Abstract
NAD(P)H/quinone acceptor oxidoreductase (QR1, NQO1, formerly DT-diaphorase; EC 1.6.99.2) protects animal cells from the deleterious and carcinogenic effects of quinones and other electrophiles. In this paper we report the apoenzyme structures of human (at 1.7-Å resolution) and mouse (2.8 Å) QR1 and the complex of the human enzyme with the substrate duroquinone (2.5 Å) (2,3,5,6-tetramethyl-p-benzoquinone). In addition to providing a description and rationale of the structural and catalytic differences among several species, these structures reveal the changes that accompany substrate or cofactor (NAD) binding and release. Tyrosine-128 and the loop spanning residues 232–236 close the binding site, partially occupying the space left vacant by the departing molecule (substrate or cofactor). These changes highlight the exquisite control of access to the catalytic site that is required by the ping-pong mechanism in which, after reducing the flavin, NAD(P)+ leaves the catalytic site and allows substrate to bind at the vacated position. In the human QR1-duroquinone structure one ring carbon is significantly closer to the flavin N5, suggesting a direct hydride transfer to this atom.
Keywords: DT diaphorase, cancer, chemoprotection, chemotherapy
NAD(P)H:Quinone acceptor oxidoreductase type 1 (QR1, NQO1; EC 1.6.99.2) is a flavoenzyme (homodimer of 273 residues, one FAD per monomer) that catalyzes the obligatory two-electron reduction of quinones to hydroquinones (1–3). This reaction prevents the one-electron reduction of quinones by cytochrome P450 reductase and other flavoproteins, resulting in oxidative cycling of deleterious radical species. The enzyme is inducible by a wide variety of Michael reaction acceptors and other electrophiles (4, 5). In addition to its possible role in the detoxification of dietary quinones, the enzyme has been shown to catalyze the reductive activation of quinolic chemotherapeutic compounds such as mitomycins (6), anthracyclines, and aziridinyl-benzoquinones. Overexpression of QR1 in many tumors, including those of lung, colon, liver, and breast, makes this enzyme an ideal target for the development of additional activatable cytotoxic compounds.
We previously reported the crystal structures of rat liver quinone reductase (rQR1) in complex with NADP+ and the ternary complex with the inhibitor Cibacron Blue (CB) and the substrate duroquinone (2,3,5,6-tetramethyl-1,2-benzoquinone; DQ) (7). These structures have revealed insight into many biochemical and physiological properties of QRs, as well as the mechanism of quinone reduction. These structures have provided a wealth of information, including the overall fold, the nature of the dimer, the interactions of the FAD cofactor, and the possibility of hydride transfer between the NADH and FAD cofactors and from FADH2 to the quinone substrate. Nonetheless many important aspects of QR1 function remain elusive. This is caused in part by the lack of structural information about the apoenzyme (apo) containing only the FAD prosthetic group (apo QR1) or of QR1 with substrate (i.e., with no inhibitor). In this paper, we report the structures of the apo forms of the mouse QR1 (mQR1) and the human QR1 (hQR1), as well as the structure of the complex of hQR1 with DQ, a QR1 substrate. In addition to providing a detailed account of the movements that occur upon substrate and cofactor binding and release, these structures provide a rationale for the biochemical differences among the QR1s of three species: human, mouse, and rat. The enzymes of these three species show small, but significant, differences in their fine specificity for the electron acceptor substrate. For example, although QR1 from all three species can reduce menadione (vitamin K3), the rates of the human and mouse enzymes are slower by a factor of 2 than that of the rat enzyme (8, 9). rQR1 is also more effective in activating the chemotherapeutic agents mitomycin C, EO9 (10), and CB1954 (11) than is the human enzyme. Substitution of a single amino acid in the rat sequence (Tyr-104 to Gln) makes the mutated rat enzyme behave like the human and mouse wild-type enzymes (8). These differences can now be explained by comparing the crystal structures of the enzymes.
Methods
Crystallization and Data Collection.
