillespie M, Frendewey D. Mol Gen Genet. 1995;247:698–708. doi: 10.1007/BF00290401. [DOI] [PubMed] [Google Scholar]
- 8.Abou Elela S A, Igel H, Ares M., Jr Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 9.Kufel J, Dichtl B, Tollervey D. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanfreau G, Abou Elela S, Ares M J, Guthrie C. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abou Elela S A, Ares M., Jr EMBO J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seipelt R, Zheng B, Asuru A, Rymond B C. Nucleic Acids Res. 1999;27:587–595. doi: 10.1093/nar/27.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chanfreau G, Rotondo G, Legrain P, Jacquier A. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanfreau G, Legrain P, Jacquier A. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 16.Qu L-H, Henras A, Lu Y-J, Zhou H, Zhou W-X, Zhu Y-Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J-P. Mol Cell Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arluison V, Buckle M, Grosjean H. J Mol Biol. 1999;289:491–502. doi: 10.1006/jmbi.1999.2789. [DOI] [PubMed] [Google Scholar]
- 18.Conway L, Wickens M. Methods Enzymol. 1989;180:369–379. doi: 10.1016/0076-6879(89)80112-0. [DOI] [PubMed] [Google Scholar]
- 19.Zuker M. Methods Mol Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
- 20.Chelladurai B, Li H, Zhang K, Nicholson A W. Biochemistry. 1993;32:7549–7558. doi: 10.1021/bi00080a029. [DOI] [PubMed] [Google Scholar]
- 21.Heus H A, Pardi A. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 22.Legault P, Li J, Mogridge J, Kay L E, Greenblatt J. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 23.Ryter J M, Schultz S C. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krinke L, Wulff D L. Nucleic Acids Res. 1990;18:4809–4815. doi: 10.1093/nar/18.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattoccia E, Baldi I M, Gandini-Attardi D, Ciafre S, Tocchini-Valentini G P. Cell. 1988;55:731–738. doi: 10.1016/0092-8674(88)90231-0. [DOI] [PubMed] [Google Scholar]
- 26.Reyes V M, Abelson J. Cell. 1988;55:719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- 27.Scharl E C, Steitz J A. EMBO J. 1994;13:2432–2440. doi: 10.1002/j.1460-2075.1994.tb06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rotondo G, Huang J, Frendewey D. RNA. 1997;3:1182–1193. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou D, Frendewey D, Lobo Ruppert S M. RNA. 1999;5:1083–1098. doi: 10.1017/s1355838299990726. [DOI] [PMC free article] [PubMed] [Google Scholar]