Expression of human and mouse recombinant QR1 in Escherichia coli and subsequent purification were carried out by methods as described (12). Crystals of native hQR1 were grown by the hanging drop vapor diffusion method from a solution containing 10–15 mg/ml (≈500 μM) protein in 25 mM Tris⋅HCl, pH 8.0, 5 μM FAD, mixed with equal volumes of reservoir solution consisting of 30% of PEG 3350, 200 mM sodium-acetate, 100 mM sodium-tricine at pH 8.5, and 12–24 μM FAD, and equilibrated at a temperature of 25°C with 1 ml of reservoir solution. mQR1 crystals were obtained by using the same conditions. hQR1-DQ crystals grow under similar conditions with the addition of DQ (8.9 mM) and NADH (25 μM). Crystals of apo hQR1 and hQR1-DQ belong to the triclinic space group P1 (a = 55.7 Å, b = 57.0 Å, c = 97.5 Å, α = 76.7°, β = 77.0°, and γ = 86.2°) with two physiological dimers in the asymmetric unit. At low resolution, this crystal form shows an apparent monoclinic symmetry (C2 with pseudo cell dimensions of 76.0, 81.0, 95.0 Å, and monoclinic angle β = 106°) that breaks at high resolution. mQR1 crystallizes in the space group P21 (a = 55.0 Å, b = 161.7 Å, c = 67.1 Å and monoclinic angle β = 123.3°), also with two physiological dimers in the asymmetric unit. The mQR1 x-ray diffraction data were collected at room temperature, whereas hQR1 data were collected at 100 K after freezing the crystal in their mother liquor with liquid nitrogen. The structure of hQR1 was determined from data collected at beamline X4A at the National Synchrotron Light Source (NSLS) with a R-Axis IV Image Plate detector. The native data set used in the refinement was obtained at beamline X25 of NSLS by using a wavelength of 1.1 Å with a Brandeis charge-coupled device. hQR1-DQ and mQR1 data were collected on a rotating anode generator (Cu target), using an R-Axis IV image plate detector. All data were reduced with the hkl software package (13).
Structure Determination and Refinement.
Initial structures of the apo hQR1 and mQR1 crystals were obtained by molecular replacement with the package amore (14) by using a monomer of rQR1 (86% sequence identity, Fig. 1; code 1QRD) as the search molecule. The orientation and position of each monomer were found independently. Rotation function searches with a single monomer showed independent peaks 4 σ above the next higher peak for all monomers in the asymmetric unit. All searches gave unambiguous solutions. For example, in the case of native hQR1 the final correlation in the translation search for the four monomers (two physiological dimers) was 61% with an R value of 35.4%, after rigid body fitting. Cycles of positional and B-factor refinement followed by simulated annealing refinement were carried for all the structures. For the hQR1-DQ complex the structural model of apo hQR1 was used. Noncrystallographic symmetry restraints were applied with different weights for main-chain and side-chain atoms. These restraints were not used in the refinement of native hQR1 because of the high resolution of the data.
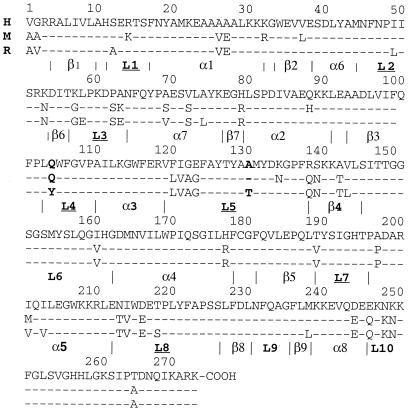
Figure 1.
Sequence alignment of hQR1 (H), mQR1 (M), and rQR1 (R). Secondary elements are named between the vertical lines that mark their beginning and end residues.
Fo−Fc electron density maps of the hQR1-DQ calculated before inclusion of the substrate showed well-defined density for the DQ in all monomers. DQ was built into this density and refined by using parameters and topology generated with related compounds from the Cambridge Structural Database (15).
All structures were refined by using the program cns (16) and manual rebuilding was performed by using the program o (17). The atomic coordinates of the native hQR1 (code 1D4A), the hQR1-DQ complex (code 1DXO), and the mQR1 (code 1DXQ) were deposited in the Protein Data Bank. Figures were drawn with the programs molscript (18) and raster3d (19).
Results
Structure Determination and Refinement.
This paper reports three QR1 structures: apo hQR1, apo mQR1, and hQR1 in complex with DQ, a QR1 substrate. The structure of apo hQR1, determined at 1.7-Å resolution, is the highest-resolution QR1 reported to date. All three structures were refined to R values lower than 21% (Table 1), with excellent stereochemistry (rms deviation from the ideal lower than 0.001 Å for bond lengths and less than 1.3o bond angles). Ramachandran diagrams place more than 82% of the residues in the most favored regions. Final maps show excellent density for most portions of the polypeptide chains and the FAD (Fig. 2), as well as for the bound DQ (Fig. 2B).
Table 1.
Summary of the crystallographic data
| Protein sequence | Human | Mouse | |
|---|---|---|---|
| Substrate | Apo | DQ | Apo |
| Resolution, Å | 1.7 | 2.5 | 2.8 |
| Unique reflections | 98,473 | 27,749 | 18,909 |
| Completeness | 78.6% | 92% | 80% |
| Rsym* | 5.2 | 4.4 | 10.0 |
| Crystallographic refinement | |||
| No. atoms of model | 9,496 | 9,036 | 8,924 |
| No. atoms of solvent | 592 | 84 | 0 |
| Rvalue (Rfree) | 20.9 (25.3) | 20.9 (26.1) | 20.6 (27.6) |
| 〈B〉† (Å2) | 23.8 | 25.6 | 29.4 |
Rsym = ΣhΣfj|Ifj − 〈Ih〉|∖ΣhΣfj|Ihj|, where h represents an unique reflection and j means symmetry equivalent indices. I is the observed intensity, 〈I〉 is the mean value of I of the reflection.
†Average temperature factor.
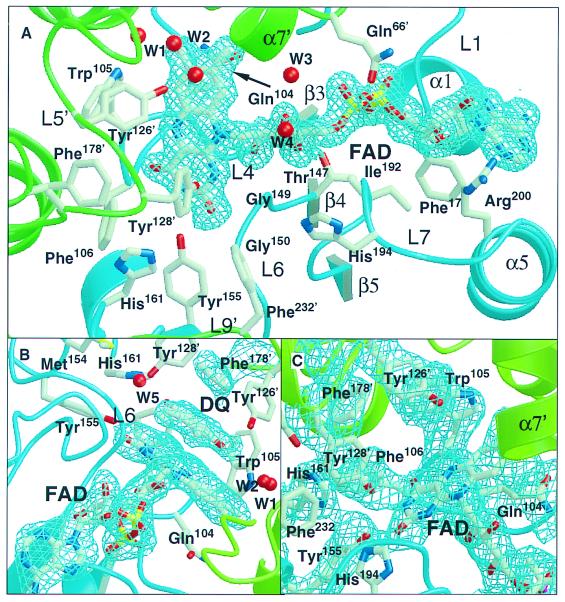
Figure 2.
Portions of the electron density map (2Fo−Fc) of the three QR1 crystals. (A) apo hQR1 binding site showing the density around the FAD prosthetic group. (B) hQR1-DQ complex showing the density around the substrate. (C) apo mQR1 density in the active site.
Structure Description.
Overall structure.
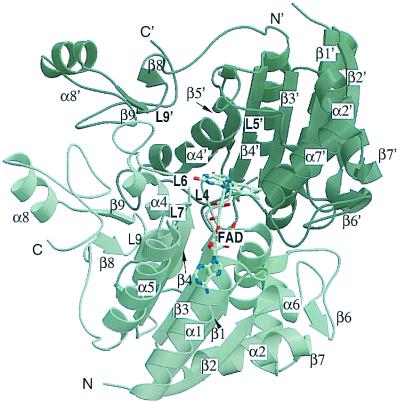
QR1 is a homodimer made of two interlocked monomers related by a 2-fold axis (Fig. 3). Each monomer has two separate domains: a catalytic domain with an α/β fold similar to that of flavodoxin (residues 1–220‖) and a small C-terminal domain that forms part of the binding site for the adenosine portion of the NAD. The catalytic domain includes a core of five parallel β-strands (β1–β5) alternating with five helices (α1–α5). Between strand β2 and helix α2 there is an insertion of a small region (residues 41–82) that includes two helices (α6 and α7) and two antiparallel strands (β6 and β7). Loops L1–L8 connect these different elements of secondary structure. The C-terminal domain includes a portion of L8, β8, L9, β9, α8, and the terminal loop region L10. Two equivalent catalytic sites, formed by portions of both monomers, are present at the dimer interface. Residues from loops L1 (residues 11–16), L4 (residues 104–110), L6 (residues 150–164), and L7 (residues 190–197) of one monomer, and L3′ (residues 58–66) and L5′ (residues 121–140) of the other monomer** bind each of the two FADs in a dimer. These residues together with L9′ (residues 230–236) form each of two equivalent catalytic sites per dimer.
Figure 3.
Schematic representation of the QR1 dimer. The numbering of the secondary element is indicated. The bound FAD is shown in one of the two sites.
The catalytic site has three distinct regions: one that binds FAD, one that binds the adenine ribose portion of NAD(P), and one that binds either the hydride acceptor (substrate) or the hydride donor [the nicotinamide moiety of NAD(P)H]. The overlap of the hydride acceptor and donor sites is consistent with the ping-pong kinetics of QR (20). These three regions will be described separately.
FAD binding site.
The FAD prosthetic group is tightly bound to hQR1 and mQR1 and does not come off the enzymes readily under native conditions. The three structures reported here contain 12 independent views of the FAD. In each it binds to the enzyme in the same manner. As some of the water molecules in this site were modeled only in the higher-resolution structures, we will refer to the structure of apo hQR1 when describing these molecules.
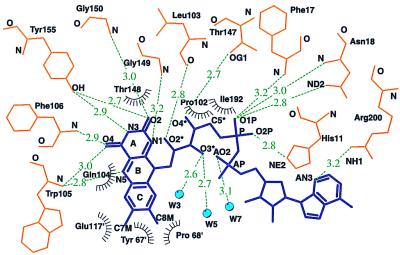
FAD-QR1 interactions involve primarily residues from one of the two monomers of the dimer (Fig. 4). The isoalloxazine moiety of the FAD of hQR1 and mQR1, which is planar in most of the monomers, is centered over loop L4, contacting residues of loop L6. It has extensive contacts with L4, specially with main-chain atoms and with the aliphatic side-chain atoms of Leu-103 and Gln-104. All four available groups of the isoalloxazine ring A make hydrogen bonds with groups of the protein. The ribitol, the diphosphate, and the adenosine moieties lie in the cleft between the N-terminal regions of helices α1 and α5.
Figure 4.
Hydrogen bonding and van der Waals interaction observed between FAD and protein in hQR1. Open radiated circles indicate hydrophobic interactions. Hydrogen bonds are represented by dashed green lines; water molecules are shown as blue filled circles.
Most of the FAD-enzyme interactions in h(m)QR1 are highly homologous to those described in the structures of rQR1 (7). However, there are significant differences involving Gln-104, which replaces a tyrosine in rQR1, and several water molecules not present in rQR1. The aliphatic portion of the Gln-104 side chain in h(m)QR1 is in contact with C5A and C6 on the re face of the isoalloxazine. On the other face, two highly conserved water molecules (W1 and W2 in Fig. 2 A and B), seen in the eight independent copies of the hQR1, form short contacts with the methyl groups of ring C (Fig. 2). W1 forms hydrogen bonds with the side-chain Nɛ1 of Trp-105 as well as with carbonyl oxygens in residues of the second monomer—Glu-117′ and Phe-120′—and with W2. W2 is coordinated by another water molecule (not shown in the figure) that is hydrogen-bonded to the main-chain NH of Gly 122′ and to the side-chain OH of Ser-71′. Two additional water molecules are hydrogen-bonded to the O3 of ribitol; one of the two (W3 in Fig. 2A) is also hydrogen-bonded to another water molecule (W4 in Fig. 2A), which in turn is hydrogen-bonded to Nɛ1 of Gln-104.
Empty NAD adenosine site.
The residues that make up the adenosine and the diphosphate binding sites were identified in the rQR1/NADP+ structure we reported earlier (7). The adenosine binding site is a cleft between loop L9 and residues 128–130 in L5. Leu-230, Asn-231, Phe-232, and Phe-236 of L9 make van der Waals contacts with the adenosine moiety of NAD. The adenine lies over Leu-230 and the main chain of Asn-231. The main-chain NH of Phe-232 provides a hydrogen bond to the 2′-phosphate of the NADP+.
In the human and mouse apo structures, residues in L9 shift their positions by several Å, closing the cleft. An inter monomer hydrogen bond between the side-chain OH of Tyr-132′ and Nδ2 of His-161 anchors the C terminus of L5′. By fixing the conformation of L5′, this hydrogen bond locks Tyr-132′ and Glu-123′ into conformations that fall outside the allowed regions of the Ramachandran plot. In mQR1 and hQR1, residues 128 and 129 recede from the cleft, thereby widening it relative to rQR1 by 0.56–0.95 Å in the mouse and 0.5 Å in the human.
Empty substrate (donor/acceptor) binding site.
The unoccupied substrate binding pockets of the apo human and mouse structures are 10 Å wide, 9 Å deep, and 4 Å high (360 Å3). Phe-106, the main-chain carbonyl of Gly-174′, Phe-178′, and Trp-105 form the internal wall, Tyr-126′ and Tyr-128′ form the roof, and the isoalloxazine ring of FAD makes up the floor of the cavity. His-161 and two water molecules (W1 and W2) form the opposite side of the pocket. The entrance to the pocket is limited by glycines 149 and 150, His-194, and Pro-68′ from the N terminal of helix α7′ of the second monomer.
In hQR, the density of Tyr-128 is not well defined and was interpreted as showing multiple conformations. However, in the absence of substrate (or inhibitor), both hQR1 and mQR1 show that the most occupied conformer of Tyr-128′ is one in which the side chain is close to Phe-232 and His-161 of the other monomer, shielding the region from solvent and from possible reaction with O2.
Substrate binding site with bound acceptor (DQ).
DQ binds to the active site through a series of contacts involving the flavin and several hydrophobic and hydrophilic residues (the total accessible surface area buried by binding DQ is 427 Å2). Five aromatic residues (Trp-105, Phe-106, Phe-178′, Tyr-126′, and Tyr-128′), Gly-149, and the central portion of the isoalloxazine of FAD provide most of the contacts. In addition, the quinone is hydrogen-bonded to a water molecule not present in the apo enzyme (W5 in Fig. 2B) that bridges the Nɛ of His-161 and the OH of Tyr-128′. The plane of the quinone ring stacks 3.5 Å from the isoalloxazine plane with the C2-CH3 bond perpendicular to the plane of the indole of Trp-105. The distances from the flavin N5 (hydride donor) to the possible acceptors of the hydride in the quinone are 4.5 Å to O1, 4.1 Å to C1, and 3.55 Å to C2.
The O1 of DQ makes an almost perpendicular hydrogen bond with the OH of Tyr-126′ and a more regular hydrogen bond with W2 (Fig. 5). The methyl group at C5 points toward the outside of the cavity and contacts Gly-150.
Figure 5.
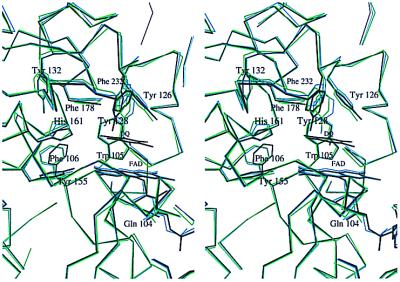
Stereo view of the changes that take place upon binding of DQ to hQR1. The catalytic sites of apo hQR1 (blue), hQR1-DQ (black), and apo mQR1 (green) are shown for comparison. Only the FAD, DQ, water molecules, and nonglycine residues that participate in binding are shown.
The 1,088 α-carbons of the two dimers of apo hQR1 and those of the hQR1-DQ can be aligned with an rms deviation of only 0.3 Å. However, there are small, but significant, differences in the positions of some binding site residues between the DQ-bound and DQ-free enzyme (Fig. 5). Comparison of the two structures shows changes in the positions of Phe-106, His-161, Phe-232′, and Tyr-128′ (Fig. 5) upon substrate binding. Tyr-128′ shows the largest change in the active site, resulting in 2-Å difference in the OH position.
Discussion
Comparison of hQR1, mQR1, and rQR1.
An important feature of the human and mouse crystals is that they were grown in the absence of the dye CB, which was needed to stabilize the crystals of the rat enzyme. The structures of rQR1 have shown that CB, a nicotinamide nucleotide analog, binds to a site that fully overlaps that of NADP+. Thus, in both rat structures the adenosine binding site was occupied either by NADP+ or CB. As a consequence, no information was available on the forms of the enzyme in which the NAD site or the substrate site is empty.
hQR1, mQR1, and rQR1 have a high degree of sequence identity (mouse/human 86%; mouse/rat 94%; human/rat 86%) and structural similarity. When the dimers of the three structures are aligned and the positions of the 544 α-carbons are compared, the rms deviation between the human and mouse structures is 0.47 Å, between mouse and rat is 0.76 Å, and between rat and human 0.98 Å. If only the catalytic domains are aligned, the equivalent rms differences are 0.44 Å (220 α-carbons), 0.60 Å (219 α-carbons), and 0.55 Å (220 α-carbons), respectively. Clearly, even though the mouse and rat enzymes show the highest degree of sequence identity, the structures of human and mouse apo QR1 are more similar to each other than to the rat structure. The major differences between rQR1 and the other two enzymes are concentrated in the binding site, in particular in loop L9, which in the rQR1 structures interacts with either the adenine portion of NADP+ or CB. These changes are more a consequence of cofactor binding than of sequence differences between species. On the other hand, more subtle changes in the middle portion of L5 and the position of the prosthetic group seem to be a product of differences in sequence among human, mouse, and rat. These two kinds of structural changes and their consequences will be discussed in the following sections.
Conformational Changes During Catalysis.
Binding of the cofactor [NAD(P)].
During NAD(P)H (or CB) binding the largest structural changes occur in loops L5 and L9. These changes, best seen by comparing native (apo) hQR1 with rQR1-DQ/CB (Fig. 6) and rQR1-NADP+, correspond to the closing and opening of the catalytic site during catalysis. Binding of the cofactor (or CB) opens the cleft and results in a movement of L9 and Tyr-128; the maximum displacement (2.7 Å) is shown by the α-carbon of Phe-232. In the rat complexes, Tyr-128 participates in a stacking interaction with either the triazine ring of CB or the ribose of the NADP, which results in a large displacement of the side chain. The apo structures show that this displacement reverts in the absence of substrate or inhibitor, such that Tyr-128 occupies the vacant space. A smaller, but significant, movement is experienced by the main chain of residues 128–130 (≈0.4 Å) (Fig. 6).
Figure 6.
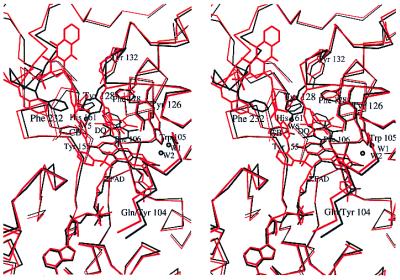
Stereo view of the overlap of the two DQ complexes known to date. The ternary complex rQR1-CB/DQ is colored red and hQR1-DQ black. Only the FAD, DQ, water molecules, and nonglycine residues that participate in binding are shown.
Binding of substrate (quinone).
Analysis of the apo hQR1 and mQR1 structures shows that the substrate/nicotinamide binding site is a highly hydrophobic pocket with three potential hydrogen-bonding residues (Tyr-126′, Tyr-128′, and His-161) suitable for binding large quinolic compounds. In the hQR1-DQ structure, DQ is bound to this site by hydrophobic interactions, as well as by a water-mediated (W5) bridge from O4 to His-161. In these structures, C2 of the quinone lies almost on top of the N5 of FAD (3.5 Å apart), suggesting a Michael addition of the hydride from the FADH2 to this position of the substrate.
Comparison of all of the available QR1 structures shows that the side chain of Tyr-128 can adopt many different conformations. The different conformations result in opening and closing of the binding site, as well as in different interactions with substrates. For example, comparison of the structures of hQR1 and hQR1-DQ shows that Tyr-128 moves inside the cavity and away from the FAD to make room for the DQ (Fig. 5). In this new position, Tyr-128′ is hydrogen-bonded to a water molecule that, in turn, makes hydrogen bonds to His-161 and O4 of the DQ. It appears that these interactions are an important aspect of the catalytic mechanism.
Smaller, but significant, movements are seen for the side chains of residues Phe-106, His-161, and Phe-232′. Main-chain atoms of loops L5′ and L9′ also show small shifts in position. These changes appear to adapt the binding site for maximizing the interaction with substrates.
The binding site conformation observed previously in the rQR1-CB/DQ complex (7) differs in detail from those of hQR1 and mQR1. The presence of CB in the rQR1-CB/DQ structure may contribute to differences in the position of the DQ, as parts of the binding site (including the region occupied by Tyr-128 in hQR1) are occupied by the triazine ring of the CB. In the rat structure, Tyr-128 swings outward, making hydrophobic contact with the triazine ring of CB, thereby providing a hydrogen bond to the O1 of DQ. In the human structure, Tyr-128 is inside the cavity, forcing the quinone 2.5 Å closer to the central ring of the isoalloxazine (Fig. 6). The change in position of the flavin that results from the Tyr-104 to Gln substitution described below also may contribute to the differences in the mode of binding of DQ.
In the human and mouse enzymes, the side chain of Phe-106 appears rotated with respect to the position observed in rQR1/CB/DQ (Fig. 6), making a contact with the methyl group at C3. These changes in the position of Phe-106 appear to be ideal for accommodating different substituents in the quinone ring of the substrate while maintaining the overall placement of the ring. Movements of Phe-106 will have the effect of making the pocket defined by Phe-178′, Ile-175′ and Trp-105 and Phe-106 itself, accessible or inaccessible to a given substrate.
Active Site Differences Among Species.
FAD binding site.
The most significant sequence difference between human (or mouse) and rat QR1 is the replacement of rat Tyr-104 by Gln (Fig. 1). Comparison of the structures shows that replacing a bulky aromatic residue (tyrosine) by a smaller amphipathic residue (glutamine) increases the space in the FAD binding pocket of the mouse and human enzymes, allowing the dimethylbenzene ring of the flavin to move 0.5 Å deeper into the cavity (Fig. 6). Because the substrate (and the nicotinamide portion of NAD) participate in a stacking interaction with the flavin, this change in the position of the flavin affects the position of the substrates.
It is well known that residues surrounding the active site of redox enzymes play important roles in the electron transfer reactions by, among other things, modulating the reduction potential of the redox centers. In solution, the isoalloxazine moiety of FAD adopts two different conformations depending on the oxidation state: the oxidized form is planar, whereas the reduced form is bent at the center by approximately 40° (butterfly conformation). In all QR1 structures the FAD-enzyme interactions appear to stabilize the flat oxidized form. There are differences, however, between rQR1 on the one hand and hQR1 and mQR1 on the other.
A detailed analysis of the FAD contacts suggests then the binding site of the human and mouse QR1 might accommodate a certain degree of bending of the isoalloxazine around its central axis; in these two enzymes, the shortest van der Waals contacts occur at the center of the isoallozaxine ring and relax to longer contacts toward the extremes. In rQR1, Tyr-104 is held in position by Pro-102 with one side of its aromatic side chain against the isoallozaxine ring (maximum distance to FAD atoms of 3.7 Å) preventing any bend of the FAD. This additional steric constraint present in rQR1 is absent in the human and mouse enzymes in which residue 104 is a glutamine. In these enzymes (human and mouse), the plane of the carboxyamide of Gln-104 makes an angle of 12° with the plane of the isoalloxazine. The shortest distance between a Gln-104 atom and an FAD atom is 3.2 Å (glutamine α-carbon to FAD N5), but the distances grow toward the end of the side chain, up to 4.9 Å in the amide group. These distributions of interactions must be important when the isoalloxazine is reduced and conjugation in the central ring is lost because bending of the cofactor may be accommodated more easily in hQR1 and mQR1 than in rQR1. This should result in a more negative redox potential in the case of rat compared with the human and mouse enzymes. Although experimental redox potentials are not available for comparison, this observation may correlate with the higher rate of hydride transfer by the rat enzyme for almost all electron acceptors.
NAD(P) binding site.
The only sequence difference in this region between rQR1 and the mouse and human enzymes is the change of Thr (in rat) to Ala (in human and mouse) at position 130 (Fig. 1). This substitution reduces the steric hindrance caused by the two additional side-chain atoms of this threonine with Thr-127 and with the residue 134 (Lys in hQR1 or Asn in mQR1), causing the main chain of residues 128–130 to recede from the cleft by a small, but significant, distance (approximately 0.6 Å in hQR1 and 0.9 Å in mQR1), a movement opposite to that observed in loop L9 associated with the release of the cofactor. Moreover, in mQR1 Asn-134 makes an additional hydrogen bond with Ser-225, pulling L5 further away from the cleft and moving the side chain of Tyr-128 toward the cleft. The L5 shift makes the less flexible portion of the adenosine binding cleft wider in hQR1 and mQR1 than in rQR1. Although these changes are small, the widening of cleft may loosen the binding of the adenosine in a region with hydrophobic interactions. These observations correlate with the reduction in apparent affinity (by half) for NADH observed in the human and mouse enzymes (8).
Substrate binding site.
Although in apo hQR1 and mQR1 Tyr-128 is in the same conformer, the small changes in L5 described above, amplified by the long side chain of Tyr-128, result in a significant displacement: the OH of Tyr-128, which is involved in substrate binding, shifts by about 2 Å between hQR1 and mQR1. These changes and the ones described above can account for the fine specificity of human, mouse, and rat for menadione, mitomycin C, and CB1954 (11).
Other differences.
Analysis of the aligned sequences (Fig. 1) shows a region close to the active site, which has a relatively high number of differences between human and rodent (rat and mouse): Ser-72′ is substituted by Val, Ala-122′ by Gly, Gly-123′ by Glu, and Leu-120′ by Phe. Close to the N-terminal of α7′, the changes of Ser-72′ to a larger Val and of Ala-122′ to a smaller Gly seem to complement each other. Replacement of Leu-120′ by a bulky aromatic residue has no effect on the nearby residues (Trp-105 and Phe-126′). The side chain of Val-122′ points toward the protein core and substitution by Ile has no effect on the size or the lining of the binding site. Ser-72′ to Val is the only substitution that results in loss of a potential hydrogen bond to the ligand. Interestingly, water molecules W1 and W2 separate the overall region of the substitutions from the binding site.
Conclusions
Taken together, the results indicate that, as part of the catalytic cycle, QR1 undergoes local conformational changes associated with binding and release of cofactor and substrate. These changes open and close the catalytic site during the exchange between cofactor and substrate necessary as part of the ping-pong mechanism. As a result, in the apo structures the pockets occupied by the NAD(P)H and the quinone are partially occluded by groups of the enzyme in such a way that the flavin and the protein groups involved in substrate and cofactor binding have reduced exposure to solvent and molecular oxygen when the sites are unoccupied.
In addition, the plasticity of the substrate binding portion of the site could be involved in allowing the site to accommodate a large variety of quinone substrates, including molecules of such different sizes as benzoquinone, menadione, mitomycin C, and streptonigrin. This plasticity may provide significant advantages in targeting QR1 for the activation of chemotherapeutic compounds by allowing the design of activatable compounds of different sizes and varied chemical properties.
Although very similar in structure and in primary sequence, hQR1, mQR1, and rQR1 exhibit fine specificity features that can be correlated with the structural differences that result from replacement of a small number of amino acids.
Acknowledgments
This article is dedicated to the memory of Professor Lars Ernster in whose laboratory QR1 (DT-Diaphorase) was first isolated in 1958. Synchrotron data were collected at the National Synchrotron Light Source (Brookhaven National Laboratory) line X25 and Howard Hughes Medical Institute beamline X4A. We thank Drs. Lonny Bergman, Hal Lewis, and Craig Ogata for help and guidance at the lines. This work was supported by Grant GM51362 from the National Institute of General Medical Sciences to L.M.A., Grant CA 51210 from the National Cancer Institute to D.R., and Grant CA 44530 from the National Cancer Institute to P.T.
Abbreviations
- DQ
duroquinone
- QR
quinone reductase
- QR1
NAD(P)H:quinone acceptor oxidoreductase type 1
- rQR1
rat QR1
- hQR1
human QR1
- mQR1
mouse QR1
- CB
Cibacron Blue
- apo
apoenzyme
Note Added in Proof
Skelly et al. (21) just reported a lower-resolution structure of hQR1. No comparisons are possible yet, because their coordinates have not been released.
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1D4A, 1DXO, and 1DXQ).
This work was presented in abstract form at the annual meeting of the American Crystallographic Association, May 22, 1999, Buffalo, NY.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050585797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050585797
Residue numbers in this paper are those used in the Protein Data Bank coordinates, accession number 1D4A; they are shifted down by one residue with respect to those in other publications.
When referring to the catalytic site, secondary structure elements and residues will be primed (second monomer) or unprimed (main monomer). As the two monomers are equivalent, primes will be used only when distinction is needed.
References
- 1.Ernster L, Navazio F. Acta Chem Scand. 1958;12:595. [Google Scholar]
- 2.Ernster L, Ljunggren M, Danielson L. Biochem Biophys Res Commun. 1960;2:88–92. [Google Scholar]
- 3.Ernster L, Danielson L, Ljunggren M. Biochim Biophys Acta. 1962;58:171–188. doi: 10.1016/0006-3002(62)90997-6. [DOI] [PubMed] [Google Scholar]
- 4.Prochaska H J, De Long M J, Talalay P. Proc Natl Acad Sci USA. 1985;82:8232–8236. doi: 10.1073/pnas.82.23.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talalay P. Adv Enzyme Regul. 1989;28:237–250. doi: 10.1016/0065-2571(89)90074-5. [DOI] [PubMed] [Google Scholar]
- 6.Siegel D, Gibson N W, Preusch P C, Ross D. Cancer Res. 1990;50:7483–7489. [PubMed] [Google Scholar]
- 7.Li R, Bianchet M A, Talalay P, Amzel L M. Proc Natl Acad Sci USA. 1995;92:8846–8850. doi: 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S, Knox R, Wu K, Deng P S, Zhou D, Bianchet M A, Amzel L M. J Biol Chem. 1997;272:1437–1439. doi: 10.1074/jbc.272.3.1437. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Wu K, Zhang D, Sherman M, Knox R, Yang C. Mol Pharmacol. 1999;56:272–278. doi: 10.1124/mol.56.2.272. [DOI] [PubMed] [Google Scholar]
- 10.Beall H, Murphy A, Siegel D, Hargreaves R, Butler J, Ross D. Mol Pharmacol. 1995;48:499–504. [PubMed] [Google Scholar]
- 11.Chen S, Knox R, Lewis A D, Friedlos F, Workman P, Deng P S, Fung M, Ebenstein D, Wu K, Tsai T M. Mol Pharmacol. 1995;47:934–939. [PubMed] [Google Scholar]
- 12.Chen S, Clarke P, Martino P A, Deng P S, Yeh C-H, Lee T D, Prochaska H J, Talalay P. Protein Sci. 1994;3:1296–1304. doi: 10.1002/pro.5560030816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otwinoswski Z, Minor W. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 15.Allen F H, Kennard O. Chem Des Automation News. 1993;8:31–37. [Google Scholar]
- 16.Brunger A, Adams P, Rice L. Curr Opin Struct Biol. 1998;8:606–611. doi: 10.1016/s0959-440x(98)80152-8. [DOI] [PubMed] [Google Scholar]
- 17.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 19.Merrit E A, Murphy E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 20.Hall J M, Lind C, Golvano M P, Rase B, Ernster L. In: Structure and Function of Oxidation Reduction Enzymes. Åkenson Å, Ehrenberg A, editors. Oxford: Pergamon; 1972. pp. 443–443. [Google Scholar]
- 21.Skelly J V, Sanderson M R, Suter D A, Bauman U, Read M A, Gregory D S, Bennett M, Hobbs S M, Neidle S. J Med Chem. 1999;42:4325–4330. doi: 10.1021/jm991060m. [DOI] [PubMed] [Google Scholar]